Abstract
Insulin-like growth factor 1 (IGF1) and IGF binding protein 3 (IGFBP3) play an important role in the development and progression of renal cell carcinoma (RCC). We evaluated the association of functional polymorphisms in IGF1 and IGFBP3 with susceptibility and prognosis of RCC. We genotyped nine potentially functional polymorphisms in IGF1 and IGFBP3 and assessed their association with risk of RCC in a two-stage case-control study compromising 1027 cases and 1094 controls, and with prognosis in a cohort of 311 patients. We found rs5742714 in the 3′-UTR of IGF1 was significantly associated with risk and prognosis of RCC. In the combined set, the rs5742714 GC/CC genotypes were significantly associated with decreased risk of RCC compared with the GG genotype (OR = 0.82; 95% CI = 0.68–0.98, P = 0.002). Furthermore, patients with the rs5742714 GC/CC genotypes showed improved survival than those with the GG genotype (Log-rank P = 0.025, HR = 0.36, 95% CI = 0.14–0.93). Besides, the rs5742714 GC/CC genotypes were associated with significantly decreased expression of IGF1 mRNA and lower IGF1 serum levels. Moreover, the luciferase reporter assays revealed the potential effect of rs5742714 genotype on the binding of microRNAs to IGF1. Our findings suggest that the IGF1 polymorphism rs5742714 may be a genetic predictor of susceptibility and prognosis of RCC.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is the predominant form of kidney malignancy, accounting for more than 80% of all malignant kidney tumors1,2. Approximately 25% of patients show developed metastases at diagnosis and it is likely that 30% of the remaining patients will develop metastases even following surgical treatment3,4. Patient prognosis is generally poor following metastasis3. Currently, the prediction of RCC prognosis largely depends on conventional prognostic factors such as pathological tumor stage and grade.
In recent years, different genetic variations have been identified and associated with the development and prognosis of RCC in several candidate gene studies5,6,7,8 as well as a large genome-wide association study9. For instance, genetic polymorphisms in angiogenesis-related genes, which play a crucial role in the pathogenesis of RCC, were proved susceptibility loci or prognostic predictors of RCC5,7. Our previous studies have found that polymorphisms at the VHL and HIF1A genes could jointly influence RCC progression and survival6. However, these findings were not consistent across studies in different populations, which may indicate differences in the genetic architecture of ethnic groups, and investigating genetic variations in other candidate genes is still required.
The insulin-like growth factor (IGF) system plays a crucial role in regulating cell proliferation, differentiation, and apoptosis10,11. The system consists of IGF1 and IGF2 as well as their cell surface receptors (IGF1R and IGF2R), and six specific IGF-binding proteins (IGFBPs). IGFBP3, which is the predominant IGFBP, binds to the majority of circulating IGF1 to regulate its biological activity. Aberrant expression or regulation of IGF1 and IGFBP3 has been demonstrated to facilitate carcinogenesis and have an effect on cancer prognosis10. Chuang et al. found, using a combination of cDNA microarrays, western blot, and immunohistochemistry, that IGFBP3 was a marker for clear cell RCC and that increased IGFBP3 expression was associated with a higher Fuhrman grade12. In addition, it was reported that serum insulin-like growth factor-1 was an independent predictor of prognosis in patients with RCC13.
Findings from several studies have consistently found that genetic polymorphisms in IGF1 and/or IGFBP3 are associated with the risk or prognosis of various cancers, including colorectal cancer14, lung cancer15, prostate cancer16, and breast cancer17. However, the number of studies that have tested polymorphisms at the IGF1 or IGFBP3 locus for evidence of association with RCC has been sparse. A polymorphism in IGFBP3 (rs2854744) was found to be a risk factor for RCC; however, the study had a small sample size consisting of 158 patients and 316 controls18. In light of the critical role of IGF1 and IGFBP3 in RCC, it is possible that genetic variants from these two genes will have an effect on the risk and/or prognosis of RCC. Therefore, in our present study, we selected nine potentially functional polymorphisms in IGF1 (rs6214, rs6218, rs35767, rs5742612, and rs5742714) and IGFBP3 (rs2132572, rs2854744, rs2854746 and rs282734), and evaluated their association with risk and prognosis of RCC in a two-stage case-control study.
