Abstract
It is highly possible that copy number variations (CNVs) in susceptible regions have effects on chronic obstructive pulmonary disease (COPD) development, while long noncoding RNA (lncRNAs) have been shown to cause COPD. We hypothesized that the common CNV, named nsv823469 located on 6p22.1, and covering lncRNAs (major histocompatibility complex, class I, A (HLA-A) and HLA complex group 4B (HCG4B)) has an effect on COPD risk. This association was assessed through a two-stage case-control study, and was further confirmed with COPD and pulmonary function-based family analyses, respectively. The copy number loss (0-copy/1-copy) of nsv823469 significantly decreased risk of COPD compared with normal (2-copy) (OR = 0.77, 95% CI = 0.69–0.85). The loss allele, inducing copy number loss of nsv823469, has a tendency to transmit to offspring or siblings (P = 0.010) and is associated with forced expiratory volume in 1 second (FEV1) (P = 0.030). Furthermore, the copy number loss of nsv823469 in normal pulmonary tissue decreases the expression levels of HCG4B (r = 0.315, P = 0.031) and HLA-A (r = 0.296, P = 0.044). Our data demonstrates that nsv823469 plays a role in COPD and pulmonary function inheritance by potentially altering expression of HCG4B.
Similar content being viewed by others
Introduction
Multiple association studies, especially genome-wide association studies (GWASs), have revealed several susceptible loci for chronic obstructive pulmonary disease (COPD) and pulmonary function1,2,3. However, these loci by no means cover the complete genetic predisposition of COPD, because they mainly focus on single nucleotide polymorphism (SNP). Copy number variation (CNV)4,5 also contributes to COPD heritability6,7. CNVs can hold greater influence over covering genes than SNPs do, because CNVs can cause changes in a large DNA fragment while SNPs can only cause one base change8. Thus, it is highly possible that the CNVs located on susceptible regions to COPD have contributed to COPD development.
Long non-coding RNAs (lncRNAs) have been documented that play important roles in COPD9,10. Altered expression of lncRNAs, such as LINC00882, LINC00883 and PVT1, was observed in lung tissues of COPD patients11,12. A study showed that one CNV that caused disruption of LINC00299 contributed to human developmental disorders13. Overall, we hypothesized that the CNVs covering lncRNAs in COPD susceptible regions have effect on COPD risk.
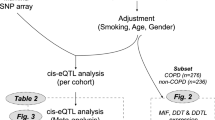
Among the susceptible loci of COPD, only the 6p21 region was discovered by the GWAS conducted in the East Asian population14. The 6p21 region was included at a ~3.8 Mb interval on chromosome 6p21.32–22.1 with long-range linkage disequilibrium, and was found to have association with lung function in GWAS15. In reference to the published East Asian CNVs data16, we only found one common CNV named nsv823469 with altered copy number frequency (ACNF) > 5% located on the 6p22.1 region and covering lncRNAs. Thus, in the current study, we conducted a two-stage case-control study to find the correlation between the CNV and COPD risk, and further proved the association in two family-based analyses. The function of this CNV was further assessed by quantitative real time PCR.
Results
Association between the nsv823469 and risk of COPD
Significantly lower frequencies of the loss genotypes (0-copy/1-copy) were observed in cases than in controls in both the southern (P < 0.001) and the eastern (P = 0.005) Chinese population (in Table 1). According to the genetic model selection strategy based on the smallest Akaike Information Criterion (AIC) value17, the additive genetic model was best fitting for analysis on the effect that nsv823469 holds in COPD susceptibility. Compared to the 2-copy, the loss genotypes (0-copy/1-copy) conferred a significantly decreased risk of COPD in southern Chinese population (adjusted odds ratios (OR) = 0.77, 95% confidence interval (95% CI) = 0.68–0.87) and east Chinese population (adjusted OR = 0.76, 95% CI = 0.65–0.91). Through merging the two populations (Breslow-Day test: P = 0.767) in order to increase our study power, the COPD risk among the loss genotypes carriers was decreased by 23% in comparison to the 2-copy carriers (adjusted OR = 0.77, 95% CI = 0.69–0.85). Data from the stratification analysis further showed there was no significant difference among the two or three stratum-ORs (Breslow-Day test: P > 0.05 for all); meanwhile, no significant interaction was observed among all surrounding factors and the CNV on decreasing COPD risk (P > 0.05 for all; Supplementary Table S1).
