Abstract
Meta-analyses of randomized controlled trials (RCTs) suggest calcium could have adverse effects on cardiovascular disease, although these findings are controversial. To clarify, we assessed whether people with genetically higher calcium had a higher risk of coronary artery disease (CAD), myocardial infarction (MI) and their risk factors. We used a two-sample Mendelian randomization study. We identified genetic variants (single nucleotide polymorphisms (SNPs)) that independently contributed to serum calcium at genome-wide significance which we applied to large extensively genotyped studies of CAD, MI, diabetes, lipids, glycaemic traits and adiposity to obtain unconfounded estimates, with body mass index (BMI) as a control outcome. Based on 4 SNPs each 1 mg/dl increase in calcium was positively associated with CAD (odds ratio (OR) 1.49, 95% confidence interval (CI) 1.02–2.17), MI (OR 1.58, 95% CI 1.06–2.35), LDL-cholesterol (0.21 standard deviations, 95% CI 0.01–0.4), total cholesterol (0.21 standard deviations, 95% CI 0.03-0.38) and possibly triglycerides (0.19 standard deviations, 95% CI −0.1–0.48), but was unlikely related to BMI although the estimate lacked precision. Sensitivity analysis using 13 SNPs showed a higher risk for CAD (OR 1.87, 95% CI 1.14–3.08). Our findings, largely consistent with the experimental evidence, suggest higher serum calcium may increase the risk of CAD.
Similar content being viewed by others
Introduction
Calcium is widely seen as part of a healthy diet that promotes bone health. Recent meta-analysis of randomized controlled trials (RCTs) has suggested that calcium has little benefit for fracture prevention1. Unexpectedly, some meta-analyses of these RCTs also suggest that calcium may increase the risk of cardiovascular events, myocardial infarction (MI) and stroke2,3,4,5, although not all meta-analyses of RCTs concur on this point6. Controversy over calcium supplements arose with the Auckland Calcium Study (ACS) reporting calcium supplements increased cardiovascular disease (CVD) events in a secondary analysis of a large RCT in 20087. However, this was a hypothesis generating study because CVD events were not one of the original study outcomes. Re-analyzing data from the US Women’s Health Initiative (WHI), in a sub-group analysis of those not taking calcium supplements at randomization, the same authors of the ACS found that calcium supplementation increased the risk of MI2. Incorporating the results in a meta-analysis, the use of calcium supplementation increased the risk of MI, by 21% 2. This meta-analysis only included a small number of trials (n = 7) and compliance was low especially in the calcium arm due to adverse effects on gastrointestinal disorders, which might attenuate the potential impact on CVD events. Moreover, none of these trials were specifically designed to include primary CVD endpoints, and thus CVD events were not collected in a standard and systematic manner. In 2015, another meta-analysis of RCTs found the risk of coronary heart disease (CHD) in older women was not increased by calcium supplements6. An increase in CVD events in earlier studies was attributed to selection bias6, however, all these studies relied on adverse event reporting, whose classification is open to biases, because CVD events may not be considered as intervention-related and were not reported in some trials8. Another two recent meta-analysis of RCTs demonstrated calcium supplements (with or without vitamin D) increased risk of MI by 24–28% 4,9. The discrepancy beteen meta-analyses may be due to the inclusion of controversial RCTs, such as trials with participants who differed on CVD risk between study arms at baseline10. Moreover, meta-analysis of RCTs also suggest inconsistent effects on cardiovascular disease risk factors, specifically that calcium reduces the risk of type 2 diabetes (T2DM)2,3,4,5 and improves glucose metabolism but also increases lipids11,12. These contradictory findings for different cardiovascular risk factors again make interpretation uncertain. A large RCT with cardiovascular events as the primary endpoint and its risk factors as secondary outcomes would be definitive, but would take several years and might be difficult to justify given the lack of convincing evidence for benefits of calcium supplementation, even for bone mineral density1.
