Abstract
Cbx3/HP1γ is a histone reader whose function in the immune system is not completely understood. Here, we demonstrate that in CD8+ T cells, Cbx3/HP1γ insufficiency leads to chromatin remodeling accompanied by enhanced Prf1, Gzmb and Ifng expression. In tumors obtained from Cbx3/HP1γ-insufficient mice or wild type mice treated with Cbx3/HP1γ-insufficient CD8+ T cells, there is an increase of CD8+ effector T cells expressing the stimulatory receptor Klrk1/NKG2D, a decrease in CD4+ CD25+ FOXP3+ regulatory T cells (Treg cells) as well as CD25+ CD4+ T cells expressing the inhibitory receptor CTLA4. Together these changes in the tumor immune environment may have mitigated tumor burden in Cbx3/HP1γ-insufficient mice or wild type mice treated with Cbx3/HP1γ-insufficient CD8+ T cells. These findings suggest that targeting Cbx3/HP1γ can represent a rational therapeutic approach to control growth of solid tumors.
Similar content being viewed by others
Introduction
The heterochromatin protein 1 (HP1) family of proteins are histone readers that associate with modified histones, and are involved in epigenetic regulations1,2. Cbx3/HP1γ interacts with the methyl groups of histone H3 at lysine 9 (H3K9Me3) and with methyl transferases as well as other proteins3,4,5,6. It has been shown to associate with both heterochromatin and euchromatin suggesting it may participate in transcriptional repression and activation, respectively7,8. Additionally, Cbx3/HP1γ controls gene expression through two intimately related processes: transcriptional elongation and co-transcriptional splicing of nascent RNA transcripts9,10,11,12. Homozygous germline deletion of Cbx3/HP1γ in mice causes embryonic lethality13,14. Previously we have shown that Cbx3/HP1γ haploinsufficiency is sufficient to regulate the high-affinity antibody response to protein antigens15. However, it has not been determined how Cbx3/HP1γ does so.
After encountering infected or malignant cells, naïve CD8+ T cells become activated and differentiate into effector cells to clear the infectious agent or control tumor growth16. Majority of effector cells die, but a small number will become memory cells. However, under persistent antigen exposure as in cancer or chronic infections, a subset of CD8+ effector T cells can enter an altered differentiation program known as T-cell exhaustion (TEX)17. CD8+ TEX cells express unique transcription factors as well as those shared with effector/memory cells, among them are Tbx21/T-bet, Eomesodermin (Eomes) and Prdm1/Blimp-1. However, these factors interact with distinct partners in addition to being differentially expressed in the two populations17,18. More importantly, CD8+ TEX cells can be reversed to become functional. Thus, identifying novel mechanisms that can reinvigorate exhausted CD8+ T cells would provide means to control tumor growth.
The ability of CD8+ effector T cells to kill targets requires the production of lytic molecules, perforin (encoded by Pfr1) and granzyme B (encoded by Gzmb), and the inflammatory cytokine interferon γ (INFγ, encoded by Ifng). Induction of Pfr1, Gzmb and Ifng is regulated in part by the transcription factor Runt-related 3 (Runx3)19,20.
Cytotoxic CD8+ T cells detect malignant-self either directly through interaction of the T-cell receptor (TCR) on CD8+ T cells with tumor antigens or indirectly in the form of so-called “danger signals” such as the retinoic acid early transcript-1 (RAE-1) and related ligands. Nonetheless, most activated cytotoxic CD8+ T cells (human and mouse) share the expression of the natural killer group 2, member D receptor (NKG2D) encoded by Klrk1, which upon binding to RAE-1 or related ligands, induced on tumor cells, stimulates effector responses that are independent of TCR21. For many tumors, tumor infiltrating Klrk1/NKG2D+ CD3+ CD8+ T cells or T cells expressing chimeric Klrk1/NKG2D receptor were shown to have promising anti-tumor efficacy22,23,24,25,26,27.
The elimination of CD4+ FOXP3+ Treg cells by T cells expressing the chimeric Klrk1/NKG2D receptor in the ovarian tumor microenvironment inhibited tumor growth and increased survival in mice27. Similarly, treatment of melanoma tumor-bearing mice with monoclonal antibody specific for CTLA4 led to the selective depletion of CD4+ Treg cells and a concomitant increase of CD8+ effector T cells within tumor lesions resulting in increased survival28,29. These results suggest that finding new means to manipulate the ratio of effector CD8+ T cells over CD4+ Treg cells is a viable strategy to control cancer.
Neuroblastoma (NB) is the most common extracranial solid tumor of childhood and the third most common cause of pediatric cancer death30. Despite the use of multimodal therapy, patients with high-risk NB have a poor prognosis due to high rate of relapse. Treatment with anti-GD2, a ganglioside expressed on a subset of NB tumors cells, is the current standard of care for high-risk NB31,32,33. However, this therapy causes severe pain requiring the use of continuous opiate infusions because GD2 is also expressed on pain fibers, and not all patients respond to this therapy34. Therefore, there is a need to explore novel approaches including immunotherapies to control high-risk NB.
Here we find that adoptive transfer of Cbx3/HP1γ-insufficient CD8+ effector T cells alone into wt tumor-bearing mice greatly limits tumor growth. Within the tumor microenvironment of Cbx3/HP1γ-insufficient mice or wt mice treated with Cbx3/HP1γ-insufficient CD8+ T cells, we detect an increase of Klrk1/NKG2D+ infiltrating CD8+ effector T cells, a concomitant decrease in CD4+ Treg cells. We propose that together these changes may have provided the anti-tumor immunity observed in mice.