Transmission mode of nsv823469 among COPD and pulmonary function pedigrees
According to number of the mutant loss alleles, the 2-copy, 1-copy, and 0-copy genotypes were defined as wild genotypes (two normal alleles), mutant heterozygote (one normal allele and one loss allele) and mutant homozygote (two loss alleles), respectively. Family based association test (FBAT) showed that the distribution of different genotypes of nsv823469 was in accordance with that of Mendelian inheritance in families of COPD and pulmonary function18,19,20. The transmission disequilibrium test and sibship disequilibrium test (TDT & SDT) conducted on the 157 COPD families showed that nsv823469 had a preferential transmission of the loss allele from parents to healthy offspring or siblings under the additive genetic model (P = 0.010). Moreover, the loss of nsv823469 genotypes was a significant protective factor on COPD in additive genetic model (OR = 0.50, 95% CI = 0.34–0.73) as is shown in Table 2. Consistently, The quantitative transmission disequilibrium test (qTDT) conducted on the 391pulmonary function families also showed that the loss allele of nsv823469 has a tendency to transmit to offspring or siblings with relatively high forced expiratory volume in 1 second (FEV1) (P = 0.030; Table 3). However, no such genetic predisposition was observed for forced vital capacity (FVC) (P = 0.254), FEV1/FVC (P = 0.362) and FEV1/FEV1-predicted (P = 0.110).
Effect of nsv823469 on FEV1
Basing on the transmission mode of nsv823469 introduced above, we also tested the correlation between genotypes of nsv823469 and FEV1 in all subjects of 391pulmonary function families as well as in sub-groups stratified according to categories of sex, age, smoking status, drinking status and using biomass as fuels. As shown in Table 4, the values of FEV1significantly increased along with the number of loss allele (mean ± standard deviation: 2-copy, 2.34 ± 0.81 vs. 1-copy, 2.35 ± 0.81 vs. 0-copy, 2.69 ± 0.85; K-W test: P = 0.001). Moreover, this trend was observed in almost all sub-groups with statistical significance except for pack-years smoked ≥ 20 packs and ever drunk due to the limited sample size.
Effect of nsv823469 on expression of HCG4B, HLA-H, and HLA-A
Because nsv823469 covers the sequence of major histocompatibility complex, class I, H (HLA-H), major histocompatibility complex, class I, A (HLA-A), and HLA complex group 4B (HCG4B) (Supplemental Figure S1)16, we further tested the effect of nsv823469 on the three genes. As is shown in Fig. 1a,b, significant deviations in mRNA levels of HCG4B and HLA-A were observed in the samples of normal pulmonary tissue with different genotypes of nsv823469 (P = 0.002 for HCG4B and P = 0.043 for HLA-A). After controlled factors of sex, age and smoking by partial correlation analysis, the expression of HCG4B (r = 0.315, P = 0.031) and HLA-A (r = 0.296, P = 0.044) were still significantly positively correlated with the copy number of nsv823469. However, no significant association was observed in HLA-H with nsv823469 (P = 0.950, Fig. 1c). It means that copy number loss of nsv823469 significantly decreased the expression of HLA-A and HCG4B. Furthermore, we found that the expression of HCG4B was significantly correlated with that of HLA-A (r = 0.448, P = 0.001; Fig. 1d), while HCG4B was not significantly correlated with major histocompatibility complex, class I, F (HLA-F), major histocompatibility complex, class I, G (HLA-G), and major histocompatibility complex, class I, J (HLA-J) (P > 0.05 for all; Supplementary Figure S2a–c). In addition, the CNV has no effect on HLA-F, HLA-G and HLA-J expressions, as is expected (P > 0.05 for all; Supplementary Figure S2d–f).
(a) Effect of the CNV copy number on HCG4B; (b) Effect of the CNV copy number on HLA-H; (c) Effect of the CNV copy number on HLA-A; Bars = SD. P value was inferred with the Kruskal-Wallis test. As shown, significant deviation was observed for HCG4B and HLA-A but not HLA-H between different copy number tissues; (d) Correlation between the expression of HCG4B and HLA-A. A significant correlation between HCG4B and HLA-A was observed. The spearman rank correlation test was used.