In this situation assessing coronary artery disease (CAD), MI and their risk factors according to genetically determined serum calcium, i.e., Mendelian randomization (MR), may provide insight. Since genetic endowment is randomly allocated at conception MR studies provide genetic randomization analogous to the randomization in RCTs, and so are less vulnerable to confounding and reverse causation. MR has been successfully used in cardiovascular research to investigate potential etiological mechanisms, prioritize drug targets and increase understanding of current therapies13. Here, we took advantage of genome wide association studies of calcium and large extensively genotyped studies of CAD, MI, T2DM and other CVD risk factors to obtain less confounded estimates of the effect of serum calcium on CAD, MI and their risk factors. Given calcium has no effect on body mass index (BMI) in meta-analysis of RCTs14,15, we used BMI as a control outcome, because in an unbiased analysis we would expect to see no association of calcium with BMI.
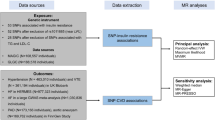
Methods
Data sources
Serum calcium
From a large meta-analysis of genome wide associations studies (GWAS) including 20,611 individuals of European ancestry16, we obtained single nucleotide polymorphisms (SNPs) independently contributing to serum calcium at genome wide significance (p < 5 × 10−8). We assessed correlation (linkage disequilibrium) between SNPs using SNP Annotation and Proxy (SNAP) Search system (http://www.broadinstitute.org/mpg/snap/ldsearchpw.php) for the same reference catalog and population17. When the correlation coefficient between SNPs was high (R2 ≥ 0.8) we discarded the SNP with the larger P value, when the correlation was lower we kept all SNPs but took into account their correlation matrix. We identified pleiotropic effects of these SNPs from Ensembl (Homo sapiens – phenotype) (http://grch37.ensembl.org/Homo_sapiens/Info/Index), a comprehensive genotype to phenotype cross-reference. We used SNPs that are approximately independent as determinants of calcium (i.e., R2 < 0.01) as the main analysis. As a sensitivity analysis we further included SNPs with low correlation (0.01 ≤ R2 < 0.8), identified from the linkage disequilibrium correlation matrix (Supplementary Table 1).
Coronary artery disease and its risk factors
Association of SNPs with the phenotypes were extracted from publicly available consortia. Data on coronary artery disease/myocardial infarction have been contributed by Coronary ARtery DIsease Genome wide Replication and Meta-analysis (CARDIoGRAM) plusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG18. As no calcium-related SNPs can be identified from the CARDIoGRAMplusC4D Metabochip, all summary data on the gene-CAD association were obtained from the CARDIoGRAM GWAS, a meta-analysis of 22 GWAS studies of European descent imputed to HapMap 2 involving 22,233 cases and 64,762 controls. As sensitivity analysis, we also use CARDIoGRAMplusC4D 1000 Genomes-based GWAS, a meta-analysis of GWAS studies of mainly European, South Asian, and East Asian, descent imputed using the 1000 Genomes phase 1 v3 training set with 38 million variants19. The study interrogated 9.4 million variants and involved 60,801 coronary artery disease (CAD) cases and 123,504 controls, and 43,676 myocardial infarction (MI) cases and 128,199 controls19. Data on T2DM was contributed by the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM, http://diagram-consortium.org/downloads.html), which includes 12,171 cases and 56,862 controls in Stage 1 GWAS20 and 26,488 cases and 83,964 controls in the Trans-ethnic GWAS meta-analysis21,22. Genetic associations with high density lipoprotein (HDL) cholesterol, low density lipoprotein (LDL) cholesterol, triglycerides, and total cholesterol in 188,577 people have been contributed by Global Lipids Genetics Consortium (GLGC) investigators and have been downloaded from http://csg.sph.umich.edu/abecasis/public/lipids2013/ 22 . Genetic associations with fasting insulin (n = 38,238), fasting glucose (n = 46,186), log-transformed homeostatic model assessment insulin resistance (log-transformed HOMA-IR, n = 46,186) have been contributed by Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) investigators and have been downloaded from http://www.magicinvestigators.org/, which relates to people of European ancestry without diabetes23.
