Abstract
Whilst locoregional control of head and neck cancers (HNCs) has improved over the last four decades, long-term survival has remained largely unchanged. A possible reason for this is that the rate of distant metastasis has not changed. Such disseminated disease is reflected in measurable levels of cancer cells in the blood of HNC patients, referred to as circulating tumour cells (CTCs). Numerous marker-independent techniques have been developed for CTC isolation and detection. Recently, microfluidics-based platforms have come to the fore to avoid molecular bias. In this pilot, proof of concept study, we evaluated the use of the spiral microfluidic chip for CTC enrichment and subsequent detection in HNC patients. CTCs were detected in 13/24 (54%) HNC patients, representing both early to late stages of disease. Importantly, in 7/13 CTC-positive patients, CTC clusters were observed. This is the first study to use spiral microfluidics technology for CTC enrichment in HNC.
Similar content being viewed by others
Introduction
Head and neck cancers (HNCs) account for the 7th most common tumour type globally. Whilst there have been improvements in locoregional control of HNC, distant metastasis remains a challenge1. Circulating tumour cells (CTCs), are rare cancer cells shed into circulation, representing metastatic seeds and providing a window into metastasis. CTCs have the potential to provide critical information on the metastatic cascade, tumour heterogeneity and chemoresistance2,3,4,5. Whilst CTCs have been well studied in metastatic breast, prostate and colorectal cancer patients3,6, the HNC CTC field remains in its infancy1,7,8,9. Epidermal growth factor receptor (EGFR), which has been shown to be amplified in HPV-negative tumours compared to HPV-positive tumours, has previously been characterized in HNC CTCs7,9,10,11,12,13,14.
CTCs, when present, are extremely rare in comparison to the plethora of white blood cells in circulation15. The CellSearch system (Janssen Diagnostics) has FDA-approval for CTC enumeration in a number of tumour types and has been used to demonstrate the clinical relevance of EpCAM-positive CTCs6. This system has been robust in CTC enumeration through marker based CTC capture6,16. Numerous marker-based assays using microfluidic technologies are available and have been previously reported17,18. However, it has been shown that affinity based platforms have limitations due to poor sensitivity19,20. This is further compounded by varying levels of cell surface marker expression, CTC heterogeneity and specific processes such as epithelial-to-mesenchymal transition (EMT)1,21,22. The current view is that CTCs can show an intermediate EMT phenotype2,20,23 as well as express varying degrees of other markers such as PD-L1, CXCR4 and Plastin-32,24. Based on the heterogeneity found among CTCs, compounded with sometimes low-EpCAM expression, critical subpopulations may not be detected. Therefore, various marker-independent CTC enrichment strategies need to be tailored for each tumour type18,19,20. Marker-independent enrichment relies on the physical properties of CTCs such as size, deformability, charge and density1,15,19,25,26.
Recently, there has been a demand in the field for the isolation of viable CTCs on which to perform functional assays. There has been a shift toward methodologies that allow for (i) marker-independent CTC capture (ii) CTC propagation (iii) minimal pre-enrichment processing and (iv) processing larger volumes of blood in a short time period1,2,27,28,29. To encompass the above, the spiral microfluidics technology developed by Warkiani et al.15,27,30,31,32. was evaluated. The spiral chip is a marker-independent and high-throughput device that utilises hydrodynamic forces present in curvilinear microchannels for size-based cell sorting15,27. The spiral chip has previously been reported for use in breast and lung cancers32. CTC separation from lysed whole blood occurs based on CTC deformability and size. In this study, we enriched blood from 24 HNC patients using the spiral microfluidics platform and enumerated CTCs using immunocytochemistry. Our evaluation is the first study using the spiral technology for CTC enrichment from patients with HNCs, and shows positive results. Future, multi-centre trials are required and warranted before this technology can be implemented in a clinical setting.
Results
Spiral Chip enrichment
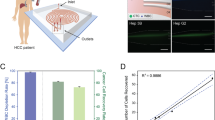
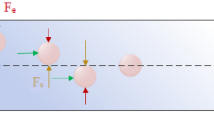
The spiral chip utilizes the inherent Dean vortex flows present in curvilinear microchannels under continuous flow, along with inertial lift forces, which focus the CTCs near the microchannel inner wall while driving the smaller hematologic cells toward the microchannel outer wall, allowing an efficient separation at the outlet (Fig. 1). The cells are separated as a function of deformability and size, as cells occupy different lateral positions away from the walls of the microchannel, allowing for separation at the bifurcation position27. Typically, cells larger than 14 μm flow into the “CTC outlet” and cells smaller than this size flow into the “waste outlet.”