Prediction of mechanism on HCG4B mediating HLA-A
Because some similarities were found between the mRNA sequence of HCG4B and that of HLA-A, bioinformatics analysis was performed to deduce a possible molecular mechanism through the website miRcode (http://mircode.org/index.php), which identifies putative target sites base on seed complementarity and evolutionary conservation21. As is shown in Supplementary Table S2, HCG4B might act as a competing endogenous RNAs (ceRNA) spongingmiR-122 and miR-1352 to increase the HLA-A expression.
Discussion
Based on a two-stage case-control study and two family based analyses, nsv823469 was identified to be associated with decreased risk of COPD in Chinese, and the loss allele has a tendency to transmit to health offspring/sibling and those with relatively high FEV1. Functional analysis further showed the CNV has effect on HLA-A and lncRNA HCG4B expression, meanwhile HCG4B could regulate the expression of HLA-A.
Now the function of HCG4B remains unknown. In the current study, we found that the expression of HCG4B was positively correlated with that of HLA-A, which suggested that HCG4B may regulate the expression of HLA-A. The molecular mechanism may be that HCG4B acts as a competing endogenous RNAs (ceRNA) sponging miR-122 and miR-1352. HLA-A plays an important role in COPD development with respect to immune function, and has been identified to have high expression in alveolar epithelial type II cells (ATII cells) and higher frequency in peripheral blood lymphocytesin COPD patients to mediate the development of COPD22,23,24. Moreover, evidences have supported that HLA-A participates in CD8 T cell involving in apoptosis of lung cells in the pathological process of COPD24. Based on the foregoing evidence, it is functionally possible that the loss copies of nsv823469 conferred a decreased risk of COPD.
Consistently, both the case-control study and COPD family-based analysis demonstrated significantly decreased risk in subjects with loss copies in comparison to those with 2-copy. Furthermore, nsv823469 had a preferential transmission of loss allele towards health children or siblings from parents. Moreover, the pulmonary function-based family analysis showed that the loss allele has a tendency to transmit to offspring/siblings with relatively high FEV1. Altogether, nsv823469 contributes to COPD predisposition and phenotypic pleiotropy. In addition, the family-based designs gave high credibility to our findings, because these designs could effectively control the confounding effects caused by various confounders in the case-control studies widely seen in association studies25.
As of now, only three studies have examined the associations between genomic CNVs and COPD risk4,26,27, and all of them focused on coding genes. In our study, we paid close attention to lncRNA CNVs that are located in the susceptible regions of COPD. We further found this CNV’s association with COPD risk in Chinese through the mechanism of regulating the expression of the lncRNA HCG4B and followed HLA-A. To our best knowledge, this is the first study investigating CNVs crossing with lncRNA on COPD risk.
There are also some limitations to the current study. Firstly, based on case-control study and family-based analyses, biases, such as selection bias and information bias, cannot be completely ruled out. Secondly, limited by the capabilities of the various technologies, we did not reveal the molecular function of the lncRNA on COPD development. We also did not substantiate the exact mechanism of how HCG4B influences the HLA-A expression. Nevertheless, all studies here exerted consistent results that the CNV has functional association with COPD risk and lung function, and it strongly suggests such association is not achieved by chance.
In summary, our study identified a putatively functional CNVnsv823469 that conferred declined risk of COPD and was beneficial to pulmonary function. The CNV underlies a biological mechanism that it could induce a low expression of HCG4B and followed HLA-A. Taken together, the CNV nsv823469 might be a genetic biomarker to predict risk of COPD in Chinese.
Methods
Case-control study
As described in previously published studies5,28,29, a two-stage case-control study was conducted in the southern Chinese and eastern Chinese population. In brief, 1025 COPD patients and 1061 controls were enrolled from Guangzhou city; 486 COPD patients and 616 controls were recruited from Suzhou city. COPD was diagnosed according to the Global initiative for chronic Obstructive Lung Disease (GOLD) criterion of FEV1/FVC < 70% after inhalation of 400 μg salbutamol30. The controls with FEV1/FVC > 70% were age (±5 years) and sex frequency-matched with the cases. The subjects donated 5-mL peripheral blood after giving their informed consents, and were interviewed using a structured questionnaire to provide data on demographic variables and risk factors. Their frequency distributions in case and control groups have been described in our previous publication28 (Supplementary Table S3). This study was approved by the institutional review boards of Guangzhou Medical University and Soochow University.