Materials and Methods
Study population
This ongoing study was started in May 2004 and was approved by the institutional review board of Nanjing Medical University. The details of the inclusion criteria were described previously6,19. In brief, all subjects were genetically unrelated ethnic Han Chinese individuals. Those individuals who received chemotherapy or radiotherapy before surgery, or had a different type of cancer, were excluded from the present study. The control subjects were genetically unrelated to the patients and they had no individual history of cancer. The first (original) set of patient and control cohorts tested in the study included 355 patients and 362 cancer-free controls that were recruited from May 2004 to October 2009. The patients in this set were followed up prospectively every 6 months from the time of enrollment until death or until last time of follow-up. Of this cohort, 41 patients (11.5%) were excluded because of a lack of adequate information for follow-up, and 3 additional cases (0.8%) were excluded due to low DNA quality. The second, validation set comprised of 672 RCC cases and 703 controls. In the both sets, controls were frequency-matched to cases by age (±5 years) and sex. Each participant donated 5 ml venous blood collected in an EDTA tube after providing written informed consent. This study was approved by the Local Ethics Committees of the First Affiliated Hospital with Nanjing Medical University and were carried out in accordance with the approved guidelines. At recruitment, the written informed consent was obtained from each subject.
SNP selection and genotyping
Polymorphisms in IGF1 and IGFBP3 were selected using genotype data from unrelated Han Chinese individuals in Beijing from the HapMap database (HapMap Data Rel 24/phaseII Nov08, on NCBI B36 assembly, dbSNP b126). Polymorphisms were considered from 2 kb upstream of the IGF1 and IGFBP3 locus to 2 kb downstream. We identified potentially functional polymorphisms according to the following criteria: (1) located in the 5′ flanking region, the 5′ untranslated region (UTR), the 3′-UTR, or the coding region causing an amino acid change and (2) a minor allele frequency (MAF) of greater than 5% in the Chinese population. Five polymorphisms in IGFI (rs6214, rs6218, rs35767, rs5742612, and rs5742714) and four in IGFBP3 (rs2132572, rs2854744, rs2854746, and rs282734) were selected. Genotyping was performed using the TaqMan SNP genotyping method, as we previously described20.
Quantitative measurement of IGF1 and IGFBP3 serum levels
Serum samples were available from 100 RCC cases and 100 controls. Collected serum was stored to clot for 30 minutes at 4 °C before centrifugation at 2000 rpm for 10 minutes at 4 °C. Serum was isolated and stored at −80 °C before use. IGF1 and IGFBP3 concentrations were measured using a sandwich enzyme immunoassay (Quantikine immunoassay kit, R&D Systems Inc, Minneapolis, MN, USA) and calculated using a standard curve according to the manufacturer’s instructions.
Analysis of IGF1 mRNA expression
Forty-six surgically removed paratumor renal samples were used to measure IGF1 mRNA levels in vivo. All tissue samples were stored in liquid nitrogen immediately following collection. RNA extraction and cDNA preparation was described previously21. IGF1 mRNA was measured using quantitative real-time reverse transcription (RT)-PCR on an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA, USA). The ACTB gene was used as an internal reference. The primers used for IGF1 were 5′-GCTCTTCAGTTCGTGTGTGGA-3′ (sense) and 5′-GCCTCCTTAGATCACAGCTCC-3′ (antisense). Amplification conditions were as described previously21.
Cell lines
HEK-293 and renal cell adenocarcinoma cell line 786-o were provided by Dr. Z. Zhang (Department of Molecular and Genetic Toxicology, School of Public Health, Nanjing Medical University, Nanjing, China) and were used as previously reported22.
Construction of IGF1 3′-UTR reporter plasmids
IGF1 3′-UTRs (bps) containing either the G allele or C allele of rs5742714 were inserted into the XbaI site of pGL3 promoter vectors (Genscript, Nanjing, China). The accuracy of the constructed plasmids was verified by DNA sequencing.