Recovery Studies
FaDu, CAL27, RPMI2650 and MDA-MB-468 cell line cells, which all have a mean diameter above 15 μm (Fig. 2), were used for spike-in experiments. The percentage recovery of spiked-in cells (Fig. 3i) ranged from 60–70% when experiments were performed with 100–150 cells spiked into 10 mL blood, trending to lower recover rates (40–60%) when less cells were spiked in. The overall recovery of the spiked cell lines were comparable to each other. We also assessed the decrease in background leukocytes and recovery of cancer cells after repeat passages through the spiral chip. When the samples were collected from the “CTC outlet” and run through the chip again, a 30% decrease in leukocytes was observed, however there was a 10–15% decrease in cancer cell recovery which was accounted for in the dead volumes of the setup. More than 1 repeat run did not further purify the sample. Single cells and cell clusters were able to be enriched by the spiral chip (Fig. 3ii). Similarly, bioreactor-cultured spheroids could also be enriched by the spiral chip (Fig. 4).
(i) The mean % recovery and standard deviation for spiked cell lines (n = 3). (ii, a) Presentation of spike-in and recovered tumour cells FaDu cells into blood of a normal healthy volunteer. Tumour cells: cytokeratin (red), DAPI (blue) White blood cells: CD45 (green), DAPI (blue). Scale bar represents 10 μm (b) RFP-labelled tumour cells from whole blood. Scale bar represents 50 μm.
Patient Characteristics
A total of 24 patients and 8 normal healthy controls were included in this study. Patient characteristics are shown in Table 1. The median age was 62 years (range 36–89); including men (n = 22), women (n = 2). The most distinctive histology was mucosal oral cavity SCC (63%). The majority of patients presented with tumour grading T2/T3 (71%) and either no nodal spread N0 (50%) or advanced nodal spread N2a-c (50%). Patients were assessed for distant metastases by CT/PET-CT and MRI scans. All patients were found to be radiographically M0 upon assessment. CTC status was determined at the same time point. The clinicopathological patient findings are presented in Table 2.
Enrichment of CTCs from controls and patients
10 mL blood samples were obtained from 8 healthy volunteers and 24 head and neck cancer patients and processed using the spiral chip. All samples were enumerated for CTCs. Putative CTCs were identified on the stained cytospots as positive for: pan-CK, DAPI and negative for CD45. Further characterisation of head and neck CTCs was performed by EGFR ICC and DNA FISH. Leukocytes were identified as positive for CD45, DAPI and negative for pan-CK. No CTC-like events were observed in the 8 normal healthy volunteers (Fig. 5). CTCs were detected in 13/24 (54%) (Range 1–12 CTCs/10 ml blood sample) of the HNC patients screened in the study. CTC clusters were readily detectable in 7/13 (53.8%) of the CTC-positive patients (Fig. 6). The CTCs were further molecularly characterised and found to have EGFR gene amplifications (Fig. 7). Further observations made in the patient samples include double positive pan-CK and CD45 cells in 2/20 samples and 27 CK-negative, CD45-negative cells in 1 patient presenting with a CTC cluster. These events were not considered for enumeration in the study.
(a) 100x magnification showing the CTC cluster in the top of the cytospot. 1000x images of (b) DAPI (c) Cytokeratin/DAPI composite (d) DAPI (e) Cytokeratin (f) Cytokeratin/DAPI composite. Scale bar represents 50 μm. Figure 6 continued. Patient “micro_12”; CTC doublet detected in blood. (g) DAPI (h) Cytokeratin/DAPI composite (i) EGFR/DAPI composite. Patient “micro_03” single CTCs detected in blood. (j) DAPI (k) Cytokeratin/DAPI composite (l) EGFR/DAPI composite. Scale bar 10 μm (g–i). Scale bar 50 μm (j–l).
