Abstract
The whitefly Bemisia tabaci (Genn.) is a pest and vector of plant viruses to crop and ornamental plants worldwide. Using RNA interference (RNAi) to down regulate whitefly genes by expressing their homologous double stranded RNAs in plants has great potential for management of whiteflies to reduce plant virus disease spread. Using a Tobacco rattle virus-derived plasmid for in planta transient expression of double stranded RNA (dsRNA) homologous to the acetylcholinesterase (AChE) and ecdysone receptor (EcR) genes of B. tabaci, resulted in significant adult whitefly mortality. Nicotiana tabacum L. plants expressing dsRNA homologous to B. tabaci AChE and EcR were constructed by fusing sequences derived from both genes. Mortality of adult whiteflies exposed to dsRNA by feeding on N. tabacum plants, compared to non-dsRNA expressing plants, recorded at 24-hr intervals post-ingestion for three days, was >90% and 10%, respectively. Analysis of gene expression by real time quantitative PCR indicated that whitefly mortality was attributable to the down-regulation of both target genes by RNAi. Results indicated that knock down of whitefly genes involved in neuronal transmission and transcriptional activation of developmental genes, has potential as a bio-pesticide to reduce whitefly population size and thereby decrease virus spread.
Similar content being viewed by others
Introduction
Management approaches to reduce the negative effects caused by insect damage to agricultural crops have undergone a considerable shift as the result of a number of recent technological advancements in pest control. An important insect of cotton such as Helicoverpa armigera has been controlled worldwide by expressing an insecticidal protein in planta obtained from Bacillus thuringiensis(Bt)1. Transgenic crops that offer protection from insect pests have aided the agriculture sector by reducing damage to crops resulting from insect damage2. However, there are limitations to the Bt technology including off-target effects to beneficial insects such as natural/biological predators of harmful pests and pollinators3 and its ineffectiveness against sap-sucking insects belonging to the order Hemiptera4. The great potential for using transgenic plants to control phloem-feeding insects belonging to the order, Hemiptera has not been realized, in part, owing to the need to identify and optimize strategies that target and undermine key insect receptors.
RNA interference (RNAi) technology is based on the expression of double stranded RNA (dsRNA) that shares nearly 100% sequence homology with a desired target gene for optimal silencing5 and has been widely used in functional genomics studies carried out on insects6. RNAi is a natural mechanism present in eukaryotes that serves as a defense mechanism against viruses and is also involved in the regulation of gene expression depending upon internal and external environmental conditions7. This phenomenon in animals was first demonstrated in Caenorhabditis elegans by silencing of unc-22 gene in a specific and systemic manner8. Primarily, the mechanism of RNAi mediated gene silencing is based on the exogenous production of short interfering RNAs/microRNAs (siRNAs/miRNAs) by an organism to control the expression of genes. The process can also be triggered in insect cells either by expression of dsRNA or by dsRNA imported into the cells by a transporter protein such as systemic RNA interference defective-1(SID-1)9. The long dsRNA is cleaved by an RNaseIII type nuclease known as Dicer into siRNAs ranging in size from 21–25 bp with 2 nucleotide 3′ overhangs. The siRNAs are recruited by the RNA-induced silencing complex (RISC), a multi-protein complex where one strand is cleaved and degraded, referred to as the passenger, while the other serves as the guide strand. The passenger strand is targeted for degradation upon cleavage, and guide strand integration directs the RISC complex to bind the specific target messenger RNA. When RISC detects the target mRNA with the aid of the guide strand, it attaches to the target, which is degraded by the RNase component of the RISC complex, known as, Argonaute10. A number of different applications for RNAi have been reported, including its use to cause mortality in insects11. Double-stranded RNA has been shown to be persistent in the plasma hemolymph of Blattella germanica, making it lucrative for experimental RNAi studies12. Injection of dsRNA into the pea aphid, Acyrthosiphon pisum, resulted in the silencing of the genes Ap-crt and Ap-cath-L that encode calreticulin and cathepsin-L, respectively13. RNAi-mediated knockdown of midgut genes (Nlsid-1 and Nlaub) of Nilaparvata lugens demonstrated a promising role of RNAi for controlling phytophagous insects in the field14. Suppression of two chitin synthase genes (AgCHS1 and AgCHS2) present in African malaria mosquito (An. gambiae) larvae reduced larval chitin content, which resulted in increased vulnerability to the insecticide, diflubenzuron15.