Results
Cbx3/HP1γ-insufficient CD8+ T cells are endowed with enhanced effector capacity
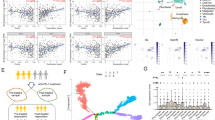
Recently, we showed that in C57BL/6 mice, haploinsufficiency of Cbx3/HP1γ impairs the lymphoid-tissue germinal center (GC) reaction and high-affinity antibody response against a thymus (T)-dependent antigen (Ag) conjugated to chicken gamma globulin (NP-CGG)15. The effects were CD8+ -T-cell-intrinsic. Additionally, Cbx3/HP1γ protein was induced in CD8+ T cells after activation, and its level was greatly reduced in insufficient cells (Fig. S1A). These results prompted us to ask what CD8+ T-cell functions are controlled by Cbx3/HP1γ and by what mechanisms. Quantitative polymerase chain reaction (qPCR) and Western blot analyses demonstrated that on day 2 after activation, Cbx3/HP1γ-insufficient CD8+ T cells expressed more Bcl6, Prdm1 and Tbx21 compared to control cells (Fig. 1A,B). On day 5 of activation, Prdm-1 expression was sustained at a high level in Cbx3/HP1γ-insufficient CD8+ T cells while that of Runx3 and Bcl6 was unaltered (Fig. 1A). By contrast, Eomes expression was repressed in Cbx3/HP1γ-insufficient CD8+ T cells on both days compared to control cells (Fig. 1A,B). Expression of Prf1, Gzmb and Ifng was upregulated in Cbx3/HP1γ-insufficient CD8+ T cells compared to controls (∼1.5 fold, 2 fold and 1.5 fold increase, respectively) (Fig. 1C–E). Correspondingly, a higher percentage of IFNγ+ cells was detected in Cbx3/HP1γ-insufficient cultures (50%) compared to wt controls (22%), and insufficient CD8+ T cells also produced a higher amount of IFNγ protein (Fig. 1D). A phenotypic survey of wt and insufficient activated CD8+ T cells showed they expressed similar levels of IL-2, CD25, CD127 (IL-7Rα) and CD132 (common γ chain or IL-2Rγ) (Fig. S1B–D). Similar frequency of effector (CD44hiCD62Llo) and central (CD44hiCD62Lhi) memory CD8+ T cells was detected in wt and Cbx3/HP1γ-insufficient mice suggesting that activation did not appear to favor the expansion or reduction of either population in Cbx3/HP1γ-insufficient mice (Fig. S1E,F). To determine if the augmented production of perforin, granzyme B and IFNγ would endow Cbx3/HP1γ-insufficient CD8+ T cells to better induce apoptosis than wt cells, in vitro-activated CD8+ and CD4+ T cells were co-cultured with target neuroblastoma NB-9464 tumor cells35, and tumor-cell apoptosis assessed by measuring cleaved/activated caspase 3 (CC3) production. At a ratio of 5:1 (5 effector cells to 1 target cell) more CC3 was detected in tumor cells co-cultured with Cbx3/HP1γ-insufficient CD8+ effector T cells compared to control co-cultures (Fig. 1F). Wild type and insufficient CD4+ activated T cells induced similar CC3 level. These results show that after activation, Cbx3/HP1γ-insufficient CD8+ T cells could differentiate into effector cells armed with enhanced killing capacity to induce apoptosis in tumor cells.
(A) Bcl6, Prdm1/Blimp, Runx3, Eomes and Tbx21 mRNA levels were assessed by RT-qPCR from in vitro-activated CD8+ T cells; p = 0.05 for Eomes day 5. (B) Eomes and T-bet proteins were detected by Western blots using total lysates from cells generated as in (A); lo: low rIL-2 concentration (10 U/ml), hi: high IL-2 (100 U/ml. (C) Gzmb, Ifng and Prf1 mRNA levels were determined by RT-qPCR using cells as in (A). (D) The frequency of IFNγ+ population was determined using intracellular FACS with cells as in (A). Numbers in FACS plots represented percent cells. Histograms indicated IFNγ protein expression levels. (E) Granzyme B protein expression was detected by Western blots using cells as in (A). (F) Effector CD4+ and CD8+ T cells were co-cultured with target NB-9464 cells at a 1:1 or 5:1 effector to target ratio for 24 hrs. Apoptosis, indicated by the presence of cleaved caspase 3, was assessed with Western blots using total NB-9464 cell lysates from co-cultures. All results were representative of 3–5 independent experiments. For (A and C), results represented fold difference; unit 1 indicated no change (n = 10 of each genotype). Full-length Western blots are shown in Supplementary Information. For A and C, statistical analysis was performed with GrathPad unpaired student t-test.
Cbx3/HP1γ deficiency limits tumor growth in mice
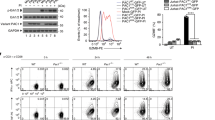
CD8+ effector T cells can directly kill tumor cells by releasing perforin, granzyme B and IFNγ. We reasoned that Cbx3/HP1γ-insufficient CD8+ T cells armed with heightened killing capacity would be more efficient in controlling tumor growth compared to wt cells. To exclude the contribution of non-immune cells in tumor killing, we performed fetal liver reconstitution of Rag2−/−cγ−/− mice, which have no T, B, NKT or NK cells. At week 10, mice reconstituted with wt littermate, Cbx3/HP1γ+/− or Cbx3/HP1γ−/− E17.5 fetal livers were implanted subcutaneously (sc) with NB-9464 tumor cells. Tumor growth was visible in mice reconstituted with wt fetal livers compared to those receiving Cbx3/HP1γ+/− or Cbx3/HP1γ−/− fetal livers (Fig. 2A). Concordantly, tumor volume was reduced in mice reconstituted with Cbx3/HP1γ+/− or Cbx3/HP1γ−/− fetal livers compared to those receiving wt fetal livers (Fig. 2B). As expected, CD8+ T cells from Cbx3/HP1γ+/− and Cbx3/HP1γ−/− fetal liver chimeras expressed significantly less and no Cbx3/HP1γ, respectively (Fig. 2B). To confirm that tumor volume reduction occurred in our Cbx3/HP1γ-insufficient mice as well, NB-9464 tumor cells were implanted in wt and Cbx3/HP1γ-insufficient mice. At day 30 after implantation, tumors from Cbx3/HP1γ-insufficient mice were smaller than those from wt mice, and ∼50% of tumor-bearing wt mice suffered tumor invasion into their peritoneal cavity, which was not observed in Cbx3/HP1γ-insufficient mice (Fig. 2C). Median volume of day 30 tumors from Cbx3/HP1γ-insufficient mice was 2.4 fold lower than that of wt mice (Fig. 2D). NB-9464 tumor-bearing Cbx3/HP1γ-insufficient mice survived longer than controls (Fig. S2). Tumors from Cbx3/HP1γ-insufficient mice exhibited extensive lymphocytic infiltration, decreased tumor-cell proliferation (less KI-67+ tumor cells) as well as large areas of necrosis, and increased tumor-cell apoptosis (higher levels of CC3) (Fig. 2E–G). By contrast, tumors excised from wt or Cbx3/HP1γ-insufficient mice expressed similar levels of the stimulatory RAE-1 and inhibitory programmed cell death ligand 1 (PD-L1) ligands, both of which were induced in tumor cells (Fig. 2H,I). These results demonstrate that Cbx3/HP1γ-insufficient mice are as efficient in controlling tumor growth as Cbx3/HP1γ−/− fetal liver chimeras.
