Abstract
Although the SLE risk gene loci of HLA-DR and HLA-DQ within the major histocompatibility complex (MHC) region has been gradually revealed by recent Genome-Wide Association studies (GWAS), the association of HLA-DP polymorphisms with SLE was minimally reported. Considering that the variants in rs3077 and rs9277535 in the HLA-DP region could influence the immune response by affecting antigen presentation of HLA class II molecules to CD4+ T cells, the present study aimed to explore the role of HLA-DP polymorphisms in SLE. In total, samples from 335 SLE patients and 635 healthy controls were collected and genotyped by a polymerase chain reaction-high resolution melting (PCR-HRM) assay. A significant positive correlation was observed between the SNP rs3077, rs9277535 of HLA-DP and SLE susceptibility (rs3077, OR = 0.74, 95%CI = 0.60–0.91, P = 0.004; rs9277535, OR = 0.72, 95%CI = 0.59–0.88, P = 0.001). Rs3077 polymorphism was corelated to IL-17, INF-γ and cutaneous vasculitis (P = 0.037, P = 0.020 and P = 0.006, respectively). Additionally, rs3077 AA genotype carriers showed lower concentration of inflammatory cytokines and lower cutaneous vasculitis incidence than did the other two genotype. No significant association was observed between rs9277535 and cytokines or any clinical features. In conclusion, HLA-DP polymorphisms (rs3077 and rs9277535) were associated with SLE susceptibility and the levels of some inflammatory cytokines in SLE patients.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disorder characterized by a lack of tolerance to self-antigens and the hyper production of multiple pathogenic autoantibodies and immune complexes, which result in systemic inflammation and damage to multiple organ systems1. With significant morbidity, disability, and mortality rates, the quality of life and life expectancy in SLE patients are notably influenced1,2,3. The estimated incidence rates of SLE distribute differently in Asia, America and Europe, varying from 0.001% to 0.097%4,5. In China, the prevalence of SLE is 0.03% and it’s one of the most common chronic diseases6. However, the etiology of SLE has not yet been clarified. Among the well-known predisposing variables such as environmental, infection and hormonal factors, it has been established that genetic factors have pivotal effects on susceptibility to SLE7,8,9.
With the application of genome-wide association and independent replication studies, more than 40 robust genetic associations with SLE have been identified, and the gene variants in the major histocompatibility complex (MHC) region are believed to contribute the greatest genetic risk for SLE10,11,12,13. MHC region is about 4 Mb, divided into three subregions, namely class I (HLA-A, B, C), class II (HLA-DR, DP, DQ) and class III (C2 et al.) region. The genes which encode glycoproteins that process and present peptides for T cells recognition fall under classes I and II, while other immune genes like C4A and C4B located within the class III region. Although the long-range linkage disequilibrium (LD) within the MHC region makes the disease susceptibility contribution of each component gene difficult to assess, overwhelming evidence confirmed that the genetic variants in HLA-DR and HLA-DQ are predisposed to SLE14,15. Through transancestral mapping of the MHC region, allele variants in HLA-DPB1 were also shown associated with SLE16. Besides, HLA-DPB1*05:01 was reported relating to the presence of autoantibodies in Japanese SLE patients17. However, these studies neither have found the role of allele variants on HLA-DPA1 nor have explored the further association between the reported SNPs and the process of SLE.
Additionally, considering that a number of studies have revealed that HLA-DP genes were associated with susceptibility to many autoimmune diseases, including Wegener’s granulomatosis, systemic sclerosis, multiple sclerosis and others18,19,20, HLA-DP polymorphisms are likely to play an important role in SLE. In addition, recent Genome-Wide Association studies (GWAS) validated that rs3077 and rs9277535 genetic variants in the HLA-DP locus were significantly associated with HBV infection in Asian population21. Due to the fact that HLA-DP genes encode antigen-binding sites and exon 2 of HLA-DPs is highly polymorphic, the variants of rs3077 and rs9277535 may regulate the immune responses to infection by means of affecting the function of antigen presentation of HLA class II molecules in immune cells.