The ability to express dsRNA in transgenic plants for RNAi-mediated control of insects offers an effective way to control pests of economic importance, as plants retain RNAi machinery which imparts in them a natural ability for cross-kingdom gene silencing16. Many reports have demonstrated the potential for transgenic plant-mediated pest control by expressing dsRNA homologous to genes essential for insect survival11. For example, transgenic Arabidopsis thaliana targeting the Rack1 gene and transgenic Nicotiana benthamiana targeting the MPC002 gene of Myzus persicae have been reported to adversely affect aphid life span17. Silencing of the CYP6AE14c gene impaired Helicoverpa armigera larval resistance to gossypol in Gossypium hirsutum L.18 and Western corn rootworm was reported to be effectively controlled by transgenic Zea mays L. expressing dsRNA against the V-ATPase-A gene19.
Among phloem-feeding insect pests of agricultural importance, the Bemisia tabaci (Genn.) sibling species group is one of the most damaging, causing losses in agronomic and horticultural crops, nearly worldwide20. Collectively, B. tabaci has a broad host range consisting of approximately 500 plant species, worldwide. It causes yield losses by feeding damage, and through the transmission of plant viruses that undermine plant growth and productivity21. Outbreaks caused by cotton leaf curl disease (CLCuD) have resulted in reduced cotton and vegetable crop production, with losses as high as 100%22. A greater understanding of the status and dynamics of B. tabaci in Pakistan cropping systems is needed to more effectively combat CLCuD. Previous studies have identified B. tabaci haplotypes and biotypes23 from Pakistan, including the exotic, invasive MEAM I haplotype (also known as the B biotype) that groups in the North Africa-Mediterranean-Middle East (NA-MED-ME) clade (also, Middle East Asia Minor 1) that was present in only Sindh Province, and two endemic cryptic species that group in the Asia 1 and Asia II 1 clades. The Asia 1 cryptic species was identified in Punjab and Sindh Province, whereas, Asia II 1 was present only in Punjab Province24,25,26. In a recent study that analyzed 593 whitefly collections from 255 locations of Pakistan, the presence of the aforementioned two cryptic species, was confirmed, including Asia II-5 from cotton and Asia II-7 from cotton and non-cotton hosts, and an additional previously uncharacterized species was reported from cotton plants, referred to as ‘Pakistan’25.
The increased invasiveness of certain B. tabaci, has been attributed to higher fecundity, and to the development of resistance to organophosphate, pyrethroid and neonicotinoid insecticides27,28,29,30. Whitefly management has therefore become of great concern in agricultural production systems. Whitefly mortality has been achieved by expressing dsRNA in transgenic tobacco plants targeting the V-ATPase A gene, resulting in a significant reduction in the whitefly population size within 8 days, post-exposure31. Another recent report has demonstrated the potential of RNAi technology for Bemisia control by down-regulating midgut specific genes involved in osmoregulation32.