(A) Fetal liver reconstitution was performed with fetal liver cells from wt littermate, Cbx3/HP1γ+/− and Cbx3/HP1γ−/− day 17.5 embryos (E17.5) into Rag2−/−cγ−/− recipient mice (n = 5-6 recipients for each genotype). Ten weeks after reconstitution, chimeric mice were implanted subcutaneously (sc) with NB-9464 tumor cells. Red arrows indicated tumors. (B) Tumor volume was measured on day 21 and every 2 days until day 29. Total Cbx3/HP1γ protein expression was assessed in day 5 in vitro-generated CD8+ effector T cells from chimeric mice. (C) Tumor implantation in wt and Cbx3/HP1γ-insufficient mice was performed as in (A). Red arrows and red outlines indicated tumor size and tumor in the peritoneal cavity of wt mice; n = 8 for each genotype. (D) Tumor volume was measured. (E) Tumor morphology and lymphocyte infiltration was assessed by hematoxylin and eosin (H&E) stain on paraffin sections of day 30 tumors. L = lymphocyte, T = tumor cells, red outlines indicated lymphocyte regions. (F) KI-67 was detected by immunohistochemistry on paraffin sections of day 30 tumors. Brown stain indicated KI-67 positivity, white unstained areas showed necrosis. For (E and F), images were shown as 100X (left, 10X ocular and 10X objective lens) and 400X (right, 10X ocular and 40X objective lens); 25 μm scale bar. (G) Day 30 tumors were excised and tumor cells were lysed. Cleaved caspase 3 was detected by Western blots. (H) RAE-1 protein expression was determined by Western blots in total lysates of day 30 tumors excised from wt and Cbx3/HP1γ-insufficient mice. (I) Relative Pd-l1 mRNA expression was measured by RT-qPCR using day 30 tumor cells. All results were representative of 3 independent experiments with 4 different tumors. Full-length Western blots are shown in Supplementary Information. Statistical analysis was performed with the Graphpad Two-Way Anova (B and D) and student t-test (I).
Cbx3/HP1γ insufficiency increases CD8+ effector T cells and reduces CD4+ Treg cells in tumor microenvironment
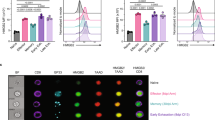
The successful control of tumor is in part dictated by the simultaneous activation of immune cells including CD8+ effector T cells and the elimination of immune suppressive elements in the tumor microenvironment. Within the microenvironment of NB-9464 tumors from Cbx3/HP1γ-insufficient mice, we detected an increased frequency of CD122+ CD44+ CD8+ (2.8 fold higher) T cells expressing the stimulatory receptor Klrk1/NKG2D (2.5-fold higher; Fig. 3A,B). The frequency of CD122+ IFNγ+ T cells among the CD8+ NKG2D+ population was 5 fold higher in tumors from Cbx3/HP1γ-insufficient mice compared to wt mice (Fig. 3C). The frequency of CD25+ CD4+ T cells expressing the inhibitory receptor CTLA4 was ∼2.6-fold lower in tumors from Cbx3/HP1γ-insufficient mice (Fig. 3D). Additionally, there was a ∼3-fold decrease in the frequency and number of CD25+ CD4+ FOXP3+ T cells (Fig. 3E and S3) as well as a reduction in Foxp3 and Ctla4 expression (Fig. 3F). It is plausible that NK and NKT cells, known to also participate in tumor clearance21,36, might have caused tumor growth reduction seen in our Cbx3/HP1γ-insufficient mice. However, we observed no differences in the frequency of NKT or Klrk1/NKG2D+ NKT cells in tumors from wt or Cbx3/HP1γ-insufficient mice (Fig. S4A,B). No discernable differences were observed in the frequency of tumor-infiltrating CTLA4+ CD8+ T cells from either wt or Cbx3/HP1γ-insufficient mice (Fig. S4C). Similar frequency of tumor-infiltrating ICOS+ CD4+ and ICOS+ CD8+ T cells was recovered from wt and Cbx3/HP1γ-insufficient mice (Fig. S4D). Tumor-infiltrating NK cells were not detected in tumors from either wt or Cbx3/HP1γ-insufficient mice (Fig. S4E). Tumors from wt and Cbx3/HP1γ-insufficient mice contained similar frequency of CD80+ and CD86+ cells (Fig. S4F). Relatively little differences were observed for other immune cells including B220+ B cells, Gr1+ and Mac-1+ myeloid cells in tumors from wt and Cbx3/HP1γ-insufficient mice (Fig. S4G,H). Cbx3/HP1γ-insufficient CD4+ T cells did not have the propensity to adopt a TH1 phenotype under non-polarizing condition (Fig. S4I). Moreover, Cbx3/HP1γ insufficiency did not affect the generation of natural CD4+ Treg cells in tumor bearing mice (Fig. S4J). Together, these results show that in tumors from Cbx3/HP1γ-insufficient mice there is an increase in CD8+ effector T cells and a decrease of CD4+ Treg cells as well as CD4+ CD25+ CTLA4+ T cells.
Flow cytometry was performed to identify tumor-infiltrating lymphocyte populations within day 30 tumors excised from wt and Cbx3/HP1γ-insufficient mice (n = 8). Graphs compiled FACS data. (A) NK1.1−CD122+ CD44+ CD3+ CD8+ T cells were gated from total CD8+ T-cell population in day 30 tumor-cell suspensions. Percent NK1.1−CD122+ CD44+ CD3+ CD8+ T cells were calculated from total CD8+ T cells, **p = 0.0023. (B) NK1.1−NKG2D+ CD44+ CD3+ CD8+ T cells were gated from total CD8+ T-cell population in day 30 tumor-cell suspensions. Percent NKG2D+ CD44+ CD3+ CD8+ T cells were calculated from total CD8+ T cells, **p = 0.0067. (C) CD122+ IFNγ+ cells were gated from CD8+ NKG2D+ T cells. Percent was calculated from total CD8+ NKG2D+ T cells, ****p < 0.0001. (D) CD25+ CTLA4+ T cells were gated from total CD4+ T-cell population within day 30 tumors from wt or Cbx3/HP1γ+/− mice. Percent CD25+ CTLA4+ T cells were calculated from total CD4+ T cell-population, ****p < 0.0001. (E) CD25+ FOXP3+ cells were gated from CD4+ T cells. Percent was calculated from total CD4+ T-cell population, **p = 0.003. (F) Ctla4 and Foxp3 mRNA expression was detected by RT-qPCR in day 30 tumors (4 tumors from each mouse strain). Results represented fold difference; unit 1 indicated no change. Each symbol represented an individual mouse; bars represented group median. Statistical analysis was performed with the GraphPad unpaired student t-test.