Meanwhile, antigen presentation is related to CD4+ T cells directly. And active CD4+ T cells, which could differentiate into disparate T cells subsets, play a critical role in development of SLE by secreting all kinds of cytokines, such as inflammatory cytokines (IL-1β, IL-6, INF-γ, TNF-α and IL-17), or the inhibitory cytokine IL-10. Serum INF-γ was primarily derived from Th1 cells, IL-6 and IL-10 were mainly from Th2 cells and IL-17 were from Th17 cells. Some studies have shown that these cytokines influenced SLE22.
Therefore, in the present study, we aim to explore the association of the HLA-DP polymorphisms rs3077 and rs9277535 with SLE susceptibility and further investigate the influence of HLA-DP polymorphisms rs3077 and rs9277535 on critical serum cytokine levels and clinical features in SLE in Chinese Han nationality.
Results
Characteristics of the included subjects
The primary demographic and clinical characteristics were illustrated in Table 1. A total of 335 SLE patients and 635 health people participated in the study. The age and sex of the participants from two groups were matched (P = 0.795 and 0.480, respectively). The average age of SLE patients and health controls were 35.77 and 36.28 years old, respectively. In SLE patients, the average disease duration was 36.00 (12.00–96.00) months and the average SLEDAI score was 11.76 ± 5.53. Among the SLE group, ANA, anti-dsDNA and anti-Sm antibody positive frequencies were 91.94%, 35.82% and 28.66%, respectively; while the average serum C3 concentration was 0.67 ± 0.30 g/L, and the average C4 was 0.15 ± 0.08 g/L. Additionally, the prevalences of cutaneous vasculitis, arthritis, albuminuria, rash, pleuritis, pericarditis and neurological disorder were 9.55%, 39.40%, 74.63%, 29.85%, 1.49%, 1.49% and 9.85%, respectively.
Genotyping and LD evaluation
All participants were genotyped via the PCR-HRM methods for SNPs rs3077 and rs9277535 in the HLA-DP region. The veracity of the results was confirmed by direct sequencing of PCR products from randomly selected samples. The direct sequencing results were consistent with all of the corresponding genotyping results. All genotypes were distributed in concordance with the Hardy-Weinberg equilibrium (HWE), as determined at the 0.05 significance level.
Haploview was applied to perform linkage disequilibrium evaluation. As shown in Fig. 1, rs3077 and rs9277535 in HLA-DP were in slight linkage disequilibrium.
Association analysis of HLA-DP polymorphisms with SLE susceptibility
There was a significant difference between the SLE group and the healthy controls in the genotypic distributions of rs3077 and rs9277535. Genotyping and allele modeling showed considerable difference in rs3077. We observed that the rs3077 GA-genotype and the rs9277535 non-GG genotype associated with lower SLE susceptibility (dominant model, rs3077, OR = 0.65, 95%CI = 0.50–0.84, P = 0.001; rs9277535, OR = 0.58, 95%CI = 0.44–0.76, P < 0.001, respectively). This was supported by the further allele model in which rs3077 allele A and rs9277535 allele A was also showed associated with lower SLE susceptibility (rs3077, OR = 0.74, 95%CI = 0.60–0.91, P = 0.004, statistical power = 0.8029; rs9277535, OR = 0.72, 95%CI = 0.59–0.88, P = 0.001, statistical power = 0.8970, respectively) (Table 2).
Association analysis of HLA-DP polymorphisms with serum cytokine levels
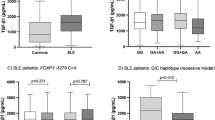
Analysis of the influence of the two HLA-DP polymorphisms on serum cytokine characteristics in SLE patients is illustrated in Table 3. Rs3077 was associated with IL-17, IL-1β, and INF-γ (IL-17: P = 0.037, recessive model, P = 0.011; IL-1β recessive model, P = 0.049; INF-γ: P = 0.020, recessive model, P = 0.008, respectively) and rs3077 AA genotype had lower IL-17, IL-1β and INF-γ concentrations than the other two genotypes. Meanwhile, serum C3 and C4 concentrations were also affected by rs3077 and rs9277535 polymorphisms (rs3077: C3, P = 0.003; rs9277535: C4, recessive model, P = 0.019, respectively). Nevertheless, there was no significant association between rs9277535 and IL-17, IL-1β, INF-γ and C3 (IL-17, P = 0.949; IL-1β, P = 0.569; INF-γ, P = 0.895; C3, P = 0.299, respectively). And neither rs3077 nor rs9277535 showed association with IL-4, IL-6, IL-10, or IL-23.