Discussion
HNC patients often present with advanced metastatic spread, predominantly to the lungs, liver and bone1,33. The ability to assess “intact” metastatic precursors in the form of tumour cells in blood holds remarkable potential as a liquid biopsy. The number of CTCs, and molecular and genetic changes in the CTCs could be monitored in patients following therapy and non-responders determined at an earlier time point4,34,35. Moreover, patients whose disease will potentially respond to specific therapies could be identified. Recent studies have shown that PD-L1 is frequently expressed on CTCs and that these are present in patients where anti-PDL1 therapies could be effective24,36,37. Numerous CTC enrichment platforms have been developed in recent years, each with its own advantages and limitations1,21,38,39,40. In this study, we tested and evaluated the spiral microfluidics chip for CTC enumeration and characterization. This platform has features which are of value to clinical research; CTCs are found in suspension and not immobilized allowing for functional assays and CTC clusters can be readily isolated with a leukocyte depletion of up to 4 log15. Limitations of the spiral technology are that “small CTCs” or CTCs ( < 12 μm) in size will be lost15,41.
Four cell lines representing a mean cell diameter range between 16–28 μm were chosen for the spiking experiments to get a decent coverage of known head and neck cancer cells. In our hands, the percentage recovery of the spiral chip is similar to that of other microfluidic CTC isolation platforms19,21. By repeat passages of the spiral chip, a decrease of 30% background leukocytes was found at the expense of 10–15% recovery. However, repeated passages were not performed with patient samples so as not to risk losing CTCs. The range of detected CTCs in HNC patients remains low1,13,42,43. Moreover, it was observed that the same population of leukocytes was present after 2 or more repeat runs and this could not be further purified. Furthermore, the spiral chip successfully sorted for cell clusters and tumour spheroids in the CTC outlet.
This pilot study evaluated the use of a spiral chip for CTC enrichment in HNCs. HNC patients ranged from early stage (T1N0M0) to advanced stage patients (T4N2b). Patients with early stage disease present with low rates of distant metastasis and a favourable prognosis. Whilst patients with locally advanced disease are more likely to have CTCs44. CTCs were observed in 50% of the HNC samples, ranging from 1–12CTCs/10 mL sample. This finding is consistent with previously published studies using marker-independent enrichment platforms7,13,45. Notably, post spiral enrichment involves transferring rare cells onto a glass slide and immunocytochemistry which can lead to cell loses34. Therefore, the actual CTC count per HNC patient could be much higher.
Over half of the CTC positive patients presented with clusters of cancer cells. These multicellular CTC clusters have shown 23–50 fold increased metastatic potential compared to single CTCs in breast cancers and associations with adverse clinical outcomes1,46,47,48,49, however the clinical significance in HNC is yet to be determined1,50,51,52. Moreover, these cell clusters have been reported to have a shorter circulation half-life in blood, consistent with rapid entrapment within the capillaries of distant organs46. Initial studies have shown that CTC clusters are potent initiators of distant organ metastasis, highlighting the need to develop strategies which disrupt cell-cell or cell-matrix adhesions in these clusters to reduce metastatic potential53. Further investigation is required to determine the clinical implications of CTC clusters in HNC patients. Importantly, CTCs were found upon presentation to clinic in 3/3 patients presenting with queried lung lesions by the MDT clinic (Fig. 8). These lung lesions were to be investigated by ultrasound guided fine-needle biopsy to determine the extent of distant metastasis. In these patients, a window for treatment escalation could become a possibility if the presence of CTCs predicts for patients at-risk of developing metastasis. Further studies assessing CTCs during the course of treatment and timing of blood sampling are warranted to investigate the role of monitoring CTCs in HNC50.
Conclusion
We present the first study in HNCs using a microfluidics based approach for CTC enrichment. 24 HNC patient blood samples were enriched using the spiral microfluidics chip. We were able to identify single and clusters of CTCs and further characterise these cells by ICC and DNA FISH. This study highlights the use of the spiral microfluidic technology in HNC and provides future clinical use in the field of CTCs.
Materials and Methods
Patient cohort
Ethics approval was obtained from the Metro South and North Health Service District Human Research Ethics Committee in accordance with the National Health and Medical Research Council’s guidelines (HREC/12/PHAH381) to collect blood from the Royal Brisbane and Women’s Hospital (RBWH). All methods were performed in accordance with these ethical guidelines and regulations. This study also has Queensland University of Technology ethics approval (1400000617). After written informed consent was obtained from the participants, 10 mL blood samples were collected from patients presenting to the HNC clinic.