In this study, we have targeted using RNAi two genes involved in signaling pathways important for whitefly development and neural transmission, hypothesizing that interference with these essential pathways could be lethal. Acetylcholinesterase (AChE) is involved in hydrolysis of the neurotransmitter acetylcholine into its acetyl-CoA and acetate components, thereby clearing the residual neurotransmitter molecules from the synaptic cleft that regulate normal behavior. Blockage of AChE leads to increased acetylcholine levels, causing the continuous stimulation of muscles and glands, and resulting in muscular dysfunction, paralysis, and death. AChE also has a vital role in neurite outgrowth and synapse formation33,34. For example, RNAi mediated silencing of AChE delivered by dsRNAs ingested by Helicoverpa armigera and Blattella germanica, underscores the potency of this target for gene knock downs in insects, much as has been previously achieved with commercial insecticides35,36. The ecdysone receptor gene, EcR, and second target examined in this study, is involved in the steroid signaling pathway whose activation induces a cascade of ecdysone responsive genes that are critical for insect development, and is also important for molecular structure and function of a number of gene products. In recent studies, dsRNA expression to down-regulate EcR function in adult Drosophila melanogaster, demonstrated that EcR decreased expression can perturb reproduction, courtship behavior, and stress resistance37,38, as well as adversely affecting survival and sex-mediated signaling in adults39.
Here, tobacco plants transient expression of dsRNA homologous to sequences of the B. tabaci acetylcholinesterase and ecdysone receptor gene, respectively, were tested separately using Tobacco rattle virus (TRV)-induced gene silencing (VIGS) vector (TRV-VIGS) for dsRNA expression in N. tabacum. The TRV-VIGS consists of the modified viral genomic component, TRV RNA1 that encodes viral movement, and TRV RNA2, which is involved host plant gene silencing, a system first designed to study transiently functional genomics of viral-plant interactions40, without requiring stable plant transformation. Results indicate that transiently, as well as transgenically expressed dsRNA induced RNAi, resulted in whitefly mortality in adult whitefly, post-dsRNA ingestion from tobacco plants.
Results
DNA sequencing and analysis of AChE and EcR sequences for dsRNA design
Twenty sequences of 200 bp were obtained by PCR for 20 different field collections of B. tabaci from Pakistan, and have been deposited in the GenBank database as Accession numbers KU307111 – KU307130 for AChE and KU307151 – KU307170 (sequences not released) for EcR. Multiple sequence alignment of the AChE and EcR from Pakistan, was carried out with MegAlign software using the ClustalW method. Consensus sequences were generated from the alignments and the results revealed that both the sequences have +60 bp nucleotides conserved among T. vaporariorum and two B. tabaci, MEAM I species (representative mtCOI accessions: KX675904 – KX675908) and Asia II (representative mtCOI accessions: KX675909 – KX675918) present in Pakistan (Table S1 and Figure S1). The sequence distance matrix generated using SDT v1.2 showed that AChE sequences were 100% conserved among the Pakistan-endemic B. tabaci species (Fig. 1a). The EcR sequences also demonstrated 100% conservation among T. vaporariorum and B. tabaci (Fig. 1b). One isolate of Asia II I showed 98% similarity with the rest of the sequences in EcR clade.
Whitefly bioassay on plants transiently expressing dsRNA
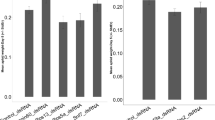
The target genes were transiently expressed in N. tabacum plants using the TRV-VIGS vector system because of its usefulness as a VIGS vector. The experiment was performed to assess the efficacy of dsRNA expression against whitefly target genes before proceeding to stably transforming plants. Significantly higher adult whitefly mortality, at 40%, was recorded 48 hrs post-plant exposure, and reached 98% after four days for tobacco plants transiently expressing dsRNA homologous to AChE. And, greater than 50% mortality was observed after six days of whiteflies feeding on tobacco plants transiently expressing dsRNA against whitefly EcR. Mortality was 100% by nine days post-plant exposure to EcR dsRNA transiently expressed in tobacco plants. Statistical analysis of mortality data (Fig. 2) demonstrated significantly higher mortality in whiteflies exposed to dsRNA of both target genes, compared to the negative controls, for which only 13% mortality was recorded after ten days. A p-value < 0.05 indicated that results were highly significant.