Dual-luciferase reporter assay
MiRNASNP v2.0 analysis was used to discover a seven-nucleotide complementary sequence between miR-580 and the IGF1 3′UTR23. A wild type (wt) IGF1 3′UTR sequence (5′-ACTTACAGACACTGAATTAATTTCCCCTGCTACTTTGAAACCAG AAAATATGACTGGCCATTCGTTACATCTGTCTTAGTTGAAAAGCATATTTTTTATTAAATTAATTCTGATTGTATTT GAAATTATTATTCAATTCACTTATGGCAGAGGAATATCAATCCTAATGACTTCTAAAA ATGTAACTAA TTGAATCATT ATCTTACATT-3′) in addition to a mutant type (mut) IGF1 3′UTR sequence with a rs5742714 different miR-580-binding sequence.
HEK-293 and 786-o cells were used for cell transfection and luciferase assays. Cells were seeded into culture medium-containing (100 μL/well) 96-well plates at a cell concentration of 1.5 × 104 cells/well, followed by 24-hour incubation (37 °C, 100% humidity, 5% CO2). The cells were allocated into four groups: Group A, which were to transfeced a non-targeting negative control RNA with pGL3 IGF-G allele; Group B, which were to transfeced a non-targeting negative control RNA with pGL3 IGF-C allele;Group C, which were transfected with miR-580 mimics and pGL3-IGF1-G; and Group D, which were transfected with miR-580 mimics and pGL3-IGF1-C, using Lipofectamine 2000 (Invitrogen Corp, CA, USA). As an internal standard, all plasmids were cotransfected with pRL-TK, which contained the Renilla luciferase gene. After transfection for 48 hours, luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega). Independent triplicate experiments were performed for each plasmid construct.
Statistical Analysis
Differences in the distribution of demographic characteristics, selected variables, and frequencies of genotypes between cases and controls were evaluated using the Student’s t-test (for continuous variables) or the chi-square test (for categorical variables). Differences in serum levels of IGF1 and IGFBP3 among different genetic groups were evaluated using the Student’s t-test or one-way analysis of variance (ANOVA). Allele frequencies of each polymorphism were tested for deviations from Hardy-Weinberg equilibrium using a chi-square goodness-of-fit test before analysis. Associations between polymorphisms and risk of RCC were estimated using computing odds ratios and 95% confidence intervals (CIs) from unconditional logistic regression analysis with adjustment for possible confounders. Survival time was calculated from the date of RCC diagnosis to the date of death or last follow-up. Different survival times according to demographic characteristics, clinical features, and IGF1 or IGFBP3 polymorphisms were estimated using the Kaplan–Meier method and compared using the Log-rank test; univariate or multivariate Cox regression analysis was performed to determine predictive factors of RCC survival by estimating the hazard ratios (HRs). Differences in luciferase reporter gene expression among different promoter constructs as well as mRNA levels from tissues with different genotypes were evaluated using Student’s t-test or ANOVA. All analyses were performed using SAS 9.1.3 (SAS Institute, Cary, NC) with two-sided P values. A P value of less than 0.05 was considered statistically significant.
Results
Characteristics of RCC patients and controls
Frequency distributions of selected characteristics of the cohorts from both the original and validation sets are presented in Table 1. There were no significant differences between cases and controls with regard to age, gender, and drinking status in both sets of cohorts (all P > 0.05). However, there were more smokers, patients with hypertension, and diabetics among the group of cases compared to that of controls. We also found that the percentage of patients from the original cohort with stage I, II, III, and IV disease was 65.3%, 19.5%, 7.1%, and 8.1%, respectively, while patients from the validation set showed percentages of 21.6%, 51.1%, 20.7%, and 6.5%, respectively.