Cell lines and culture
The cell lines FaDu (ATCC-HTB43), CAL27 (ATCC-CRL2095), RPMI2650 (ATCC-CCL30) were obtained from ATCC (Manassas, VA). UD-SCC9 HNC cells (ATCC-CRL1629) were a generous gift from A/Prof Nick Saunders (UQDI). The MDA-MB-468 breast cancer cells were originally from the ATCC transferred from the Lombardi Cancer Center and fluorescently tagged with pCherryN1, a modification of the Clonetech vector, kindly provided by Dr Alpha Yap, (UQ). Cells were cultured under standard conditions in humidified incubators at 37 °C, 5% CO2 in RPMI-1640-Glutamax (Life Technologies, Inc) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin (Pen/Strep).
3D cell culture (bioreactor)
Cells were spun down to obtain cell pellets post-trypsinization. The cell pellets were resuspended in pre-warmed 1x Happy Cell media (Biocroi Ltd, Dublin, Ireland) supplemented with 10% FBS and 1% Pen/Strep. Optimal seeding densities for the bioreactors were determined by titrating cells from 10,000–500 cells/ml. 1,500 cells/ml, resuspended in 1x Happy Cell media were layered over 2x Happy Cell media in the bioreactor. This gradient ensured that the cells remained in suspension over the incubation periods. The cells were incubated at 37 °C, 5% CO2 and imaged every 3 days. To collect the spheroids, the cell suspension was transferred to a 96 well format and an inactivation buffer was used to collapse the hydrogel matrix for 1 hour at 37 °C.
Spiking of cell lines into bloods collected from normal healthy controls
Blood was collected from normal healthy volunteers in 10 mL EDTA (BD-Plymouth, UK) tubes. The cell lines used for spiking were labelled with CellTracker™ Green (Life Technologies) as per manufacturer’s instructions. The collected blood was spiked with clinically relevant numbers (10–150) of cells within 2 hours of collection. Cell lines for spiking were harvested by trypsinisation for 5 mins at 37 °C and washed in the culture media. The cell suspension was centrifuged at 300 × g for 5 mins, the supernatant removed and cells resuspended in PBS buffer. Cell counts were performed with the Countess II FL automated Cell counter (ThermoFisher Scientific). The appropriate spike-in volume of cells was added to the blood.
Preparation of blood sample for the spiral Chip
To reduce the cellular components passing through the spiral chip, an initial red blood cell (RBC) lysis was performed, which has been shown to have minimal effects on the recovered cells15. Briefly, RBC lysis buffer (Astral Scientifix) was added to the blood sample, after which cells were centrifuged and the pellet resuspended in 10 mL of sheath buffer (1xPBS, 2 mM EDTA, 0.5% BSA).
Setup and running of the spiral Chip
Tygon® tubing was inserted into the inlet/outlets of the spiral chip, and the inlet tubing connected to a syringe pump. The spiral chip was positioned and fixed onto a phase contrast microscope (Olympus IX71). The outlet tubing was connected to two sterile 15 mL collection BD Falcon tubes. An initial priming run was performed using the sheath buffer at a flow rate of 2.0 mL/min for 5 minutes. To test patient samples, the sample was loaded carefully into a 10 ml syringe (Terumo) and pumped through the spiral chip using the syringe pump at a flow rate of 1.7 mL/min for all experiments. The outputs were collected and spun down at 300× g for 5 mins.
Immunostaining
Prelabeling and immunocytochemistry (ICC) were used for the detection of spiked tumour cells and CTCs enriched by the spiral microfluidic chip. Enriched cells were cytospun onto a glass slide and CTCs identified by ICC using the CellSearch antibody cocktail (Cytokeratin/CD45/DAPI; Janssen Diagnostics). Cells were further characterised for cell surface EGFR using an anti-EGFR antibody (AY13, BioLegend, San Diego). Briefly, the cytospun sample was air dried overnight and incubated with a mixture of CellSearch reagents (20 ul staining reagent, 20 μl DAPI, 20 μl permeabilization buffer, 20 ul fixation buffer in 120 μl PBS). The slide was incubated for 1 hour at room temperature, washed 3 times in PBS, coverslipped and imaged under a fluorescent microscope. The CellSearch criteria for CTCs were used for CTC identification: intact, cytokeratin-positive, DAPI-positive, CD45-negative cells that were larger than 4 um were considered a CTC. Cells staining positive for CD45 and DAPI and negative for pan-cytokeratin were determined to be white blood cells.