Adoptive transfer of Cbx3/HP1γ-insufficient CD8+ effector T cells inhibits tumor growth in mice
To formally demonstrate that CD8+ T cells cause inhibition of tumor growth, adoptive transfer experiments were performed. NB-9464 tumor cells were implanted in wt mice, on day 12 or 14 after implantation, mice were treated with in vitro-activated wt or Cbx3/HP1γ-insufficient CD8+ T cells, and tumor growth was monitored until day 31. Treatment of wt tumor-bearing mice with Cbx3/HP1γ-insufficient CD8+ activated T cells alone was successful in inhibiting NB tumor growth (Fig. 4A). By contrast, tumor growth continued in mice treated with wt CD8+ effector T cells. Analyses of TILs recovered from treated mice revealed 2-fold more CD122+ CD44+ CD8+ effector T cells in tumors from mice treated with Cbx3/HP1γ-insufficient CD8+ effector T cells compared to those treated with wt cells (Fig. 4B). Similarly, a higher frequency (3.8-fold more) of Klrk1/NKG2D+ CD8+ T cells were present in tumors from mice treated with Cbx3/HP1γ-insufficient CD8+ T cells than those treated with wt cells (Fig. 4C). There was a significant reduction (4-fold less) in the frequency of CD4+ CD25+ CTLA4+ T cells and Foxp3 expression in tumors obtained from mice treated with Cbx3/HP1γ-insufficient CD8+ T cells compared to those treated with wt cells (Fig. 4D,E). Expression of Pd-l1 in tumors from mice treated with wt or Cbx3/HP1γ-insufficient CD8+ effector T cells remained similar (Fig. 4E). We observed no obvious differences in the frequency of other immune cells (Fig. S5A–C). Our results show that treatment with Cbx3/HP1γ-insufficient CD8+ effector T cells alone was sufficient to limit tumor growth in wt tumor-bearing mice.
(A) On day 14 after induction, wt tumor-bearing mice were treated with wt or Cbx3/HP1γ-insufficient CD8+ effector T cells. Tumor volume was measured on day 22 after tumor induction (day 8 after treatment) until day 31 (n = 8 per treatment group). (B) NK1.1−CD122+ CD44+ CD3+ CD8+ T cells were gated from total CD8+ T-cell population in day 31 tumor-cell suspensions of treated mice. Percent NK1.1−CD122+ CD44+ CD3+ CD8+ T cells were calculated from total CD8+ T cells, *p = 0.0104. (C) NK1.1−NKG2D+ CD44+ CD3+ CD8+ T cells were gated from total CD8+ T-cell population in day 31 tumor-cell suspensions of treated mice. Percent NKG2D+ CD44+ CD3+ CD8+ T cells were calculated from total CD8+ T cells, *p = 0.0373. (D) CD25+ CTLA4+ T cells were gated from total CD4+ T-cell population within day 31 tumors of treated mice. Percent CD25+ CTLA4+ T cells were calculated from total CD4+ T cell-population, ***p = 0.0007. (E) Foxp3 and PD-L1 mRNA expression was assessed by RT-qPCR in day 31 tumors obtained from treated mice (3 tumors from each treatment group). Fold difference was calculated, and the unit 1 indicated no change in expression levels. Each symbol represented an individual mouse; bars represented group median. Statistical analysis was performed with GraphPad unpaired student t-test.
Cbx3/HP1γ occupies Prf1, Gzmb and Ifng loci and in part modulates histone H3K9me3 deposition
It is not known if and how Cbx3/HP1γ controls CD8+ T-cell effector programming. To objectively address these questions, Chromatin Immunoprecipitation followed by deep Sequencing (ChIP-Seq) was performed using chromatin from in vitro-activated wt CD8+ T cells. Cbx3/HP1γ was distributed across the entire genome of activated mouse CD8+ T cells with less occupancy at 3′-UTRs and downstream regions compared to ChIP-Seq profile of an unrelated control antibody (Table S1 and Fig. S6). Among genes occupied by Cbx3/HP1γ were Prf1, Gzmb and Ifng, and ChIP-qPCR analyses validated Cbx3/HP1γ occupancy at these loci (Fig. 5A,B). Cbx3/HP1γ was recruited mainly to the −2, −1 kb 5′ upstream and transcription start site (TSS) regions of Prf1, while it associated predominantly with the −7, −6 kb 5′ upstream, TSS and +2 kb regions of Gzmb. Cbx3/HP1γ occupancy of Ifng locus could only be confirmed at the TSS site. By contrast, in insufficient CD8+ effector T cells, Cbx3/HP1γ recruitment to corresponding regions was significantly reduced (>2 fold reduction). Although Bcl6, Prdm1, Tbx21 and Eomes expression in Cbx3/HP1γ-insufficient CD8+ T cells was modified, we observed no distinct Cbx3/HP1γ-bound peaks within these loci (Fig. S7A–D).
(A) ChIP-Seq was performed using chromatin from activated wt CD8+ T cells. Read-density tracks of Cbx3/HP1γ ChIP-Seq peaks across Prf1, Gzmb and Ifng were in black. The y-axis represented the number of reads per million mapped per 25-bp window; x-axis marked genomic locations. (B) The recruitment of Cbx3/HP1γ to Prf1, Gzmb and Ifng loci was confirmed by ChIP-qPCR, *p = 0.0181, **p = 0.0056 (−1kb) and 0.0062 (tss), ***p = 0.0001, ****p < 0.0001. (C) Histone H3K9me3 deposition on Prf1, Gzmb and Ifng loci was assessed, *p = 0.0271, **p = 0.0011, ****p < 0.0001. All ChIP-qPCR results were representative of 4–5 independent ChIPs. TSS: transcription start site, numbers on x-axis indicated positions of primers (in kb) along Prf1 and Gzmb loci, and 150 bp products were amplified using specific primers. Statistical analysis was performed with GraphPad unpaired student t-test and One-Way Anova.