Association of IGF1 and IGFBP3 polymorphisms with risk of RCC
The associations between these polymorphisms and risk of RCC in the best genetic model are presented in Table 2, and detailed genotype distributions of the polymorphisms in cases and controls are shown in Supplement Table 1. Genotype frequencies of all these nine polymorphisms in controls conformed to Hardy-Weinberg equilibrium (P > 0.05), with the exception of rs6218 in IGF1, which was removed from further analysis. In the test set, we found that the IGF1 rs5742714 and rs6214 polymorphisms were significantly associated with a decreased risk of RCC (OR = 0.66, 95% CI = 0.48–0.92 and OR = 0.65, 95% CI = 0.45–0.86 for rs5742714 and rs6214, respectively). However, in the validation set, only the association between the IGF1 rs5742714 polymorphism and decreased risk of RCC was verified (OR = 0.79, 95% CI = 0.63–0.98, GC/CC vs. GG). When patient cohorts were combined from both sets, as shown in Table 2, we found that affected individuals with either the rs5742714 GC or CC genotype had a significantly decreased risk of RCC (OR = 0.82, 95% CI = 0.68–0.98) compared to individuals with the rs5742714 GG genotype. No significant evidence of association was found between any of the other polymorphisms from IGF1 or IGFBP3 and risk of RCC.
Effects of IGF1 and IGFBP3 polymorphisms on survival of RCC patients
We then investigated patients from the first set, who had available follow-up information, for evidence of association between these polymorphisms and survival of RCC patients using the Log-rank test and Cox regression analysis. We found that the IGF1 rs5742714 polymorphism was significantly associated with patient survival. As shown in Table 3 and Fig. 1A, compared with patients with the GG genotype, those harboring the GC or GC/CC genotypes showed improved RCC survival (HR = 0.36, 95% CI = 0.14–0.94 and HR = 0.36, 95% CI = 0.14–0.93, respectively; Log-rank P = 0.031 and P = 0.025, respectively). However, no significant evidence of association was found between the other polymorphisms tested and RCC survival (data not shown).
Cox proportional hazard analysis for overall survival of RCC
We further performed a univariate and multivariate Cox proportional hazard analysis for survival of RCC patients. As shown in Table 4 and Fig. 1, clinical stage, tumor grade, and IGF1 rs5742714 (GC/CC vs. GG) were associated with RCC survival in univariate analysis; a multivariate analysis found that clinical stage was the best prognostic factor for RCC survival, followed by tumor grade (P < 0.001, HR = 20.28; 95% CI = 8.78–46.87 and P = 0.002, HR = 4.24; 95% CI = 1.70–10.6, respectively). Interestingly, IGF1 rs5742714 (GC/CC vs. GG) was also an independent predictor of RCC survival (P = 0.035, HR = 0.36; 95% CI = 0.14–0.93).
Effect of IGF1 rs5742714 genotype on IGF1 expression
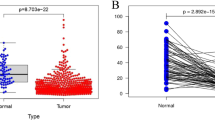
Next, we investigated IGF1 serum levels in RCC patients and controls, and tested for association between IGF1 rs5742714 genotypes and serum levels. As shown in Fig. 2A, the median serum level of IGF1 was significantly higher in cases compared to that in controls (P = 0.005). In addition, and as shown in Fig. 2B, individuals with either the GC or CC genotype had significantly lower serum IGF1 levels than individuals carrying the rs5742714 GG genotype (P = 0.018 and P = 0.049, respectively).
Serum levels of IGF1 in cases and controls.
(A) Distribution of serum IGF1 levels in 100 cases and 100 controls. The mean level of serum IGF1 in cases was significantly higher than that in controls (P = 0.005); (B) Distribution of serum IGF1 levels in controls with different IGF1 rs5742714 genotypes. The number of subjects with a GG, GC, and CC genotype was 70, 26, and 4, respectively. The mean level of serum IGF1 in IGF1 rs5742714 GC and CC groups were significantly lower than that found in the GG groups (P = 0.018 and P = 0.049, respectively).
Functional characterization of IGF1 rs5742714 polymorphism
To further explore the potential functionality of rs5742714, we investigated the effect of this polymorphism on IGF1 expression in renal tissues using real-time quantitative PCR. As shown in Fig. 3A, individuals with the GG or GC genotype had lower levels of IGF1 expression than individuals with the GG genotype, (P = 0.013 and P = 0.017, respectively). As predicted using a bioinformatics model23, the change from a G allele to a C allele at rs5742714 may create a microRNA (miRNA) binding site for hsa-mir-580. To test this finding, we constructed different pGL3 vectors which included the allele-specific binding sequences (Fig. 3B), and co-transfected with miR-508 mimics as well as controls in HEK-293 and 786-o cell lines. As shown in Fig. 3C, we observed significantly greater luciferase activity with the reporter containing the rs5742714 G allele than that containing the rs5742714 C allele. These findings indicate that the rs5742714 C allele may result in inhibition of IGF1 expression through creation of a miRNA binding site.