DNA Fluorescence in situ hybridization
Cytospots were placed in pretreatment solution for 10 mins at 98 °C, and digested with pepsin for 5 mins at 37 °C in the Dako hybridiser. After dehydration through a graded ethanol series (70%, 85%, 96%), dual colour, dual target DNA FISH assays were done with EGFR/CEN-7 FISH probe mix (DakoCytomation, Glostrup, Denmark). Cytospots were covered with 10 μL probe solution, sealed with rubber sealant and placed in the Dako Hybridizer for denaturation (5 mins, 82 °C) and hybridisation (18 hours, 45 oC). Post hybridisation stringency washes were performed at 65 oC for 10 mins. After further washing and dehydration steps, the cytospots were air dried and mounting media containing DAPI added. The cytospot was coverslipped and imaged on the Nikon Eclipse Ti inverted microscope fitted with a Nikon digital camera. EGFR was visualized as a red signal (tetramethylrhodamine isothiocyanate filter), CEP7 (fluorescein isothiocyanate filter) as a green signal and the nuclei as a blue signal with a DAPI filter. EGFR status was scored as the number of EGFR signals per nucleus and as the ratio of EGFR signals to CEP7 signals.
Additional Information
How to cite this article: Kulasinghe, A. et al. Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci. Rep. 7, 42517; doi: 10.1038/srep42517 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Kulasinghe, A., Perry, C., Jovanovic, L., Nelson, C. & Punyadeera, C. Circulating tumour cells in metastatic head and neck cancers. International journal of cancer 136, 2515–2523, doi: 10.1002/ijc.29108 (2015).
Alix-Panabieres, C. & Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer discovery 6, 479–491, doi: 10.1158/2159-8290.cd-15-1483 (2016).
Alix-Panabieres, C. & Pantel, K. Challenges in circulating tumour cell research. Nat Rev Cancer 14, 623–631, doi: 10.1038/nrc3686 (2014).
Gasch, C. et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Molecular oncology, doi: 10.1016/j.molonc.2016.07.005 (2016).
Bidard, F. C., Proudhon, C. & Pierga, J. Y. Circulating tumor cells in breast cancer. Molecular oncology 10, 418–430, doi: 10.1016/j.molonc.2016.01.001 (2016).
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical cancer research: an official journal of the American Association for Cancer Research 10, 6897–6904, doi: 10.1158/1078-0432.ccr-04-0378 (2004).
Balasubramanian, P. et al. Confocal images of circulating tumor cells obtained using a methodology and technology that removes normal cells. Molecular pharmaceutics 6, 1402–1408, doi: 10.1021/mp9000519 (2009).
Buglione, M. et al. Circulating tumour cells in locally advanced head and neck cancer: preliminary report about their possible role in predicting response to non-surgical treatment and survival. European journal of cancer (Oxford, England: 1990) 48, 3019–3026, doi: 10.1016/j.ejca.2012.05.007 (2012).
Grisanti, S. et al. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: prognostic and predictive significance. PloS one 9, e103918, doi: 10.1371/journal.pone.0103918 (2014).
Hristozova, T. et al. The presence of circulating tumor cells (CTCs) correlates with lymph node metastasis in nonresectable squamous cell carcinoma of the head and neck region (SCCHN). Annals of oncology: official journal of the European Society for Medical Oncology / ESMO 22, 1878–1885, doi: 10.1093/annonc/mdr130 (2011).
Tinhofer, I., Hristozova, T., Stromberger, C., Keilhoiz, U. & Budach, V. Monitoring of circulating tumor cells and their expression of EGFR/phospho-EGFR during combined radiotherapy regimens in locally advanced squamous cell carcinoma of the head and neck. International journal of radiation oncology, biology, physics 83, e685–690, doi: 10.1016/j.ijrobp.2012.02.009 (2012).
Zimmermann, M., Zouhair, A., Azria, D. & Ozsahin, M. The epidermal growth factor receptor (EGFR) in head and neck cancer: its role and treatment implications. Radiation Oncology 1, 1–6, doi: 10.1186/1748-717x-1-11 (2006).