Control outcome
We used BMI as a control outcome that, based on RCTs, should be unrelated to serum calcium14,15. Genetic associations with BMI (kg/m2) have been contributed by The Genetic Investigation of ANthropometric Traits (GIANT) investigators and have been downloaded from https://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files which has BMI for 152,893 men and 171,977 women of European ancestry24.
Statistical analysis
SNP-specific Wald estimates (ratio of SNP on outcome to SNP on calcium) of the effect of calcium on each outcome were combined using weighted generalized linear regression to account for correlation between the SNPs25, giving an odds ratio (OR) for CAD, MI and T2DM, and regression coefficients (β) for the other outcomes with 95% confidence interval (CI). As a sensitivity analysis we used inverse variance weighted (IVW) estimator with fixed effects26. Provided that the genetic variants are uncorrelated, the IVW estimate is asymptotically equal to the two-stage least squares estimate commonly used with individual-level data27. In IVW, the ratio estimates from each IV (or SNP) are combined in an inverse-variance weighted estimator. We also used a weighted median method to combine the SNP specific estimates for the uncorrelated SNPs25. Even after excluding SNPs with known potentially pleiotropic effects (i.e., SNPs that could influence both serum calcium and CAD/CVD risk factors), the estimates could still be biased by unknown pathways that directly link the genetic determinants of calcium to CAD, MI or other risk factors independent of the pathway via calcium. To assess this possible bias, MR-Egger regression was also used28. The same as the IVW method, MR Egger also uses an inverse-variance weighted estimator. It differs from the IVW method in that it allows the intercept (pleiotropy effect) to be non-zero. If the intercept in the regression model in MR-Egger were truly zero (or were constrained to be zero), the MR-Egger slope estimate is the same as the regression coefficient from IVW. If the intercept is zero it suggests that there is no violation of the exclusion restriction criteria (i.e., no horizontal pleiotropy); it provides an estimate of the average pleiotropic effect across all of the genetic variants, because it reflects the effect of the joint instruments on outcome (e.g., CAD/MI) when there is zero effect of the genetic variants on the risk factor (e.g. calcium). An intercept term that differs from zero suggests horizontal pleiotropy and that the IVW estimate may be biased. The weakness of the instruments was evaluated using the first-stage F-statistics calculated by  , where R2 indicates the variance explained by each genetic instrument, K indicated the number of instrument, and n indicate the sample size of the first stage29. All statistical analysis was performed using STATA 13.1 and R-software (Version 3.2.5).
, where R2 indicates the variance explained by each genetic instrument, K indicated the number of instrument, and n indicate the sample size of the first stage29. All statistical analysis was performed using STATA 13.1 and R-software (Version 3.2.5).
Results
Genetic determinants of serum calcium
GWAS gave 128 SNPs related to serum calcium with p < 1 × 10−5 16,30, from which we excluded 84 SNPs that do not reach genome wide significance (p < 5 × 10−8), 29 highly correlated SNPs (R2 ≥ 0.8), and 2 SNPs with pleiotropic effects (on bilirubin and parathyroid hormone) giving 13 genome-wide significant SNPs (rs4306808 in FAM162A, rs7336933 near DGKH/KIAA0564, rs17711722 in VKORC1L1, rs1067 in WDR5B; rs17267388 and rs11929034 in PARP9, rs4491840 in CCDC58, rs9864290 in CSTA, rs10491003 (closest gene GATA3), and rs16832956, rs17251221, rs13095172 and rs10222633 in CASR) as shown in Table 1. For the 13 selected SNPs, 9 were correlated (R2 > 0.1) among each other according to SNAP with HapMap release 22, as shown in the correlation matrix in Supplementary Table 1. Of these 13 SNPs, 4 from 4 different genes (rs7336933 (DGKH/KIAA0564), rs17711722 (VKORC1L1), rs17251221 (CASR) and rs10491003 (GATA3)) were uncorrelated R2 < 0.05 in HapMap CEU population. The first-stage F-statistics for the IV including these 4 SNPs was 52.