Association analysis of HLA-DP polymorphisms with clinical features
Comparing the distribution of genotypic frequencies of HLA-DP polymorphisms between positive and negative patients in seven specific clinical symptoms, a significant difference could be observed between rs3077 and cutaneous vasculitis (CV) (χ2= 10.132, P = 0.006) (Table 4). Moreover, as shown in Table 5, a decreased frequency of the minor allele A in CV patients (10.94%) compared with non-CV patients (28.22%) was also observed (χ2 = 8.860, P = 0.003), which suggests the relationship between the minor A allele of rs3077 and decreased susceptibility to CV in SLE patients (P = 0.003, OR = 0.312, 95%CI = 0.140–0.699). In addition, under the dominant model, the minor allele A carriers (GA + AA) exhibit lower risk for CV than those with the GG genotype (GA + AA versus GG: P = 0.001, OR = 0.245, 95%CI = 0.098–0.612).
Discussion
Systemic lupus erythematosus (SLE) is a chronic complex autoimmune disease characterized by autoantibody deposits and involvement of multiple systems23. The high heritability, high sibling relapse risk ratio and higher concordance rate in monozygotic twins confirm the genetic effect on the etiology of SLE24,25. Therefore, the exact genetic pathogenesis of SLE has long been one of the most difficult and intriguing issues.
In the present study, we found two SNPs, rs3077 and rs9277535 of HLA-DP, did associate with the SLE susceptibility and the A allele of the two SNPs are potential protective allele in SLE in the Chinese Han population. To our knowledge, HLA-DP rs3077 and rs9277535 were not reported to be related to autoimmune diseases (e.g., SLE, RA). It was our study that first investigated the association between the variants of HLA-DP and SLE susceptibility in Chinese Han population residing in Southwest China. As previous studies reported, there were some other SNPs of HLA-DPB1 significantly associated with autoimmune diseases such as rheumatoid arthritis, Wegener’s granulomatosis, systemic sclerosis and infectious diseases like chronic hepatitis B infection18,19,20,21,26,27. Our study suggested that HLA-DP region may contain some genes that have potential protective effects on SLE, which implied HLA-DP rs3077 and rs9277535 were likely to represent a shared autoimmune or infectious immune locus.
In addition, a significant positive association was observed between rs3077 with INF-γ, IL-1β and IL-17, and the rs3077 AA genotype had lower INF-γ, IL-1β and IL-17 concentration compared with other two genotypes. Moreover, rs3077 had an association with cutaneous vasculitis (χ2= 10.132, P = 0.006), with the minor allele A carriers showing lower risk of cutaneous vasculitis (P = 0.003, OR = 0.312, 95%CI = 0.140–0.699; dominant model: P = 0.001, OR = 0.245, 95%CI = 0.098–0.612). Considering that the abnormal expression of Th1, Th2 and Th17 cytokines are important in the pathogenesis of SLE28,29, it is most likely that the SNP rs3077 was not only associated with SLE susceptibility but also correlated with the levels of some inflammatory cytokines.