Kulasinghe, A. et al. Short term ex-vivo expansion of circulating head and neck tumour cells. Oncotarget, doi: 10.18632/oncotarget.11159 (2016).
The Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582, doi: 10.1038/nature14129 http://www.nature.com/nature/journal/v517/n7536/abs/nature14129.html#supplementary-information (2015).
Warkiani, M. E. et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nature protocols 11, 134–148, doi: 10.1038/nprot.2016.003 (2016).
Bidard, F. C. et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. The Lancet. Oncology 15, 406–414, doi: 10.1016/s1470-2045(14)70069-5 (2014).
Huang, Y. Y. et al. Screening and Molecular Analysis of Single Circulating Tumor Cells Using Micromagnet Array. Sci Rep 5, 16047, doi: 10.1038/srep16047 (2015).
Ozkumur, E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Science translational medicine 5, 179ra147, doi: 10.1126/scitranslmed.3005616 (2013).
Chudziak, J. et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. The Analyst 141, 669–678, doi: 10.1039/c5an02156a (2016).
Hyun, K. A. et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget, doi: 10.18632/oncotarget.8250 (2016).
Hvichia, G. E. et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. International journal of cancer 138, 2894–2904, doi: 10.1002/ijc.30007 (2016).
Bednarz-Knoll, N., Alix-Panabieres, C. & Pantel, K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer metastasis reviews 31, 673–687, doi: 10.1007/s10555-012-9370-z (2012).
McInnes, L. M. et al. Clinical Implications of Circulating Tumor Cells of Breast Cancer Patients: Role of Epithelial–Mesenchymal Plasticity. Frontiers in Oncology 5, 42, doi: 10.3389/fonc.2015.00042 (2015).
Mazel, M. et al. Frequent expression of PD-L1 on circulating breast cancer cells. Molecular oncology 9, 1773–1782, doi: 10.1016/j.molonc.2015.05.009 (2015).
Shim, S. et al. Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis. Biomicrofluidics 7, 11807, doi: 10.1063/1.4774304 (2013).
Sarioglu, A. F. et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nature methods 12, 685–691, doi: 10.1038/nmeth.3404 (2015).
Warkiani, M. E. et al. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. The Analyst 139, 3245–3255, doi: 10.1039/c4an00355a (2014).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187, doi: 10.1016/j.cell.2014.08.016 (2014).
Fischer, J. C. et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proceedings of the National Academy of Sciences of the United States of America 110, 16580–16585, doi: 10.1073/pnas.1313594110 (2013).
Hou, H. W. et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Scientific reports 3, 1259, doi: 10.1038/srep01259 (2013).
Warkiani, M. E. et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab on a chip 14, 128–137, doi: 10.1039/c3lc50617g (2014).
Khoo, B. L. et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PloS one 9, e99409, doi: 10.1371/journal.pone.0099409 (2014).
Schmidt, H., Kulasinghe, A., Perry, C., Nelson, C. & Punyadeera, C. A liquid biopsy for head and neck cancers. Expert review of molecular diagnostics 16, 165–172, doi: 10.1586/14737159.2016.1127758 (2016).
Xu, L. et al. Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PloS one 10, e0138032, doi: 10.1371/journal.pone.0138032 (2015).
Gasch, C. et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clinical chemistry 59, 252–260, doi: 10.1373/clinchem.2012.188557 (2013).
Herbst, R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567, doi: 10.1038/nature14011 (2014).
G, K., Strati, A. & A, M. et al. PDL1-expressing circulating tumor cells (CTCs) in head and neck squamous cell carcinoma (HNSCC). J Clin Oncol 33 (2015).
Lianidou, E. S., Strati, A. & Markou, A. Circulating tumor cells as promising novel biomarkers in solid cancers. Critical reviews in clinical laboratory sciences 51, 160–171, doi: 10.3109/10408363.2014.896316 (2014).
Gorges, T. M. et al. Improved Detection of Circulating Tumor Cells in Metastatic Colorectal Cancer by the Combination of the CellSearch(R) System and the AdnaTest(R). PloS one 11, e0155126, doi: 10.1371/journal.pone.0155126 (2016).
Gorges, T. M. et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clinical cancer research: an official journal of the American Association for Cancer Research 22, 2197–2206, doi: 10.1158/1078-0432.ccr-15-1416 (2016).