Association of genetically determined serum calcium concentrations with CAD risk
Table 2 shows that the estimates for the causal effect of 1 mg/dl higher serum calcium were consistently in the direction of higher risk of CAD and MI based on 4 SNPs, although sometimes the lower limit of the confidence interval included the null value in both CARDIoGRAM (OR 1.62, 95% CI 0.86 to 3.07) and CARDIoGRAMplueC4D 1000 Genomes-based GWAS (OR 1.49, 95% CI 1.02 to 2.17 for CAD, and 1.58, 95% CI 1.06 to 2.35 for MI) using IVW or using a weighted median (1.66, 95% CI 1.12 to 1.81 for CAD, and 1.65, 95% CI 1.06 to 2.56 for MI). The results were consistent based on 13 SNPs in CARDIoGRAM (OR 1.87, 95% CI 1.14 to 3.08) and CARDIoGRAMplueC4D 1000 Genomes-based GWAS using IVW (1.25, 95% CI 0.92 to 1.70 for CAD, and 1.32, 95% CI 0.94 to 1.85 for MI).
Association of genetically determined calcium with CAD risk factors
In all analyses the estimated effect of calcium on BMI was null but lacked precision. Similar observations from results using 4 SNPs were found for T2DM (OR 1.34, 95% CI 0.65 to 2.74) and glycemic parameters including fasting glucose, insulin and log HOMA-IR. In contrast, based on 4 SNPs, the estimates for the causal effect of 1 mg/dl higher serum calcium on LDL-cholesterol (0.21 standard deviation, 95% CI 0.01 to 0.40) and total cholesterol (0.29 standard deviation, 95% CI 0.09 to 0.48) were positive but in sensitivity analysis the confidence interval included the null for triglycerides (0.19 standard deviation, 95% CI −0.1 to 0.48). Using the MR-Egger method with the 4 SNPs, we could not reject the hypothesis that an association of these 4 SNPs with CAD, MI, T2DM or their risk factors was not independent of the effects on calcium (Supplementary Table 2).
Discussion
Consistent with most previous meta-analyses of RCTs2,3,4,5, we found that higher serum calcium was associated with a higher risk of CAD and probably MI, although we cannot definitively rule out the possibility of no effect. Findings for CAD risk factors, LDL- and total cholesterol were also positive, consistent with previous RCTs11,12. In addition, as expected, calcium appeared not to be associated with BMI14,15. As such, our study replicates findings from RCTs and extends them by showing the same pattern of associations for endogenous calcium in very large studies including substantial numbers of men as well as of women.
Our findings for CAD are consistent with most previous meta-analyses of RCTs largely pertaining to women2,3,4,6,9, although the estimate is higher than that suggested by these RCTs, which could be because calcium has a stronger association with MI in men. However, MR is more suitable for establishing direction than exact effect sizes, because genetically determined calcium represents lifetime exposure, whereas the RCTs were relatively short in duration and potentially biased towards the null by non-compliance, if the participants were successfully blinded. More generally, our findings are consistent with the observation that countries with higher calcium intake, such as Northern European countries, also have higher CAD mortality rates, while countries with low calcium intake tend to have low CAD mortality, such as China, Korea and Japan31. Of course, ecological evidence never proves causality, but this observation does require some explanation.
A previous Mendelian randomization study, using 17 SNPs for calcium from in and around the CASR gene region obtained from ~7000 participants in the European Prospective Investigation into Cancer and Nutrition (EPIC) -InterAct study, found calcium associated with higher fasting glucose in MAGIC32. However, our estimate, with genetically determined calcium obtained from a GWAS with a larger sample (n = 20,611)16, gave no association of calcium with fasting glucose (beta = −0.001, 95% CI −0.14 to 0.14). We also cannot completely rule out the possibility that the result on glucose was due to an insufficient sample size. A post-hoc power calculation for specifically glucose assuming a statistical confidence level of 0.05, an effect size of 0.03 (as results from the analysis using 13 SNPs), and serum calcium and glucose levels from the GWAS16,23 showed power of less than 20%, suggesting larger MR studies are necessary to further clarify the effect on fasting glucose. Our results are consistent with previous RCTs showing calcium supplementation has no effects on fasting glucose, although calcium supplements intake tends to reduce fasting insulin and improve insulin resistance11,12.