COPD family based analysis
A COPD family based analysis was conducted in the southern Han Chinese population between September 2010 and March 2015. 157 COPD probands were firstly enrolled and their immediate family members, including parents, siblings, and offspring, were asked to take a COPD diagnostic test. Excluding those who did not finish the lung function test, 293 immediate family members were ultimately enrolled, among which 44 were diagnosed with COPD while 249 were healthy. All subjects were interviewed using the above questionnaire, and donated 5-ml peripheral blood after signing the informed consent. This study was approved by the institutional review boards of Guangzhou Medical University.
Family based pulmonary function analysis
A family based pulmonary function analysis on community individuals was conducted between May 2014 and May 2015 in the southern Han Chinese. By excluding those who had no immediate family members or whose immediate family members did not complete lung function test, 391 pulmonary function relative families (n = 987) were finally recruited from annual cross-sectional surveys of COPD. Each subject donated 5 ml peripheral blood and was interviewed to provide data on the above variables after writing an informed consent. The study was approved by the institutional review boards of Guangzhou Medical University.
CNV selection and genotyping
By referring to East Asian CNVs data16, 15 CNVs were found in the region 6p21.32–22.1. Among them, only one CNV named CNVR2829.8 was prevalent in East Asian with the ACNF > 5%. The CNV was also recorded as nsv823469 in the database of genomic variants (DGV: http://dgv.tcag.ca/dgv/app/home). For the purpose of unequivocal reference, we used the label nsv823469 throughout the current study. The genotype of nsv823469 was detected by the TaqMan assay with special probes and primers (FAM labeled, cat no. Hs03587795; proprietary technology of Applied Biosystems) from the ABI by life Technology Company31 according to the standard protocol. The genotype was automatically determined by software Copy Caller 2.1 (Applied Biosystems; Supplementary Figure S3).
Detection of nsv823469 covering genes’ mRNA levels
According to the DGV database and East Asian CNVs data16, the loss of nsv823469 causes lower copies or deletion of DNA sequence that covers two lncRNAs with names of HLA-H and HCG4B, and some sequence of a coding-gene HLA-A in East Asian. The loss of nsv823469 may decrease the expressions of HCG4B, HLA-H and HLA-A with the dosage effect. Thus, we tested the mRNA levels of the above genes and a reference gene β-actin in 50 samples of normal pulmonary tissue using SYBR-Green real-time PCR, as the samples were obtained from the tumor hospital affiliated to Guangzhou Medical University with the characteristics shown in Supplementary Table S4. The expressions of HLA-F, HLA-G and HLA-J were also detected because of their genomic locations approaching to the HCG4B, while these genes might be the target genes of HCG4B. The primers for each gene presented in the Supplementary Table S5. Each sample was run in triplicate and the mean level of mRNA was calculated. Moreover, the genotype of nsv823469 for each sample was detected by the TaqMan assay.
Informed consent for Using experimental animals and human subjects
This study obtained the consents of all participants and fit in the standard moral principles of human beings and all experiments were performed in accordance with relevant guidelines and regulations. Furthermore, this study was approved by the institutional review boards of Guangzhou Medical University (Ethics Committee of Guangzhou Medical University: GZMC2007–07–0676) and Soochow University (Ethics Committee of Soochow University: SZUM2008031233).
Statistical analysis
The χ2 test was used to evaluate the consistency of loss frequency of nsv823469 in East Asian samples (11/30)16 and controls of the case-control study. The unconditional logistic regression model was applied in order to estimate the association strength between the CNV and COPD risk. The homogeneity of genetic effects among each stratum was analyzed with Breslow-Day test. Interactions between the nsv823469 and surrounding factors were assessed using the multiplicative interaction analysis. TDT & SDT of the nsv823469 among COPD families were analyzed using the FBAT software. qTDT of nsv823469 among pulmonary function families was performed on FEV1, FVC, FEV1/FVC and FEV1/ FEV1-predicted32 by the FAMILY PROCEDURE in SAS(http://support.sas.com/documentation/cdl/en/geneug/64249/HTML/default/viewer.htm#geneug_family_sect023.htm). Furthermore, the Kruskal-Wallis test and Spearman rank correlation test were performed to assess the effect nsv823469 has on pre-bronchodilator lung function traits and mRNA expressions of HCG4B, HLA-H and HLA-A. All tests were two-sided ones, and P < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Chen, X. et al. Association of nsv823469 copy number loss with decreased risk of chronic obstructive pulmonary disease and pulmonary function in Chinese. Sci. Rep. 7, 40060; doi: 10.1038/srep40060 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Cho, M. H. et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 42, 200–202, doi: 10.1038/ng.535 (2010).