HLA-DP is mainly expressed on the surface of antigen-presenting cells like macrophages, dendritic cells and B cells, and belongs to HLA α molecules, which can bind and present antigen epitopes to CD4+ T helper cells30,31,32. Rs3077 and rs9277535 are located in the 3′ untranslated regions of HLA class II genes HLA-DPA1 and HLA-DPB1, correspondingly33,34. Therefore, rs3077 and rs9277535 are most likely to affect the disease through influencing the binging and presenting antigen epitopes to CD4+ T helper cell. Though without exact evidence, this can be partly supported by the research of O’Brien and his colleagues. In O’Brien’s study, genotype and gene expression data collected and integrated from the normal liver samples obtained from 651 Europeans. They found that rs3077 allele G and rs9277535 allele G were correlated with the down-regulated level of HLA-DP mRNA, and allele A was associated with the increased level of HLA-DP mRNA in normal human liver tissue35. Therefore, considering that allele G of HLA-DP rs3077 and rs9277535 have been confirmed predisposed to chronic hepatitis B, carrying allele G of these two SNPs could down-regulate the expression of HLA-DP, which turns out to influence the antigen presentation. Compared to HBV infection, SLE is another kind of disease. However, in our present study, allele G was also found predisposed to SLE. These two consistent results suggest that though in different diseases, the role of variants of HLA-DP might be the same. Apart from that we have revealed the significant association between the two SNPs of HLA-DP and SLE, in the present study, we also found the association between rs3077 and the lower concentration of inflammatory cytokines, as well as the lower risk of cutaneous vasculitis, which might explain partly the function of rs3077 in SLE. Obviously, studies on the structure-function of the two SNPs are still needed. Therefore, the details of the mechanism, including how exactly the gene variants influence the gene expression like HLA-DP mRNA regulation, and the gene function like antigen presentation to CD4+ T cells, and so on, would be explored in our future studies. To our knowledge, because these two variants were first reported in SLE, the studies on linkage disequilibrium of HLA-DP in SLE or in autoimmune diseases are inadequate. However, we could take a sight into this issue from the reported linkage disequilibrium of the two variants in other diseases. The allele variant rs3077 in HLA-DPA1 and rs9277535 in HLA-DPB1 were thought that could be independently considered to be possible causative SNPs for HBV in a study which the linkage disequilibrium of multiple-single nucleotide polymorphism genotype data was researched by the textile plot36. But in order to clarify the role of these two variants in the numerous alleles, complex structure and the tight linkage disequilibrium of the HLA region, the relationship between these two SNPs and other reported SNPs in HLA would also be discussed in further studies.
In summary, our study firstly found that the two variants (rs3077 and rs9277535) of HLA-DP were associated with decreased SLE susceptibility, and the SNP rs3077 was correlated with the lower IL17, IL-1β and INF-γ concentrations in peripheral blood and reduced susceptibility to CV in SLE patients. This study could provide further evidence to improve our understanding of the exact function of HLA-DP in the pathogenesis of autoimmune diseases. It is worthwhile to mention that there are several limitations in our present study. The sample of patients in our study is relatively small and only Han Chinese individuals in western China were included. Therefore, further studies using large sample sizes and other ethnic populations are needed to confirm the results observed in this study.
Materials and Methods
Patients and protocol
There were 970 Chinese Han subjects recruited in the present study, including 335 SLE patients and 635 healthy controls from September 2013 to September 2014 in West China Hospital. The SLE patients enrolled met the inclusion criteria as follow: 1) diagnosed as SLE in compliance with the American College of Rheumatology classification criteria for SLE revised in 1997. The patients were all in the active stage of SLE with SLE disease activity index (SLEDAI) >4. 2) hospitalized patients without drug-induced SLE. The main clinical manifestations such as cutaneous vasculitis, arthritis, albuminuria, rash, pleuritis, pericarditis and neurological disorder were obtained retrospectively by reviewing hospital records. The healthy control should meet these inclusion criteria: without any chronic, endemic infectious or autoimmune diseases and with normal physical examination and blood tests.
All the patients have signed the informed consent to participate in this study and consented to sample collection. This study conducted in accordance with the 1975 Declaration of Helsinki and was approved by the Ethics Committee of West China Hospital.
Serological testing
Laboratory assays of SLE were analyzed by the following methods: ANA, anti-dsDNA and anti-Sm were detected with ANA, anti-dsDNA and ENA reagent kits (Euroimmun company, Germany). Complement 3(C3) and Complement 4(C4) were detected by rate nephelometry on Beckman Coulter IMMAGE 800 immunoassay (Beckman Coulter, Inc, CA, USA). All the tests were conducted according to manufacturers’ instruction.