Because Cbx3/HP1γ has been shown to interact with H3K9me3, we asked if diminished Cbx3/HP1γ levels would lead to decreased histones H3K9me3 deposition at these loci. Indeed, less H3K9m3 was positioned at sites where there was reduced Cbx3/HP1γ occupancy, specifically at 5’ upstream regions and within the gene body of Prf1 and Gzmb in Cbx3/HP1γ-insufficient CD8+ effector T cells compared to control cells (Fig. 5C). H3K9me3 deposition at Prf1 and Gzmb TSS and +2 region of the latter was not readily detectable. In Cbx3/HP1γ-insufficient CD8+ effector T cells, positioning of H3K9m3 was decreased around Ifng TSS region, coinciding with reduced Cbx3/HP1γ occupancy. These results demonstrate that the degree of histones H3K9me3 deposition at Prf1, Gzmb and Ifng loci correlates, in part, with Cbx3/HP1γ occupancy.
Runx3 occupancy and RNA Polymerase II (Pol II) recruitment/activation to Prf1, Ifng and Gzmb are regulated in part by Cbx3/HP1γ
We have previously shown that Runx3 together with Pol II in part regulate Prf1, Ifng and Gzmb expression in CD8+ effector T cells19,20. Here, ChIP-qPCR analyses showed that in Cbx3/HP1γ-insufficient CD8+ effector T cells, more Runx3 was deposited at sites where Cbx3/HP1γ and H3K9me3 occupancy was diminished (Fig. S8A). Additionally, more total Pol II was recruited to Prf1 and Ifng TSS regions and at the −7, −6 kb 5′ upstream and +2 kb regions of Gzmb in Cbx3/HP1γ-insufficient CD8+ effector T cells compared to controls (Fig. S8B). Next we asked if Cbx3/HP1γ regulates Pol II initiation [Pol (S5)]37 and elongation [Pol II (S2)]38 phases as well. ChIP-qPCR results demonstrated that the presence of Pol II (S5) at Prf1, Gzmb and Ifng loci was augmented (Fig. S8C). Specifically, more Pol II (S5) was bound at the+ 1 and+ 4 regions of Prf1 in Cbx3/HP1γ-insufficient CD8+ effector T cells. In Cbx3/HP1γ-insufficient CD8+ effector T cells, more Pol II (S5) occupied the −7 kb 5′ upstream and +2 kb regions of Gzmb, respectively; the binding pattern of Pol II (S5) and total Pol II was similar. There was also an increased in the level of Pol II (S5) assembled at the TSS region of Ifng in Cbx3/HP1γ-insufficient CD8+ effector T cells compared to control cells. There was a small increase in Pol II (S2) bound at all three loci in Cbx3/HP1γ-insufficient CD8+ effector T cells (Fig. S8D). These results show that more total and initiating/elongating Pol II are recruited to and assembled at Prf1, Gzmb and Ifng in Cbx3/HP1γ-insufficient CD8+ effector T cells.
Discussion
Here, we reveal a previously unknown and essential role for Cbx3/HP1γ in the control of tumor immunity. We show that Cbx3/HP1γ insufficiency releases the effector capacity of CD8+ T cells to control tumor growth in vivo. Under Cbx3/HP1γ insufficient condition, chromatin remodeling occurs allowing increased Runx3 and active Pol II occupancy that in part could induce Prf1, Gzmb and Ifng expression. Together, these changes may have provided the anti-tumor immunity observed in our mouse model.
Cbx3/HP1γ function is diverse. It associates with both heterochromatin and euchromatin suggesting that it may participate in transcriptional repression and activation, respectively7,8. However, it is not understood what cellular conditions invoke the repressor or activator function to control gene expression. Our results demonstrate that in activated CD8+ T cells, Cbx3/HP1γ appears to act as a repressor of the activation-induced genes Prf1, Gzmb and Infg. We propose that in normal CD8+ T cells, Cbx3/HP1γ may interact with existing H3K9me3 to maintain an unfavorable chromatin structure that limits Runx3 invasion and Pol II recruitment/activation to prevent Prf1, Gzmb and Infg excessive expression. Because Cbx3/HP1γ occupancy does not completely correspond with H3K9me3 deposition at TSS of Prf1 and Gzmb or +2 site of the latter, our data implies that Cbx3/HP1γ may interact with another protein(s) yet to be identified in CD8+ T cells. Additionally, we cannot rule out that Cbx3/HP1γ control unknown genes, which may affect Prf1, Gzmb and Infg expression.
The presence of CD8+ TEX cells has been detected in a number of tumors nonetheless their effector capacity can be reinvigorated17. It is not clear if Cbx3/HP1γ-insufficient CD8+ tumor T cells are reinvigorated TEX cells. The fact that more CD8+ T cells with effector characteristics are recovered from tumors of Cbx3/HP1γ-insufficient mice suggests that Cbx3/HP1γ insufficiency can reinvigorate the effector capacity of CD8+ T cells to control tumor growth. However, it is not yet known how Cbx3/HP1γ does so.
It is not yet understood why CD4+ Treg cells and CTLA4+CD25+CD4+ T cells are reduced. It is plausible that Cbx3/HP1γ-insufficient CD8+ effector T cells may alter the tumor microenvironment thus preventing CD4+ Treg infiltration39. Alternatively, Cbx3/HP1γ-insufficient CD8+ effector T cells may control CD4+ Treg numbers in tumors or Cbx3/HP1γ may directly control CD4+ Treg differentiation/function.
Several forms of immunotherapies are being used in the clinic with objective clinical responses. Here, we reveal an alternative novel strategy to boost tumor immunity. By reducing the levels of Cbx3/HP1γ we can increase CD8+ effector T cells at the same time eliminate CD4+ Treg immune suppression in the tumor microenvironment that may control tumor growth.
Methods
Mice and cell lines
Cbx3/HP1γ-deficient mice were generated, as described13. These mice were backcrossed to C57BL/6 for twelve generations. B6-Rag2−/−Cγ−/− mice were purchased from Taconic. When needed, female 8–12 weeks old C57BL/6 mice were purchased from The Jackson Laboratory. All mice were maintained in specific pathogen-free conditions. All mouse protocols were approved by the BIDMC Institutional Animal Care and Use Committee. All experiments were performed in accordance with relevant guidelines and regulations. The NB-9464 neuroblastoma cell line was a kind gift from Dr. Crystal MacKall at Stanford University.
Antibodies
All flow cytometry fluorochrome-conjugated antibodies were purchased from Biolegend, eBiosciences or BD Biosciences. All other antibodies were purchased as followed: anti-mouse total and phosphorylated Cbx3/HP1γ (Cell Signaling); anti-mouse RAE-1 pan-specific (R&D Systems); anti-mouse granzyme B (clone 16G6, eBiosciences); anti-mouse Eomes (Abcam); anti-mouse T-bet (clone 4B10, BioLegend); anti-mouse cleaved caspase 3 (Cell Signaling).