The heritability for total calcium is between 33% and 78% in twin studies33,34, suggesting that serum calcium levels are tightly regulated. Three major hormones are involved in the regulation of calcium homeostasis, parathyroid hormone (PTH), calcitonin and 1,25-dihydroxyvitamin D, which act on their corresponding receptors in bone, gut and kidney to maintain serum calcium concentrations35. A key regulator of the PTH release is the calcium-sensing receptor (CASR), which is mainly in the plasma membrane of chief cells of the parathyroid gland and in cells of the renal tubule36. The CASR gene encodes a protein which binds to calcium and thus plays an essential role in calcium homeostasis37. Apart from CASR, another gene, GATA3 encodes a GATA transcription factor involved in T cell lymphopoiesis, renal and vestibular morphogenesis, and parathyroid gland development38. Functional studies have shown that GATA3 haploinsufficiency causes hypoparathyroidism in populations of different ethnicities39,40,41,42. Other mechanisms exist by which calcium might contribute to CAD, for example by promoting carotid intima-media thickness43, coronary artery calcification, as occurs with calcium based phosphate binders versus non-calcium based phosphate binders44, and coagulation45. Acute induction of severe hypercalcemia in animal models reduces blood clotting time by 50%46. In vitro, increasing calcium concentrations across the physiological range reduces the clotting time of human blood47. These mechanisms could underlie effects of calcium on CAD.
Several methodologic considerations and limitations bear discussion. First, the genetic variants used for genetically determined calcium were all strongly related to calcium at GWAS level significance. No obvious reason exists for the existence of confounders of the association of these genetic variants with the outcomes considered here, for example by population stratification, because the underlying studies relate to relatively ethnically homogeneous populations of mainly European ancestry. Both calcium levels and CVD rates vary across Europe, which could be due to other factors determining calcium and CVD31, of which the calcium related genetic variants are only a marker. However, the estimate was dominated by genetic variants from the calcium-sensing receptor (CASR) gene functionally relevant to calcium, making such confounding unlikely. Second, the genetic variants used are not known to be associated with other phenotypes that might influence CAD and its risk factors, thus making biases from direct associations of SNPs with the outcomes, i.e., “pleiotropy” or violation of the “exclusion-restriction” assumption, unlikely. Moreover, we found no evidence of directional pleiotropy, i.e. that the genetic variants used to predict calcium had effects on CAD or its risk factors independent of effects via calcium. Third, we replicated established experimental evidence from meta-analysis of RCTs14,15 by showing that calcium had no effect on BMI, which gives more credence to the other estimates using the same genetic determinants of calcium. Fourth, given the use of summarized data in two samples, serum calcium levels were not measured in the sample with the outcome. However, two-sample instrumental variable analysis is more robust to chance associations than analysis of a single sample48. Fifth, it is not be possible to perform sub-group analysis or multivariable analysis as rigorously in two-sample MR as in one-sample MR using individual-level data. For example, whether the SNPs for serum calcium have different effects on CAD, T2DM or other CVD risk factors at different levels of serum calcium, by sex or at different ages could not be tested, and whether there was significant heterogeneity among sub-populations for individual instruments could not be assessed. In addition, there might be participant overlap in this two-sample MR (i.e., the same data used both for SNP selection and to calculate the IV effect)49. However, given the very large sample size of the CARDIoGRAMplusC4D consortium, assuming a 50% overlapping the sample overlap in this study is only 12%, because the dataset for deriving calcium related SNP was much smaller (n = 20,611). Thus bias from sample overlapping, if any, should not be a major concern50. Finally, both IVW and MR-Egger methods use weights that under the “NO Measurement Error (NOME) assumption”51, that is assuming the SNP-exposure associations to be known, rather than estimated. This assumption cannot be tested directly51. However, we used I2 statistics to quantify the strength of NOME violation for MR-Egger and did not find significant evidence of the violation.