Cho, M. H. et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 21, 947–957, doi: 10.1093/hmg/ddr524 (2012).
Lee, J. H. et al. DNAH5 is associated with total lung capacity in chronic obstructive pulmonary disease. Respir Res 15, 97, doi: 10.1186/s12931-014-0097-y (2014).
Janssens, W. et al. Genomic copy number determines functional expression of {beta}-defensin 2 in airway epithelial cells and associates with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 182, 163–169, doi: 10.1164/rccm.200905-0767OC (2010).
Yang, L. et al. Duplicated copy of CHRNA7 increases risk and worsens prognosis of COPD and lung cancer. Eur J Hum Genet 23, 1019–1024, doi: 10.1038/ejhg.2014.229 (2015).
Mikhail, F. M. Copy number variations and human genetic disease. Curr Opin Pediatr 26, 646–652, doi: 10.1097/MOP.0000000000000142 (2014).
Hollox, E. J. & Hoh, B. P. Human gene copy number variation and infectious disease. Hum Genet 133, 1217–1233, doi: 10.1007/s00439-014-1457-x (2014).
Moreno-De-Luca, D. & Cubells, J. F. Copy number variants: a new molecular frontier in clinical psychiatry. Current psychiatry reports 13, 129–137, doi: 10.1007/s11920-011-0183-5 (2011).
De Smet, E. G., Mestdagh, P., Vandesompele, J., Brusselle, G. G. & Bracke, K. R. Non-coding RNAs in the pathogenesis of COPD. Thorax 70, 782–791, doi: 10.1136/thoraxjnl-2014-206560 (2015).
Dey, B. K., Mueller, A. C. & Dutta, A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 5, e944014, doi: 10.4161/21541272.2014.944014 (2014).
Perry, M. M. et al. Role of non-coding RNAs in maintaining primary airway smooth muscle cells. Respir Res 15, 58, doi: 10.1186/1465-9921-15-58 (2014).
Bi, H. et al. Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm Res 64, 119–126, doi: 10.1007/s00011-014-0790-9 (2015).
Talkowski, M. E. et al. Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. Am J Hum Genet 91, 1128–1134, doi: 10.1016/j.ajhg.2012.10.016 (2012).
Kim, W. J. et al. Genome-wide association studies identify locus on 6p21 influencing lung function in the Korean population. Respirology 19, 360–368, doi: 10.1111/resp.12230 (2014).
Soler Artigas, M. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nature genetics 43, 1082–1090, doi: 10.1038/ng.941 (2011).
Park, H. et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 42, 400–405, doi: 10.1038/ng.555 (2010).
Horita, N. & Kaneko, T. Genetic model selection for a case-control study and a meta-analysis. Meta Gene 5, 1–8, doi: 10.1016/j.mgene.2015.04.003 (2015).
Horvath, S., Xu, X. & Laird, N. M. The family based association test method: strategies for studying general genotype–phenotype associations. European journal of human genetics: EJHG 9, 301–306, doi: 10.1038/sj.ejhg.5200625 (2001).
Dudbridge, F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Human heredity 66, 87–98, doi: 10.1159/000119108 (2008).
Ionita-Laza, I. et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genetic epidemiology 32, 273–284, doi: 10.1002/gepi.20302 (2008).
Jeggari, A., Marks, D. S. & Larsson, E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063, doi: 10.1093/bioinformatics/bts344 (2012).
Park, M. H. et al. Association of HLA class I antigens with diffuse panbronchiolitis in Korean patients. Am J Respir Crit Care Med 159, 526–529, doi: 10.1164/ajrccm.159.2.9805047 (1999).
Matsuzaka, Y. et al. Identification of novel candidate genes in the diffuse panbronchiolitis critical region of the class I human MHC. Immunogenetics 54, 301–309, doi: 10.1007/s00251-002-0470-8 (2002).