Effect of IGF1 rs5742714 polymorphism on IGF1 expression.
(A) Association between IGF1 expression in renal tissues and IGF1 rs5742714 genotypes. Compared to individuals with the rs5742714 GG genotype, individuals with the rs5742714 GC or CC genotype were associated with significantly decreased IGF1 mRNA levels (P = 0.013 and P = 0.017, respectively); (B) Bioinformatics analysis predicted a binding site between miR-580 and IGF1; (C) Transient transfection of reporter and mimics into HEK293 and 786-o cell lines. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega). Values are mean ± SD from more than three separate experiments that were each performed in triplicate. *Indicates a significant difference (P < 0.05). There is significant difference between Group C, which were transfected with miR-580 mimics and pGL3-IGF1-G and Group D, which were transfected with miR-580 mimics and pGL3-IGF1-C.
Discussion
In the present study, we investigated whether there was evidence of association of polymorphisms from the IGF1 and IGFBP3 loci with RCC development and survival in a Chinese population. We found that the functional polymorphism rs5742714 in the 3′-UTR of the IGF1 gene was associated with decreased risk of RCC. We also found that the IGF1 rs5742714 polymorphism was an independent prognostic predictor of RCC survival, along with clinical stage and tumor grade, in multivariate analysis. The functional role of rs5742714 was further shown by its effect on IGF1 serum levels. To our knowledge, this is the first study to demonstrate a role of IGF1 rs5742714 in the etiology and prognosis of RCC.
IGF1 and IGFBP3 have been implicated in the development and progression of various human cancers24, including RCC13,25,26. In the present study, we found that patients with RCC had higher IGF1 serum concentrations compared to controls. Genetic variations in the two genes have been extensively tested and associated with the development and progression of several types of cancer. Previous studies found that the IGF1 rs5742714 polymorphism was associated with cancer risk and prognosis. Nakao et al. found in a Japanese population that this polymorphism may have an effect on the development of pancreatic cancer in combination with obesity27. In another study performed in a Chinese population, Zhang et al. demonstrated that IGF1 rs5742714 was a genetic modifier for NSCLC prognosis, especially among patients who underwent surgery15. Similar to findings from our study, they found that patients with either the rs5742714 GC or CC genotype had a favorable overall survival (HR = 0.77, 95% CI = 0.60–0.99).
Considering the position of the rs5742714 polymorphism in the 3′-UTR of the IGF1 gene, it was predicted that the rs5742714 variant might have an effect on IGF1 expression by altering IGF1 mRNA stability and its binding ability to microRNAs (miRNAs). Interestingly, as predicted using a bioinformatics model23, substitution of the rs5742714 G allele by the C allele may create an miRNA binding site for hsa-mir-580, which was demonstrated to act as a tumor inhibitor in breast cancer, through negatively regulating TWIST1 expression28. We hypothesize that the rs5742714 C allele results in miRNA binding to the region, and subsequently, down-regulation of IGF1 expression by either mRNA cleavage or translational repression. This is in agreement with our findings from this study in which individuals with the rs5742714 GC or CC genotype showed decreased IGF1 serum levels compared to individuals with the rs5742714 GG genotype. Given the role of IGF1 in cancer development and progression, reduced levels of IGF1 because of the rs5742714 genotype may decrease cancer susceptibility and inhibit progression, which may explain our findings in the case-control study. However, the exact mechanism underlying the functional effects of this polymorphism on IGF1 expression requires further investigation.