Our study indicates that genetically higher serum calcium concentrations could have a harmful effect on MI and CAD. On the precautionary principle, given calcium does not seem as important in bone health as thought, our findings suggest reconsideration of the use of calcium supplementation and particularly fortification in the general population, especially in products used by older people who have higher risk of CAD.
Additional Information
How to cite this article: Xu, L. et al. A Mendelian randomization study of the effect of calcium on coronary artery disease, myocardial infarction and their risk factors. Sci. Rep. 7, 42691; doi: 10.1038/srep42691 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Tai, V., Leung, W., Grey, A., Reid, I. R. & Bolland, M. J. Calcium intake and bone mineral density: systematic review and meta-analysis. BMJ 351, h4183, doi: 10.1136/bmj.h4183 (2015).
Bolland, M. J., Grey, A., Avenell, A., Gamble, G. D. & Reid, I. R. Calcium supplements with or without vitamin D and risk of cardiovascular events: reanalysis of the Women’s Health Initiative limited access dataset and meta-analysis. BMJ 342, d2040, doi: 10.1136/bmj.d2040 (2011).
Bolland, M. J. et al. Effect of calcium supplements on risk of myocardial infarction and cardiovascular events: meta-analysis. BMJ 341, c3691, doi: 10.1136/bmj.c3691 (2010).
Mao, P. J. et al. Effect of calcium or vitamin D supplementation on vascular outcomes: a meta-analysis of randomized controlled trials. International journal of cardiology 169, 106–111, doi: 10.1016/j.ijcard.2013.08.055 (2013).
Wang, L., Manson, J. E. & Sesso, H. D. Calcium intake and risk of cardiovascular disease: a review of prospective studies and randomized clinical trials. American journal of cardiovascular drugs: drugs, devices, and other interventions 12, 105–116, doi: 10.2165/11595400-000000000-00000 (2012).
Lewis, J. R. et al. The Effects of Calcium Supplementation on Verified Coronary Heart Disease Hospitalization and Death in Postmenopausal Women: A Collaborative Meta-Analysis of Randomized Controlled Trials. Journal of Bone and Mineral Research 30, 165–175, doi: 10.1002/jbmr.2311 (2015).
Bolland, M. J. et al. Vascular events in healthy older women receiving calcium supplementation: randomised controlled trial. BMJ 336, 262–266, doi: 10.1136/bmj.39440.525752.BE (2008).
Ioannidis, J. A. Adverse events in randomized trials: Neglected, restricted, distorted, and silenced. Archives of internal medicine 169, 1737–1739, doi: 10.1001/archinternmed.2009.313 (2009).
Reid, I. R., Bristow, S. M. & Bolland, M. J. Cardiovascular complications of calcium supplements. Journal of cellular biochemistry 116, 494–501, doi: 10.1002/jcb.25028 (2015).
Larsen, E. R., Mosekilde, L. & Foldspang, A. Vitamin D and Calcium Supplementation Prevents Osteoporotic Fractures in Elderly Community Dwelling Residents: A Pragmatic Population-Based 3-Year Intervention Study. Journal of Bone and Mineral Research 19, 370–378, doi: 10.1359/jbmr.0301240 (2004).
Tabesh, M., Azadbakht, L., Faghihimani, E., Tabesh, M. & Esmaillzadeh, A. Effects of calcium-vitamin D co-supplementation on metabolic profiles in vitamin D insufficient people with type 2 diabetes: a randomised controlled clinical trial. Diabetologia 57, 2038–2047, doi: 10.1007/s00125-014-3313-x (2014).
Sanchez, M. et al. Oral calcium supplementation reduces intraplatelet free calcium concentration and insulin resistance in essential hypertensive patients. Hypertension 29, 531–536 (1997).