Wei, L. et al. Comprehensive analysis of gene-expression profile in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 10, 1103–1109, doi: 10.2147/COPD.S68570 (2015).
Brennan, P. Design and analysis issues in case-control studies addressing genetic susceptibility. IARC Sci Publ, 123–132 (1999).
Yang, L. et al. The Functional Copy Number Variation-67048 in WWOX Contributes to Increased Risk of COPD in Southern and Eastern Chinese. COPD 12, 494–501, doi: 10.3109/15412555.2014.948993 (2015).
Wain, L. V. et al. Copy number variation of the beta-defensin genes in europeans: no supporting evidence for association with lung function, chronic obstructive pulmonary disease or asthma. PLoS One 9, e84192, doi: 10.1371/journal.pone.0084192 (2014).
Yang, L. et al. Effects of a functional variant c.353T>C in snai1 on risk of two contextual diseases. Chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med 189, 139–148, doi: 10.1164/rccm.201307-1355OC (2014).
Huang, D. et al. Functional polymorphisms in NFkappaB1/IkappaBalpha predict risks of chronic obstructive pulmonary disease and lung cancer in Chinese. Hum Genet 132, 451–460, doi: 10.1007/s00439-013-1264-9 (2013).
Pauwels, R. A. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 163, 1256–1276, doi: 10.1164/ajrccm.163.5.2101039 (2001).
Mayo, P. et al. CNV analysis using TaqMan copy number assays. Curr Protoc Hum Genet Chapter 2, Unit2 13, doi: 10.1002/0471142905.hg0213s67 (2010).
Lake, S. L., Blacker, D. & Laird, N. M. Family-based tests of association in the presence of linkage. Am J Hum Genet 67, 1515–1525, doi: 10.1086/316895 (2000).
Acknowledgements
We thanked the approval of funds such as the National Natural Scientific Foundation of China grants (81273149, 81473040, 81402753, and 81472630), Yangcheng Scholar Grants (1201541589), Natural Science Grants of Guangdong Province (2014KZDXM046, 2014A030310091) and Shenzhen Technology Research and Development Funds (JCYJ20150403095530583). We also thanked Dr. Yuan Guo for the assistance on subject recruitment.
Author information
Authors and Affiliations
Contributions
Xiaoliang Chen performed the experiments, collected the data, and wrote the rough draft; Jiansong Chen, Di Wu, FumanQiu, HualiXiong andZihua Pan helped withthe experiments for extracting DNA and RNA, analyzing the dataand drafting the manuscript; Binyao Yang, ChenliXie,Yifeng Zhou, Dongsheng Huang, and Yumin Zhou participated in the community survey and collected data, and drafting the manuscript; Lei Yang designed the experiments, collected the data, and revised the manuscript; Xiaoxiao Lu helped with designing the study and revised the manuscript; Jiachun Lu conceived of the study and supervised all aspects of the study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
Dr. Xiaoliang Chen’s work has been funded by the National Natural Scientific Foundation of China grants (81273149, 81473040), Natural Science Grants of Guangdong Province (2014A030310091) and Shenzhen Technology Research and Development Funds (JCYJ20150403095530583). Prof. Lei Yang’s work has been funded by National Natural Scientific Foundation of China grants (81402753). Prof. Yifeng, Zhou’s work has been funded by National Natural Scientific Foundation of China grants (81472630). Prof. Jiachun Lu’s work has been funded by the National Natural Scientific Foundation of China grants (81273149, 81473040), Yangcheng Scholar Grants (1201541589), Natural Science Grants of Guangdong Province (2014KZDXM046). The other authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, X., Lu, X., Chen, J. et al. Association of nsv823469 copy number loss with decreased risk of chronic obstructive pulmonary disease and pulmonary function in Chinese. Sci Rep 7, 40060 (2017). https://doi.org/10.1038/srep40060
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40060
This article is cited by
-
Long non-coding RNA SNHG4 aggravates cigarette smoke-induced COPD by regulating miR-144-3p/EZH2 axis
BMC Pulmonary Medicine (2023)
-
LINC01414/LINC00824 genetic polymorphisms in association with the susceptibility of chronic obstructive pulmonary disease
BMC Pulmonary Medicine (2021)
-
Copy number gain of pro-inflammatory genes in patients with HBV-related acute-on-chronic liver failure
BMC Medical Genomics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.