Plaks, V., Koopman, C. D. & Werb, Z. Circulating Tumor Cells. Science (New York, N.Y.) 341, 10.1126/science.1235226, doi: 10.1126/science.1235226 (2013).
McMullen, K. P., Chalmers, J. J., Lang, J. C., Kumar, P. & Jatana, K. R. Circulating tumor cells in head and neck cancer: A review. World Journal of Otorhinolaryngology-Head and Neck Surgery, doi: 10.1016/j.wjorl.2016.05.003 (2016).
Wu, X. L. et al. Diagnostic and Prognostic Value of Circulating Tumor Cells in Head and Neck Squamous Cell Carcinoma: a systematic review and meta-analysis. Scientific reports 6, 20210, doi: 10.1038/srep20210 (2016).
Bozec, A. et al. Significance of circulating tumor cell detection using the CellSearch system in patients with locally advanced head and neck squamous cell carcinoma. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies (EUFOS): affiliated with the German Society for Oto-Rhino-Laryngology - Head and Neck Surgery 270, 2745–2749, doi: 10.1007/s00405-013-2399-y (2013).
Jatana, K. R. et al. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Archives of otolaryngology–head & neck surgery 136, 1274–1279, doi: 10.1001/archoto.2010.223 (2010).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122, doi: 10.1016/j.cell.2014.07.013 (2014).
Khoo, B. L. et al. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Science advances 2, e1600274, doi: 10.1126/sciadv.1600274 (2016).
Yu, M., Stott, S., Toner, M., Maheswaran, S. & Haber, D. A. Circulating tumor cells: approaches to isolation and characterization. The Journal of cell biology 192, 373–382, doi: 10.1083/jcb.201010021 (2011).
Aceto, N., Toner, M., Maheswaran, S. & Haber, D. A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends in Cancer 1, 44–52, doi: 10.1016/j.trecan.2015.07.006 (2015).
Jatana, K. R. et al. Effect of surgical intervention on circulating tumor cells in patients with squamous cell carcinoma of the head and neck using a negative enrichment technology. Head & neck 38, 1799–1803, doi: 10.1002/hed.24519 (2016).
Kulasinghe, A. et al. Impact of label-free technologies in head and neck cancer circulating tumour cells. Oncotarget, doi: 10.18632/oncotarget.12086 (2016).
Jatana, K. R., Lang, J. C. & Chalmers, J. J. Identification of circulating tumor cells: a prognostic marker in squamous cell carcinoma of the head and neck? Future oncology (London, England) 7, 481–484, doi: 10.2217/fon.11.19 (2011).
Hanssen, A. et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 6, 28010, doi: 10.1038/srep28010 (2016).
Acknowledgements
The authors would like to thank Prof William B Coman (Brisbane, Australia) for clinical guidance, Ms Jenny Edmunds (Clinical Trials Coordinator, RBWH), Dr. Anthony M Davies for the Happy Cell media and Dr. Christin Gasch for editorial assistance. This study was supported by the Queensland Centre for Head and Neck funded by Atlantic Philanthropies, the Queensland Government and the Royal Brisbane and Womens’ Hospital. QUT VC Fellowship for CP. QUT postgraduate research scholarship for AK.
Author information
Authors and Affiliations
Contributions
A.K., T.H.P.T.: data collection, experimental procedures, design, manuscript preparation K.O., E.T., M.E.W., T.B.: analysis, experimental design, manuscript preparation L.K., C.N., C.P.: study design, analysis, clinical accruals, manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kulasinghe, A., Tran, T., Blick, T. et al. Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci Rep 7, 42517 (2017). https://doi.org/10.1038/srep42517
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42517
This article is cited by
-
Poor patient outcome correlates with active engulfment of cytokeratin positive CTCs within cancer-associated monocyte population in lung cancer
Clinical & Experimental Metastasis (2024)
-
Liquid biopsy: early and accurate diagnosis of brain tumor
Journal of Cancer Research and Clinical Oncology (2022)
-
A short review of spiral microfluidic devices with distinct cross-sectional geometries
Microfluidics and Nanofluidics (2022)
-
Cytoplasm protein GFAP magnetic beads construction and application as cell separation target for brain tumors
Journal of Nanobiotechnology (2020)
-
3D Printing of Inertial Microfluidic Devices
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.