HLA-DP polymorphism genotyping
Genomic DNA was isolated from the peripheral blood by using Genomic DNA kit (Biotake Corporation, Beijing, China), with the concentration of DNA tested on Nanodrop 2000c spectrophotometer (Thermo Scientific, DE). The polymerase chain reaction-high resolution melting (PCR-HRM) was applied to genotype rs3077 and rs9277535 and the analysis was performed on the Light Cycler 480 (Roche Diagnostics, Penzberg, Bavaria, Germany). SNP genotyping was performed in a 20 μL reaction system which contains 10 μL Roche Master Mix (Roche Applied Science) (comprising FastStart Taq DNA Polymerase and the High Resolution Melting Dye in a reaction buffer), 2.4 μL 25 mM MgCl2, 0.2 μL 10 mmol/L Forward Primer, 0.2 μL 10 mmol/L Reverse Primer, 6.2 μL deionized water and 1 μL DNA sample as recommended by the manufacturer. The whole genotyping process was performed under the following conditions: 95 °C for 10 min in the initial denaturation step followed by 50 cycles of 95 °C for 15 s, touchdown cycling (decreasing 1 °C/cycle), 65–55 °C for 10 s, and 72 °C for 10 s in the annealing step and then denatured at 95 °C for 1 min and cooled to 40 °C for 1 min after the amplification phase. After HRM analyses, melting process and cooling, the results were analyzed by the corresponding Gene Scanning Software v1.2 (Roche Diagnostic).
Cytokine measurement
Randomly 93 samples of SLE patients were chosen to test the serum cytokine. The serum concentrations of cytokines including IL-1β, IL-4, IL-6, IL-10, IL-17, IL-23, TNF-α and INF-γ were estimated. They were quantitatively determined by Bio-Plex Pro™ Human Inflammation Assays. Procedure was performed according to the manufacturer’s instructions. Blood sampling for assessing serum levels of cytokines and other laboratory investigations were performed at the same time of clinical examination and assessment of SLE disease activity.
Statistical analysis
Statistical power was calculated by a software “PS: Power and Sample size Calculation” (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize). The Hardy-Weinberg equilibrium (HWE) was evaluated for each polymorphism independently. Mean ± SD, median and interquartile was used to describe the continuous variables with normal and skewed distribution, respectively. Student’s t test or Mann-Whitney U test were applied to compare demographic and clinical data between groups as appropriate. Allele case-control comparisons were analyzed by pearson’s chi-square test or Fisher’s exact test. Association of SNPs with susceptibility of SLE was assessed by figuring out the odds ratio (OR) and 95% Confidence Interval (CI). The following analytic methods were used when it came to compare the subjects from two groups: allelic frequency distribution of the two groups (allele A versus allele B, A as the major allele, B as the minor allele, this applied to the following methods); dominant model (AB + BB versus AA); recessive model (AA + AB versus BB). All statistical analyses were executed applying the Statistical Package for the Social Sciences (SPSS, SPSS Inc., Chicago, IL, USA), version 19.0. A two-sided P value < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Zhang, J. et al. Genetic Polymorphisms of rs3077 and rs9277535 in HLA-DP associated with Systemic lupus erythematosus in a Chinese population. Sci. Rep. 7, 39757; doi: 10.1038/srep39757 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zhu, T. Y., Tam, L. S. & Li, E. K. Labour and non-labour market productivity in Chinese patients with systemic lupus erythematosus. Rheumatology 51, 284–292, doi: 10.1093/rheumatology/ker247 (2011).
Rees, F. et al. Mortality in systemic lupus erythematosus in the United Kingdom 1999–2012. Rheumatology, kev424, doi: 10.1093/rheumatology/kev424 (2016).
Holloway, L. et al. Patient-reported outcome measures for systemic lupus erythematosus clinical trials: a review of content validity, face validity and psychometric performance. Health Qual Life Outcomes 12, 116, doi: 10.1186/s12955-014-0116-1 (2014).
Lim, S. S. et al. The Incidence and Prevalence of Systemic Lupus Erythematosus, 2002–2004: The Georgia Lupus Registry. Arthritis & Rheumatology 66, 357–368, doi: 10.1002/art.38239 (2014).
Rees, F. et al. The incidence and prevalence of systemic lupus erythematosus in the UK, 1999–2012. Annals of the Rheumatic Diseases 75, 136–141, doi: 10.1136/annrheumdis-2014-206334 (2016).