Tumor induction
Cbx3/HP1γ+/− and wild-type littermate mice were implanted subcutaneously with NB-9464 (1 × 106/mouse) tumor cells in 100 μl PBS. NB-9464 tumor volume was measured and calculated (W × L × 0.4) on day 22 and every 2 days until day 30 or 36 depending on the size of the tumor, at which time mice were sacrificed for analysis.
Fetal liver chimeras
At 17.5 days post coitum (d.p.c), pregnant female mice were euthanized and fetuses were retrieved. Genotyping of fetuses were performed. Fetal livers from fetuses of the correct genotype were dissected. Fetal liver cell suspension was prepared by passing through a 40 μm cell strainer and a 3-ml syringe in cold medium. Fetal liver cells were washed and resuspended at 1 × 107/ml in cold PBS. 200 μl of fetal liver cell suspension was injected intravenously into 7 week-old Rag2−/−Cγ−/− recipients. Ten weeks after reconstitution, chimeric mice were implanted subcutaneously with 1 × 106 NB-9464 tumor cells in 100 μl PBS. Tumor volume was measured and calculated (W × L × 0.4) on day 21 and every 2 days until day 29, at which time mice were sacrificed for analysis. Control wt fetal livers were obtained from littermate fetuses.
Adoptive T-cell therapy (ACT)
In vitro-generated CD8+ effector T cells were prepared as above. On day 0, female C57BL/6 mice were implanted subcutaneously with NB-9464 (1 × 106/mouse) tumor cells in 100 μl PBS. On day 14 after tumor induction, mice were treated with 3 × 106 CD8+ effector T cells via intravenous injection. NB-9464 tumor volume was measured and calculated (W × L × 0.4) on day 22 until day 31, at which time mice were sacrificed for analysis.
Fluorescence-activated cell sorting or flow cytometry (FACS)
FACS was performed on the BD 5-laser LSR II. Analysis was performed with FlowJo software (Tree Star, Inc.). Tumor cells and tumor infiltrating lymphocytes were prepared from NB-9464 tumors 29 or 30 days after implantation. Tumors were harvested, minced, and cell suspension was filtered through a 70 μm cell strainer (Falcon). Cells were washed and stained for appropriate surface markers as indicated in figures. Intracellular IFNγ staining was performed according to manufacturer’s protocol (Affymetrix/eBioscience). Briefly, activated or intratumoral CD8+ T cells (1 × 106) were treated with 1X cell stimulation cocktail with protein transport inhibitors (Affymetrix/eBioscience) in complete T-cell medium for 4 hrs at 37 °C. Cells were harvested and washed 2X with FACS buffer [PBS, 2% fetal bovine serum (FBS)]. Standard surface staining was performed. Cells were washed and incubated in fixation/permeabilization buffer (Affymetrix/eBioscience) for 30 minutes at room temperature (RT) or 18 hrs at 4 °C. Following incubation, cells were washed 2X with permeabilization buffer (Affymetrix/eBioscience) and stained for IFNγ using fluorochrome-antibodies. Cells were incubated for 30 minutes at RT. Cells were washed 2X with permeabilization buffer and resuspended in FACS buffer for analysis. Intracellular FOXP3 staining of intratumoral CD4+ CD25+ T cells was performed using similar protocol but without activation with stimulation cocktail.
In vitro activation and differentiation of CD8+ T cells to generate effector cells
CD8+ CD44− T cells were purified from spleen and peripheral lymph nodes using mouse CD8 FlowComp dynabeads according manufacturer’s protocol (Life Technologies/Thermo Fisher Scientific). T cells (1 × 106/ml) were activated with plate-bound anti-CD3 (clone 145-2C11, 0.25 μg/ml, BioLegend) and anti-CD28 (clone 37.51, 0.5 μg/ml, BioLegend) in T-cell medium (DMEM, 10% FBS, P/S, non-essential amino acids, HEPES, L-glutamate and sodium pyruvate) at 37 °C in 10% CO2 for 2 days. Cells were removed from CD3/CD28 activation and recultured (5 × 105/ml) in T-cell medium supplemented with IL-2 (10 U/ml or 100 U/ml) at 37 °C in 10% CO2. Cells were subcultured everyday. On day 5, cells were harvested and used for analyses or adoptive T-cell therapy.
Co-cultures of effector cells with NB-9464 tumor cell line for apoptosis induction
Target NB-9464 tumor cells were plated in 12-well plates at a density of 5 × 105 cells per well in 750 μl of RPMI with 10% fetal bovine serum (FBS). Activated CD8+ or CD4+ T cells were added. For ratio of 1:1 (effector:target), 5 × 105 CD8+ or CD4+ T cells: 5 × 105 NB-9464 cells were co-cultured in the same well; ratio 5:1, 2.5 × 106 CD8+ or CD4+ T cells: 5 × 105 NB-9464 cells. Plates were incubated at 37 °C in 5% CO2 for 24 hours. Wells were washed to remove non-adherent T cells. Adherent NB-9464 cells were collected and washed with 1 ml cold PBS.
Histology and immunohistochemistry
Hematoxylin and eosin (H&E) staining and immunohistochemistry were performed using 4 μm thick formalin-fixed, paraffin-embedded tissue sections. Briefly, slides were soaked in xylene, passed through graded alcohols and put in distilled water. Slides were then pre-treated with 1.0-mM EDTA, pH 8.0 or 1.0 mM Citrate (Zymed) in a steam pressure cooker (Decloaking Chamber, BioCare Medical) as per manufacturer’s instructions, followed by washing in distilled water. All subsequent steps were performed at room temperature in a hydrated chamber. Slides were pre-treated with Peroxidase Block (DAKO) for 5 minutes to quench endogenous peroxidase activity, followed by Serum free Protein Block (DAKO) for 20 minutes. Rabbit monoclonal antibody against mouse KI-67 (clone D3B5, Cell Signaling Technology) was applied and slides were incubated overnight at 4 °C. Slides were washed 2X in 50 mM Tris-Cl, pH 7.4 then followed with goat anti-rabbit IgG-HRP (SouthernBiotech), and incubated for 15 minutes. After further washing, immunoperoxidase staining was developed using a DAB chromogen (DAKO) and counterstained with hematoxylin. Images were acquired with the Nikon Eclipse TE2000 and were captured with the Hamamatsu Orca ER digital CCD camera and analyzed with NIS elements 4.13 software. Images were shown as 100X (10X ocular and 10X objective lens) and 400X (10X ocular and 40X objective lens).