Transgenic tobacco plants expressing dsRNA
To target the genes of whiteflies having an essential role in cellular signaling through transgenic tobacco, a construct was developed in a pJIT163 vector in such a way that partial fragments of AChE and EcR were simultaneously expressing under the control of a Figwort mosaic virus (FMV) promoter. An intron was added to separate the sense and anti-sense fragments to facilitate configuration of a hairpin mRNA upon transcription. Total DNA was isolated from the leaf tissue of the plants. Confirmation of gene insertion into the plants was established by PCR using gene specific as well as nptII gene specific primer pairs.
Expression of AChE and EcR dsRNA cassettes in transgenic tobacco plants
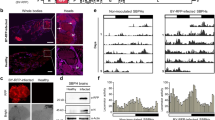
PCR positive transgenic tobacco lines G2.H3-5 and G2.H2-4 (Fig. 3) were subjected to real-time qPCR for gene expression analysis of nptII gene (Figure S2) and RNA dot blot assay for dsRNA/siRNA detection of whitefly target genes AChE and EcR. Total RNA was isolated from 3 plants of each transgenic line, and from three non-transgenic control plants. DIG-labelled RNA probes for the G2 construct, 200 bp in size for each gene fragment, were hybridized to the RNA spots on a nylon membrane (+Hybond, Amersham, UK). In-vitro transcription of cloned AChE fragment from a pTZ57R/T vector was used as a positive control and it was serially diluted to the known concentration of the transcribed RNA isolated from the transgenic tobacco lines. Color detection by NBT/BCIP revealed the presence of transcribed AChE and EcR fragments in the transgenic lines, and the accumulation was estimated using a known concentration of positive control RNA (Fig. 4).
Plant transformation and transgene analysis (a) selection of the tobacco transformants following Agrobacterium-mediated transformation with the G2 cassette and kanamycin media selection (b) Confirmation of the nptII gene for the kanamycin resistance in the transgenic lines by PCR-amplification of product size 215 bp. Loading arrangement of the samples in 1% agarose gel is, Lane 1 = positive control, Lane 2 = 1 kb DNA ladder, Lane 3 = negative control, Lane 4 –6 = transgenic line G2.H2-4, and Lane 7–9 = transgenic line G2.H3-5 (c) confirmation that construct inserted into the nuclear genome of the tobacco plants was carried out by PCR-amplification of target gene-specific primers of 400 bp product size. Loading arrangement of the samples in 1% agarose gel is as, Lane 1 = positive control, Lane 2 = 1 kb DNA ladder, Lane 3 = negative control, Lane 4–6 = transgenic line G2.H2-4, and Lane 7–9 = transgenic line G2.H3-5.
RNA dot blot showing relative expression of dsRNA in the transgenic tobacco lines (a) expressing the target gene AChE: Block 1–3 = transgenic line G2.H2-4, Block 4–6 = G2.H3-5, and Block 7–9 = negative control plants; (b) expressing the target gene EcR: Block 1–3 = transgenic line G2.H2-4, Block 4–6 = G2.H3–5 and Block 7–9 = negative control plants; and (c) serial dilution of known concentrations of the positive control RNA.
Real time PCR quantification of target gene expression
Transient expression of dsRNA in tobacco lines G2.H2-4 and G2.H3-5 were selected for insect feeding bioassays in order to evaluate the dsRNA/siRNA mediated down regulation of the target genes (AChE and EcR). Real time qPCR was performed to assess the suppression of gene expression in B. tabaci. Whiteflies were allowed to feed on the transgenic tobacco plants producing dsRNA against AChE and EcR genes of the B. tabaci. The RT-qPCR results demonstrated highly significant suppression of the expression of both target genes. The RT-qPCR results showed significantly reduced expression of both whitefly target genes compared to whiteflies allowed feeding exposure on the negative control plants, for three consecutive days. Collection of whiteflies from transgenic and control plants at three different time points was carried out for down regulation assessment. The RT-qPCR analysis revealed approximately 98% reduction in AChE and EcR mRNA expression after 24 hrs in the transgenic lines, compared to the negative control plants, respectively (Fig. 5).