Li, R. et al. Epidemiology of eight common rheumatic diseases in China: a large-scale cross-sectional survey in Beijing. Rheumatology 51, 721–729, doi: 10.1093/rheumatology/ker370 (2011).
Deapen, D. et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum 35, 311–318 (1992).
Alarcón-Segovia, D. et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis & Rheumatism 52, 1138–1147, doi: 10.1002/art.20999 (2005).
Tiffin, N., Adeyemo, A. & Okpechi, I. A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus. Orphanet J Rare Dis 8, 2, doi: 10.1186/1750-1172-8-2 (2013).
Deng, Y. & Tsao, B. P. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nature Reviews Rheumatology 6, 683–692, doi: 10.1038/nrrheum.2010.176 (2010).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature Genetics 41, 1228–1233, doi: 10.1038/ng.468 (2009).
Han, J.-W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature Genetics 41, 1234–1237, doi: 10.1038/ng.472 (2009).
Yang, W. et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 6, e1000841, doi: 10.1371/journal.pgen.1000841 (2010).
Jiang, C. et al. Differential Responses to Smith D Autoantigen by Mice with HLA-DR and HLA-DQ Transgenes: Dominant Responses by HLA-DR3 Transgenic Mice with Diversification of Autoantibodies to Small Nuclear Ribonucleoprotein, Double-Stranded DNA, and Nuclear Antigens. The Journal of Immunology 184, 1085–1091, doi: 10.4049/jimmunol.0902670 (2009).
Kim, K. et al. The HLA-DRbeta1 amino acid positions 11-13-26 explain the majority of SLE-MHC associations. Nat Commun 5, 5902, doi: 10.1038/ncomms6902 (2014).
Fernando, M. M. et al. Transancestral mapping of the MHC region in systemic lupus erythematosus identifies new independent and interacting loci at MSH5, HLA-DPB1 and HLA-G. Annals of the rheumatic diseases 71, 777–784, doi: 10.1136/annrheumdis-2011-200808 (2012).
Furukawa, H. et al. Association of increased frequencies of HLA-DPB1*05:01 with the presence of anti-Ro/SS-A and anti-La/SS-B antibodies in Japanese rheumatoid arthritis and systemic lupus erythematosus patients. PloS one 8, e53910, doi: 10.1371/journal.pone.0053910 (2013).
Wu, X. M. et al. Association of susceptibility to multiple sclerosis in Southern Han Chinese with HLA-DRB1, -DPB1 alleles and DRB1-DPB1 haplotypes: distinct from other populations. Mult Scler 15, 1422–1430, doi: 10.1177/1352458509345905 (2009).
Wieczorek, S., Holle, J. U. & Epplen, J. T. Recent progress in the genetics of Wegener’s granulomatosis and Churg–Strauss syndrome. Current Opinion in Rheumatology 22, 8–14, doi: 10.1097/BOR.0b013e3283331151 (2010).
Radstake, T. R. D. J. et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nature Genetics 42, 426–429, doi: 10.1038/ng.565 (2010).
Kamatani, Y. et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nature Genetics 41, 591–595, doi: 10.1038/ng.348 (2009).
Talaat, R. M., Mohamed, S. F., Bassyouni, I. H. & Raouf, A. A. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: Correlation with disease activity. Cytokine 72, 146–153, doi: 10.1016/j.cyto.2014.12.027 (2015).
He, S.-j. et al. Therapeutic effects of DZ2002, a reversible SAHH inhibitor, on lupus-prone NZB×NZW F1 mice via interference with TLR-mediated APC response. Acta Pharmacologica Sinica 35, 219–229, doi: 10.1038/aps.2013.167 (2013).
Ghodke-Puranik, Y. & Niewold, T. B. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J Autoimmun 64, 125–136, doi: 10.1016/j.jaut.2015.08.004 (2015).
Niewold, T. B. Advances in lupus genetics. Curr Opin Rheumatol 27, 440–447, doi: 10.1097/bor.0000000000000205 (2015).