Western blots
Cells (1 × 106) cells were lysed with Radio-Immunoprecipitation Assay (RIPA) buffer (Boston BioProducts) containing protease inhibitor cocktail (Roche) on ice for 30 minutes. Cells were centrifuged at 14,000 rpm for 15 minutes at 4 °C. Protein concentration was determined by Bio-Rad Protein Assay Kit (BioRad). 10 μg of protein extracts were denatured at 95 °C for 10 minutes, separated by SDS-PAGE, and transferred onto PVDF membranes (EMD Millipore). Membranes were probed with antibodies. Proteins of interest were detected with HRP-conjugated secondary antibodies and visualized with the Pierce ECL Western blotting substrate (Thermo Fisher Scientific). Membranes were exposed with HyBlot CL Autoradiography films (Denville Scientific, Inc.), and developed with the Kodax X-OMAT 2000 Processor. Films were scanned with the Epson XP-610 scanner using black and white setting, and images were captured by Adobe Photoshop Elements Editor.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The Bio-Rad Hard-Shell PCR Plates (BioRad) were used. In each well, 2.5 μl of sample was added in triplicates along with 7.5 μl of the primer mixture, which consisted of 0.5 μl forward primer, 0.5 μl reverse primer, 1.7 μl PCR grade water, and 5 μl of Roche LightCycler 480 SYBR Green I Master (Roche). The plate was centrifuged at 300 RCF for 1 minute and read in the Bio-Rad CFX384 Real-Time System (BioRad). The qPCR settings were as follow: 95 °C for 10:00 minutes 95 °C for 0:10 minutes, 62 °C for 0:20 minutes, 72 °C for 0:20 minutes (Read Plate), GOTO step 2 for 39 times, 92 °C for 0:05 minutes, Melt Curve 65.0 °C to 97.0 °C at increment 0.5 °C for 1:00 minutes (Read Plate), 40 °C for 0:10 minutes.
Chromatin immunoprecipitation (ChIP)
The chromatin immunoprecipitation (ChIP) was performed using the MAGnify ChIP System according to manufacturer’s protocol with minor modifications to optimize the assay (Life Technologies). Briefly, 2 × 107 CD8+ T cells were used for each ChIP experiment. The volume of Dynabeads Protein A/G, DNA Purification Magnetic Beads, and all the buffers were decreased by half of the suggested amount. Cells were lysed with 500 μl of lysis buffer containing 1X protease inhibitors and incubated on ice for 20 minutes. Chromatin was sheared using a Sonic Dismembrator (Fisher Scientific Model 120) at 50% power, at 10 seconds per cycle, for 7 cycles. The sheared chromatin was added to the Antibody-Dynabeads complex and incubated overnight at 4 °C in a rotating shaker for maximum binding. The heat source used was the Thermal Cycler from Bio-Rad (model C1000). The DNA was eluted using 80 μl of elution buffer, and the purified DNA was immediately used for qPCR.
ChIP followed by deep sequencing (ChIP-Seq)
Chromatin Immunoprecipitation
Mouse day-5-activated T cells were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer and disruption with a Dounce homogenizer. Lysates were sonicated and the DNA sheared to an average length of 300–500 bp. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, proteinase K and heat for de-crosslinking, followed by ethanol precipitation. Pellets were resuspended and the resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield. An aliquot of chromatin (30 μg) was precleared with protein G agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using cbx3+/−/HP1γ antibody (Cell Signaling). Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65 °C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
ChIP Sequencing (Illumina)
Illumina sequencing libraries were prepared from the ChIP and Input DNAs by the standard consecutive enzymatic steps of end-polishing, dA-addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on HiSeq 2500. Sequences (50 nt reads, single end) were aligned to the mouse genome (mm10) using the BWA algorithm. Aligns were extended in silico at their 3′-ends to a length of 200 bp, which is the average genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. The resulting histograms (genomic “signal maps”) were stored in BigWig files. Details of data analysis are shown in Supplementary Information.
Additional Information
How to cite this article: Sun, M. et al. Cbx3/HP1γ deficiency confers enhanced tumor killing capacity on CD8+ T cells. Sci. Rep. 7, 42888; doi: 10.1038/srep42888 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Canzio, D., Larson, A. & Narlikar, G. J. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 24, 377–386, doi: 10.1016/j.tcb.2014.01.002 (2014).
Singh, P. B. & Georgatos, S. D. HP1: facts, open questions, and speculation. J Struct Biol 140, 10–16, doi: S1047847702005361 [pii] (2002).
Hediger, F. & Gasser, S. M. Heterochromatin protein 1: don’t judge the book by its cover! Curr Opin Genet Dev 16, 143–150, doi: S0959-437X(06)00035-9 [pii] 10.1016/j.gde.2006.02.013 (2006).
Lomberk, G., Wallrath, L. & Urrutia, R. The Heterochromatin Protein 1 family. Genome Biol 7, 228, doi: gb-2006-7-7-228 [pii]10.1186/gb-2006-7-7-228 (2006).
Takada, Y. et al. HP1gamma links histone methylation marks to meiotic synapsis in mice. Development 138, 4207–4217, doi: 138/19/4207 [pii] 10.1242/dev.064444 (2011).
Takanashi, M. et al. Heterochromatin protein 1gamma epigenetically regulates cell differentiation and exhibits potential as a therapeutic target for various types of cancers. Am J Pathol 174, 309–316, doi: ajpath.2009.080148 [pii] 10.2353/ajpath.2009.080148 (2009).
Lomberk, G., Bensi, D., Fernandez-Zapico, M. E. & Urrutia, R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol 8, 407–415, doi: ncb1383 [pii] 10.1038/ncb1383 (2006).
Minc, E., Courvalin, J. C. & Buendia, B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet 90, 279–284, doi: ccg90279 [pii] (2000).
Saint-Andre, V., Batsche, E., Rachez, C. & Muchardt, C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol 18, 337–344, doi: 10.1038/nsmb.1995 (2011).
Yearim, A. et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep 10, 1122–1134, doi: 10.1016/j.celrep.2015.01.038 (2015).
Smallwood, A. et al. CBX3 regulates efficient RNA processing genome-wide. Genome Res 22, 1426–1436, doi: gr.124818.111 [pii] 10.1101/gr.124818.111 (2012).