The IGFBP3 rs2854744 polymorphism has been studied in different cancers such as esophageal squamous cell carcinoma29, breast cancer30, colorectal cancer31, and urinary bladder cancer32. The polymorphism is located in the gene promoter and may have an effect on the binding affinity of transcription factors to the promoter, which would lead to changes in gene expression. There are several lines of evidence that suggest that the IGFBP3 rs2854744 A allele may enhance promoter activity and may be associated with higher levels of circulating IGFBP333,34. However, in our study, we did not find evidence for association between this polymorphism and risk or prognosis of RCC. There was also no evidence of association between IGFBP3 serum levels and rs2854744 genotype at the IGFBP3 locus. It should be noted that this polymorphism was previously found to be associated with the development of clear cell RCC in an Iranian population18; however, the sample size of the study was relatively small (158 patients and 316 controls vs. 1027 cases and 1094 controls in our study). In addition, other factors such as different genetic backgrounds may also account for the disparity between our studies. For instance, the minor allele frequency of IGFBP3 rs2854744 in their study differed dramatically from that found in ours (A allele 40.8% vs. C allele 24.1%).
In conclusion, this is the first study demonstrating a role of the IGF1 rs5742714 polymorphism in RCC susceptibility and prognosis in a Chinese cohort. In addition, the findings from the present study also highlight the putatively functional effect of this variant on IGF1 expression by changes mediated by miRNA regulation. Our large two-stage case-control study provided the statistical power to identify polymorphisms associated with risk of RCC; however, the number of patients available for survival analysis were limited and require independent confirmation. Therefore, further validation in a larger population and functional studies are required to understand the role of IGF1 in RCC better.
Additional Information
How to cite this article: Cao, Q. et al. Genetic variation in IGF1 predicts renal cell carcinoma susceptibility and prognosis in Chinese population. Sci. Rep. 6, 39014; doi: 10.1038/srep39014 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Lipworth, L., Tarone, R. E. & McLaughlin, J. K. The epidemiology of renal cell carcinoma. The Journal of urology 176, 2353–2358 (2006).
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA: a cancer journal for clinicians 64, 9–29 (2014).
Cohen, H. T. & McGovern, F. J. Renal-cell carcinoma. The New England journal of medicine 353, 2477–2490 (2005).
Drucker, B. J. Renal cell carcinoma: current status and future prospects. Cancer treatment reviews 31, 536–545 (2005).
Kawai, Y., Sakano, S., Korenaga, Y., Eguchi, S. & Naito, K. Associations of single nucleotide polymorphisms in the vascular endothelial growth factor gene with the characteristics and prognosis of renal cell carcinomas. European urology 52, 1147–1155 (2007).
Qin, C. et al. The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 23, 981–989 (2012).
Qin, C. et al. Variants in angiogenesis-related genes and the risk of clear cell renal cell carcinoma. Mutagenesis 29, 419–425 (2014).
Zhang, Y. et al. Vascular endothelial growth factor gene polymorphisms and renal cell carcinoma: A systematic review and meta-analysis. Oncology letters 6, 1068–1078 (2013).
Purdue, M. P. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nature genetics 43, 60–65 (2011).
Khandwala, H. M., McCutcheon, I. E., Flyvbjerg, A. & Friend, K. E. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocrine reviews 21, 215–244 (2000).
Pollak, M. Insulin and insulin-like growth factor signalling in neoplasia. Nature reviews Cancer 8, 915–928 (2008).
Chuang, S. T., Patton, K. T., Schafernak, K. T., Papavero, V., Lin, F., Baxter, R. C. et al. Over expression of insulin-like growth factor binding protein 3 in clear cell renal cell carcinoma. The Journal of urology 179, 445–449 (2008).
Rasmuson, T., Grankvist, K., Jacobsen, J., Olsson, T. & Ljungberg, B. Serum insulin-like growth factor-1 is an independent predictor of prognosis in patients with renal cell carcinoma. Acta oncologica 43, 744–748 (2004).
Simons, C. C. et al. Genetic Variants in the Insulin-like Growth Factor Pathway and Colorectal Cancer Risk in the Netherlands Cohort Study. Scientific reports 5, 14126 (2015).
Zhang, M. et al. A 3′-untranslated region polymorphism in IGF1 predicts survival of non-small cell lung cancer in a Chinese population. Clinical cancer research: an official journal of the American Association for Cancer Research 16, 1236–1244 (2010).