Liao, Y. et al. Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One 9, e111677, doi: 10.1371/journal.pone.0111677 (2014).
Yu, L. et al. Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci Rep 5, 14933, doi: 10.1038/srep14933 (2015).
Lopez, P., Rodriguez-Carrio, J., Caminal-Montero, L., Mozo, L. & Suarez, A. A pathogenic IFNalpha, BLyS and IL-17 axis in Systemic Lupus Erythematosus patients. Sci Rep 6, 20651, doi: 10.1038/srep20651 (2016).
Zimmermann, M. et al. IFNalpha enhances the production of IL-6 by human neutrophils activated via TLR8. Sci Rep 6, 19674, doi: 10.1038/srep19674 (2016).
Diaz, G. et al. Functional analysis of HLA-DP polymorphism: a crucial role for DPbeta residues 9, 11, 35, 55, 56, 69 and 84–87 in T cell allorecognition and peptide binding. Int Immunol 15, 565–576 (2003).
Stevanovic, S. et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood 122, 1963–1973, doi: 10.1182/blood-2012-12-470872 (2013).
Lauterbach, N. et al. Allorecognition of HLA-DP by CD4+ T cells is affected by polymorphism in its alpha chain. Mol Immunol 59, 19–29, doi: 10.1016/j.molimm.2013.12.006 (2014).
Jiang, X., Ma, Y., Cui, W. & Li, M. D. Association of variants in HLA-DP on chromosome 6 with chronic hepatitis B virus infection and related phenotypes. Amino Acids 46, 1819–1826, doi: 10.1007/s00726-014-1767-2 (2014).
Hu, L. et al. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 55, 1426–1431, doi: 10.1002/hep.24799 (2012).
O’Brien, T. R. et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12, 428–433, doi: 10.1038/gene.2011.11 (2011).
Kumasaka, N., Nakamura, Y. & Kamatani, N. The textile plot: a new linkage disequilibrium display of multiple-single nucleotide polymorphism genotype data. PloS one 5, e10207, doi: 10.1371/journal.pone.0010207 (2010).
Acknowledgements
This work was supported by the Natural Science Foundation of China 81202354 to ZCH, the Natural Science Foundation of China 81301496 to BY, the Natural Science Foundation of China 81501816 to QN, the Natural Science Foundation of China 81601830 to JLZ and Foundation of Science and Technology bureau of Sichuan Province # 2011SZ0221 to BC. These funders supported our work including study design, decision to publish and preparation of the manuscript. We thank Evan Martin from Cleveland Clinic in U.S for the language modification which bettered our manuscript.
Author information
Authors and Affiliations
Contributions
L.L.W., J.L.Z. and W.L.Z. designed the study; Y.K.W., B.C. and L.L.W. were responsible for recruitment of subjects; J.L.Z., W.L.Z., A.N.T., L.C. and Y.L. performed experiments and conducted data management; J.L.Z., W.L.Z., Y.K.W., B.C. and L.L.W. performed statistical analyses and interpreted results; J.L.Z. and W.L.Z. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, J., Zhan, W., Yang, B. et al. Genetic Polymorphisms of rs3077 and rs9277535 in HLA-DP associated with Systemic lupus erythematosus in a Chinese population. Sci Rep 7, 39757 (2017). https://doi.org/10.1038/srep39757
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39757
This article is cited by
-
Exploration of the pathogenesis of Sjögren’s syndrome via DNA methylation and transcriptome analyses
Clinical Rheumatology (2022)
-
A shared motif of hla-dpb1 affecting the susceptibility to pr3-anca positive granulomatosis with polyangiitis: comparative analysis of a Turkish cohort with matched healthy controls
Rheumatology International (2021)
-
Exploring the etiopathogenesis of systemic lupus erythematosus: a genetic perspective
Immunogenetics (2019)
-
Genetic polymorphism of rs9277535 in HLA-DP associated with rheumatoid arthritis and anti-CCP production in a Chinese population
Clinical Rheumatology (2018)
-
Genetic polymorphisms of HLA-DP and isolated anti-HBc are important subsets of occult hepatitis B infection in Indonesian blood donors: a case-control study
Virology Journal (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.