Haycock, P. C. et al. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr, doi: 10.3945/ajcn.115.118216 (2016).
Booth, A. O., Huggins, C. E., Wattanapenpaiboon, N. & Nowson, C. A. Effect of increasing dietary calcium through supplements and dairy food on body weight and body composition: a meta-analysis of randomised controlled trials. The British journal of nutrition 114, 1013–1025, doi: 10.1017/S0007114515001518 (2015).
Abargouei, A. S., Janghorbani, M., Salehi-Marzijarani, M. & Esmaillzadeh, A. Effect of dairy consumption on weight and body composition in adults: a systematic review and meta-analysis of randomized controlled clinical trials. Int J Obes (Lond) 36, 1485–1493, doi: 10.1038/ijo.2011.269 (2012).
O’Seaghdha, C. M. et al. Common variants in the calcium-sensing receptor gene are associated with total serum calcium levels. Human molecular genetics 19, 4296–4303, doi: 10.1093/hmg/ddq342 (2010).
Johnson, A. D. et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939 (2008).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics 43, 333–338, doi: 10.1038/ng.784 (2011).
Nikpay, M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics 47, 1121–1130, doi: 10.1038/ng.3396 (2015).
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics 44, 981–990, doi: 10.1038/ng.2383 (2012).
Mahajan, A. et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature genetics 46, 234–244 (2014).
Global Lipids Genetics, C. et al. Discovery and refinement of loci associated with lipid levels. Nature genetics 45, 1274–1283, doi: 10.1038/ng.2797 (2013).
Dupuis, J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics 42, 105–116, doi: 10.1038/ng.520 (2010).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206, doi: 10.1038/nature14177 (2015).
Burgess, S., Dudbridge, F. & Thompson, S. G. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 35, 1880–1906, doi: 10.1002/sim.6835 (2016).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genetic epidemiology 37, 658–665, doi: 10.1002/gepi.21758 (2013).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genetic epidemiology 40, 304–314, doi: 10.1002/gepi.21965 (2016).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–525, doi: 10.1093/ije/dyv080 (2015).
Palmer, T. M. et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Statistical methods in medical research 21, 223–242, doi: 10.1177/0962280210394459 (2012).
O’Seaghdha, C. M. et al. Meta-analysis of genome-wide association studies identifies six new Loci for serum calcium concentrations. PLoS genetics 9, e1003796, doi: 10.1371/journal.pgen.1003796 (2013).
Shin, C. S. & Kim, K. M. Calcium, Is It Better to Have Less? Global Health Perspectives. Journal of cellular biochemistry 116, 1513–1521, doi: 10.1002/jcb.25119 (2015).
Burgess, S. et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. European journal of epidemiology 30, 543–552 (2015).
Hunter, D. J. et al. Genetic contribution to renal function and electrolyte balance: a twin study. Clin Sci (Lond) 103, 259–265, doi: 10.1042/ (2002).
Whitfield, J. B. & Martin, N. G. The effects of inheritance on constituents of plasma: a twin study on some biochemical variables. Annals of clinical biochemistry 21 (Pt 3), 176–183 (1984).
Greer, F. R., Tsang, R. C., Searcy, J. E., Levin, R. S. & Steichen, J. J. Mineral homeostasis during lactation-relationship to serum 1, 25-dihydroxyvitamin D, 25-hydroxyvitamin D, parathyroid hormone and calcitonin. The American journal of clinical nutrition 36, 431–437 (1982).
Brown, E. M. et al. Calcium-ion-sensing cell-surface receptors. The New England journal of medicine 333, 234–240, doi: 10.1056/NEJM199507273330407 (1995).
Janicic, N. et al. Mapping of the calcium-sensing receptor gene (CASR) to human chromosome 3q13. 3-21 by fluorescence in situ hybridization, and localization to rat chromosome 11 and mouse chromosome 16. Mammalian Genome 6, 798–801 (1995).