Vakoc, C. R., Mandat, S. A., Olenchock, B. A. & Blobel, G. A. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19, 381–391, doi: S1097-2765(05)01390-0 [pii] 10.1016/j.molcel.2005.06.011 (2005).
Naruse, C., Fukusumi, Y., Kakiuchi, D. & Asano, M. A novel gene trapping for identifying genes expressed under the control of specific transcription factors. Biochem Biophys Res Commun 361, 109–115, doi: S0006-291X(07)01424-6 [pii] 10.1016/j.bbrc.2007.06.161 (2007).
Abe, K. et al. Loss of heterochromatin protein 1 gamma reduces the number of primordial germ cells via impaired cell cycle progression in mice. Biol Reprod 85, 1013–1024, doi: biolreprod.111.091512 [pii] 10.1095/biolreprod.111.091512 (2011).
Ha, N. et al. HP-1gamma Controls High-Affinity Antibody Response to T-Dependent Antigens. Frontiers in immunology 5, 271, doi: 10.3389/fimmu.2014.00271 (2014).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761, doi: 10.1038/nri3307 (2012).
Pauken, K. E. & Wherry, E. J. Overcoming T cell exhaustion in infection and cancer. Trends Immunol 36, 265–276, doi: 10.1016/j.it.2015.02.008 (2015).
Doering, T. A. et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144, doi: 10.1016/j.immuni.2012.08.021 (2012).
Pipkin, M. E. et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90, doi: S1074-7613(10)00010-5 [pii] 10.1016/j.immuni.2009.11.012 (2010).
Cruz-Guilloty, F. et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206, 51–59, doi: 10.1084/jem.20081242 (2009).
Lanier, L. L. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol Res 3, 575–582, doi: 10.1158/2326-6066.CIR-15-0098 (2015).
Wang, E., Selleri, S. & Marincola, F. M. The requirements for CTL-mediated rejection of cancer in humans: NKG2D and its role in the immune responsiveness of melanoma. Clin Cancer Res 13, 7228–7231, doi: 10.1158/1078-0432.CCR-07-2150 (2007).
Zhang, B. et al. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell 148, 739–751, doi: 10.1016/j.cell.2011.12.031 (2012).
Talebian, L. et al. The natural killer-activating receptor, NKG2D, on CD3+ CD8+ T cells plays a critical role in identifying and killing autologous myeloma cells. Transfusion 54, 1515–1521, doi: 10.1111/trf.12517 (2014).
Spear, P., Barber, A., Rynda-Apple, A. & Sentman, C. L. NKG2D CAR T-cell therapy inhibits the growth of NKG2D ligand heterogeneous tumors. Immunology and cell biology 91, 435–440, doi: 10.1038/icb.2013.17 (2013).
Ullrich, E., Koch, J., Cerwenka, A. & Steinle, A. New prospects on the NKG2D/NKG2DL system for oncology. Oncoimmunology 2, e26097, doi: 10.4161/onci.26097 (2013).
Barber, A., Rynda, A. & Sentman, C. L. Chimeric NKG2D expressing T cells eliminate immunosuppression and activate immunity within the ovarian tumor microenvironment. J Immunol 183, 6939–6947, doi: 10.4049/jimmunol.0902000 (2009).
Simpson, T. R. et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 210, 1695–1710, doi: 10.1084/jem.20130579 (2013).
Twyman-Saint Victor, C. et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520, 373–377, doi: 10.1038/nature14292 (2015).
Seeger, R. C. Immunology and immunotherapy of neuroblastoma. Semin Cancer Biol 21, 229–237, doi: 10.1016/j.semcancer.2011.09.012 (2011).
Cheung, N. K. et al. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte-macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 30, 3264–3270, doi: 10.1200/JCO.2011.41.3807 (2012).
Yu, A. L. et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med 363, 1324–1334, doi: 10.1056/NEJMoa0911123 (2010).
Navid, F. et al. Phase I trial of a novel anti-GD2 monoclonal antibody, Hu14.18K322A, designed to decrease toxicity in children with refractory or recurrent neuroblastoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 32, 1445–1452, doi: 10.1200/JCO.2013.50.4423 (2014).
Suzuki, M. & Cheung, N. K. Disialoganglioside GD2 as a therapeutic target for human diseases. Expert Opin Ther Targets 19, 349–362, doi: 10.1517/14728222.2014.986459 (2015).
Kroesen, M. et al. A transplantable TH-MYCN transgenic tumor model in C57Bl/6 mice for preclinical immunological studies in neuroblastoma. International journal of cancer. Journal international du cancer 134, 1335–1345, doi: 10.1002/ijc.28463 (2014).
Raulet, D. H. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol 3, 781–790, doi: 10.1038/nri1199 (2003).
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 129–143, doi: 10.1038/nrm3952 (2015).
Jonkers, I. & Lis, J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 167–177, doi: 10.1038/nrm3953 (2015).
Facciabene, A. et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature 475, 226–230, doi: 10.1038/nature10169 (2011).
Acknowledgements
We are grateful to Dr. Anjana Rao at La Jolla Institute of Allergy and Immunology for critical reading and discussion. We thank Dr. Crystal Mackall at the Center for Cancer Research, National Cancer Institute for providing the NB-9464 neuroblastoma cell line. We thank Aliakbar Shahsafaei (Brigham and Women’s Hospital, Pathology Department) for immunohistochemistry. Thanks to Arturo Contreras for fetal liver injection. We thank Paul Labhart at Active Motif and Manoj K. Bhasin at the BIDMC Bioinformatics and Systems Biology Core for analyses of ChIP-Seq data. This work was supported by NIH-NIAID grant AI099012 (to THT), The Mayer Family Fund (to THT), and Friends for Life Research Grant (to THT).
Author information
Authors and Affiliations
Contributions
T.H.T. conceived, designed and directed the study, analyzed data and wrote the manuscript with M.T.S., N.G., M.A., M.E.P. and R.E.G. N.G. and D.H.P. performed tumor induction and measured tumor volume. M.T.S., N.G. and D.H.P. performed Western blots, qPCR, in vitro activation of CD8 and CD4 T cells, FACS analysis, ChIP-qPCR and co-culture experiments. M.F. helped with in vitro activation of CD8 T cells and ChIP-qPCR. B.S. contributed to histology studies. C.N. and M.A. provide the Cbx3/HP1γ mice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sun, M., Ha, N., Pham, DH. et al. Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8+ T cells. Sci Rep 7, 42888 (2017). https://doi.org/10.1038/srep42888
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42888
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.