Cao, Y. et al. Insulin-like growth factor pathway genetic polymorphisms, circulating IGF1 and IGFBP3, and prostate cancer survival. Journal of the National Cancer Institute 106, dju085 (2014).
Al-Zahrani, A. et al. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Human molecular genetics 15, 1–10 (2006).
Safarinejad, M. R. Insulin-like growth factor binding protein-3 (IGFBP-3) gene variants are associated with renal cell carcinoma. BJU international 108, 762–770 (2011).
Cao, Q. et al. Genetic variants in RKIP are associated with clear cell renal cell carcinoma risk in a Chinese population. PloS one 9, e109285 (2014).
Qian, J. et al. Genetic polymorphisms in IGF-I and IGFBP-3 are associated with prostate cancer in the Chinese population. PloS one 9, e85609 (2014).
Cao, Q. et al. A functional variant in the MTOR promoter modulates its expression and is associated with renal cell cancer risk. PloS one 7, e50302 (2012).
Wang, M. et al. A novel functional polymorphism C1797G in the MDM2 promoter is associated with risk of bladder cancer in a Chinese population. Clinical cancer research: an official journal of the American Association for Cancer Research 14, 3633–3640 (2008).
Gong, J. et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Human mutation 33, 254–263 (2012).
Renehan, A. G. et al. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet 363, 1346–1353 (2004).
Cheung, C. W., Vesey, D. A., Nicol, D. L. & Johnson, D. W. The roles of IGF-I and IGFBP-3 in the regulation of proximal tubule, and renal cell carcinoma cell proliferation. Kidney international 65, 1272–1279 (2004).
Rosendahl, A. H., Holly, J. M., Celander, M. & Forsberg, G. Systemic IGF-I administration stimulates the in vivo growth of early, but not advanced, renal cell carcinoma. International journal of cancer Journal international du cancer 123, 1286–1291 (2008).
Nakao M. et al. Interaction between IGF-1 polymorphisms and overweight for the risk of pancreatic cancer in Japanese. International journal of molecular epidemiology and genetics 2, 354–366 (2011).
Nairismagi, M. L. et al. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene 31, 4960–4966 (2012).
Zhao, L. et al. Polymorphisms of insulin-like growth factor binding protein-3 as a predictor for risk and patient survival in esophageal squamous cell carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 74, 148–152 (2015).
Ma, X. et al. Impact of the IGFBP3 A-202C polymorphism on susceptibility and clinicopathologic features of breast cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 71, 108–111 (2015).
Ge, W., Li, Y., Xiang, H. & Li, H. Lack of association of IGFBP-3 gene polymorphisms with colorectal cancer: evidence from 17,380 subjects. Molecular biology reports 41, 2609–2615 (2014).
Selinski, S. et al. Urinary bladder cancer risk in relation to a single nucleotide polymorphism (rs2854744) in the insulin-like growth factor-binding protein-3 (IGFBP3) gene. Archives of toxicology 86, 195–203 (2012).
Deal, C. et al. Novel promoter polymorphism in insulin-like growth factor-binding protein-3: correlation with serum levels and interaction with known regulators. The Journal of clinical endocrinology and metabolism 86, 1274–1280 (2001).
Jernstrom, H. et al. Genetic and nongenetic factors associated with variation of plasma levels of insulin-like growth factor-I and insulin-like growth factor-binding protein-3 in healthy premenopausal women. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 10, 377–384 (2001).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81402321 & 81302217).
Author information
Authors and Affiliations
Contributions
W.Z., S.P. and C.Q. managed the project. L.C., L.P., S.P., L.J., W.M., Q.C. and Z.Z. collected and prepared samples. L.C., X.J., L.Q. wrote the paper. C.Q. and H.L. performed the statistics analysis. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Cao, Q., Liang, C., Xue, J. et al. Genetic variation in IGF1 predicts renal cell carcinoma susceptibility and prognosis in Chinese population. Sci Rep 6, 39014 (2016). https://doi.org/10.1038/srep39014
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39014
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.