Debacker, C., Catala, M. & Labastie, M. C. Embryonic expression of the human GATA-3 gene. Mech Dev 85, 183–187 (1999).
Shim, Y. S., Choi, W., Hwang, I. T. & Yang, S. Hypoparathyroidism, sensorineural deafness, and renal dysgenesis syndrome with a GATA3 mutation. Annals of pediatric endocrinology & metabolism 20, 59–63, doi: 10.6065/apem.2015.20.1.59 (2015).
Okawa, T. et al. A novel loss-of-function mutation of GATA3 (p.R299Q) in a Japanese family with Hypoparathyroidism, Deafness, and Renal Dysplasia (HDR) syndrome. BMC endocrine disorders 15, 66, doi: 10.1186/s12902-015-0065-7 (2015).
Zhu, Z. Y., Zhou, Q. L., Ni, S. N. & Gu, W. GATA3 mutation in a family with hypoparathyroidism, deafness and renal dysplasia syndrome. World journal of pediatrics: WJP 10, 278–280, doi: 10.1007/s12519-014-0505-x (2014).
Nanba, K. et al. A novel GATA3 nonsense mutation in a newly diagnosed adult patient of hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. Endocrine practice: official journal of the American College of Endocrinology and the American Association of Clinical Endocrinologists 19, e17–20, doi: 10.4158/ep12186.cr (2013).
Li, S. et al. Long-term calcium supplementation may have adverse effects on serum cholesterol and carotid intima-media thickness in postmenopausal women: a double-blind, randomized, placebo-controlled trial. Am J Clin Nutr 98, 1353–1359, doi: 10.3945/ajcn.113.062844 (2013).
Liu, L. et al. The effects of non-calcium-based phosphate binders versus calcium-based phosphate binders on cardiovascular calcification and bone remodeling among dialysis patients: a meta-analysis of randomized trials. Renal failure 36, 1244–1252 (2014).
Davie, E. W., Fujikawa, K. & Kisiel, W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry 30, 10363–10370 (1991).
Hilgard, P. Experimental hypercalcaemia and whole blood clotting. J Clin Pathol 26, 616–619 (1973).
James, M. F. & Roche, A. M. Dose-response relationship between plasma ionized calcium concentration and thrombelastography. J Cardiothorac Vasc Anesth 18, 581–586 (2004).
Taylor, A. E. et al. Mendelian randomization in health research: using appropriate genetic variants and avoiding biased estimates. Economics and human biology 13, 99–106, doi: 10.1016/j.ehb.2013.12.002 (2014).
Burgess, S. & Thompson, S. G. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 42, 1134–1144, doi: 10.1093/ije/dyt093 (2013).
Burgess, S., Davies, N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. Genetic epidemiology 40, 597–608, doi: 10.1002/gepi.21998 (2016).
Bowden, J. et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol, doi: 10.1093/ije/dyw220 (2016).
Author information
Authors and Affiliations
Contributions
L.X. and S.L.L. did data analysis, L.X. and C.M.S. wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xu, L., Lin, S. & Schooling, C. A Mendelian randomization study of the effect of calcium on coronary artery disease, myocardial infarction and their risk factors. Sci Rep 7, 42691 (2017). https://doi.org/10.1038/srep42691
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42691
This article is cited by
-
Genetically mimicked effects of ASGR1 inhibitors on all-cause mortality and health outcomes: a drug-target Mendelian randomization study and a phenome-wide association study
BMC Medicine (2023)
-
Insights into Causal Cardiovascular Risk Factors from Mendelian Randomization
Current Cardiology Reports (2023)
-
The impact of serum 25-hydroxyvitamin D, calcium, and parathyroid hormone levels on the risk of coronary artery disease in patients with diabetes: a Mendelian randomization study
Nutrition Journal (2021)
-
A phenome-wide association study of genetically mimicked statins
BMC Medicine (2021)
-
Calcium intake, calcium supplementation and cardiovascular disease and mortality in the British population: EPIC-norfolk prospective cohort study and meta-analysis
European Journal of Epidemiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.