Whitefly mortality following exposure to transgenic plants expressing dsRNA
Approximately, 200 whiteflies were allowed to feed on transgenic as well as control tobacco plants grouped in separate cages. More than 30% mortality was observed after one day of feeding exposure to the AChE and EcR transgenic lines, but no significant mortality was observed in whiteflies exposed to the negative control tobacco plants. At two days post-feeding exposure, whitefly mortality on transgenic tobacco plants was 40–45% for the transgenic lines G2.H2-4 and G2.H3-5. After three days post-feeding exposure, greater than 90% mortality was observed in both transgenic lines, while insignificant mortality was observed in negative control tobacco plants (Fig. 6).
Discussion
The whitefly B. tabaci poses a threat to agricultural production worldwide. An alternative to chemical pesticides involves the development of whitefly resistance using plant breeding approaches, but successes have been minimal, while also being time and labor intensive41. Introduction of transgenic Bt cotton has helped farmers for more than two decades to control lepidopteran insect pests, resulting in decreased pesticide use and lowered management costs42. However, to the present Bt has not been effective against sap-sucking insects especially whitefly. Another solution involves the use of RNAi that can selectively target the genes belonging to particular insect species, thereby avoiding the targeting of beneficial insects. Over the past few years, this technology has been shown effective for insect gene silencing in functional genomics studies. The methods adopted for RNAi in insects including dsRNA spray, micro-injection and artificial diet based feeding were, although successful, are not yet suitable for field applications. However, the expression of dsRNAs in transgenic plants to silence target genes has been demonstrated to be economic for insect control at the field level11,41.
The whitefly B. tabaci is considered one of the most damaging among agricultural insect pests in tropical to temperate regions of the world and also a host of different mites43,44. Phylogenetic studies have identified several different endemic cryptic species in Pakistan that belong to the Asia I and II major clades, as well as the exotic MEAM I haplotype (B biotype), which has been widely established nearly globally in agricultural systems45. The analysis of AChE and EcR genes as prospective RNAi targets that result in mortality of B. tabaci demonstrated great promise for target design that takes into account conserved regions of genes that are likewise conserved across the B. tabaci sibling species group, and the greenhouse whitefly T. vaporariorum. Thus the results reported here illustrate the utility of targeting homologous regions of insect genes for RNAi, to achieve broad-spectrum whitefly control.
Pesticides, such as organophosphates, pyrethroids and carbamates have become less effective for whitefly control, and their use at high concentrations worsens environmental conditions and health of non-target organisms28. In this study, whiteflies exposed to tobacco plants transgenically expressing dsRNA showed significant down-regulation of the target genes, AChE and EcR, that lead to a high rate of mortality. This was accomplished by fusing the partial fragments of both the genes, separated by a 115 bp intron, expressed under control of the FMV promoter, which drives viral gene expression in various plant tissues and organs, including the phloem. Analysis by RNA dot blot analysis of the transgenic tobacco plants, compared with serial dilutions of the positive control, indicated that the dsRNA/siRNA relative concentration ranged from 10 to 100 pg. Moreover, down regulation of the target genes in the transgenic plants was corroborated in bioassays in which whiteflies given feeding exposure to tobacco plants, showed significantly higher mortality, compared to the non-transgenic control tobacco plants. Results from the RT-qPCR confirmed that early mortality was associated with the down-regulation of the AChE and EcR genes in whiteflies exposed to dsRNA expressing tobacco plants, and that whiteflies feeding on non-transgenic control plants showed significantly higher expression of the target genes, compared to exposed to transgenic lines expressing the respective genes. The results from these experiments confirmed the vital role of the two target genes (AChE and EcR) in life processes. Specifically, knock downs of AChE and EcR interfered with whitefly mating and oviposition before it resulted in mortality. The latter genes also are targets of insecticides such as Imidacloprid46 and benzoyl hydrazine47,48 that affect the neuronal and steroid functions of insects. Previously RNAi technology has been shown effective in down-regulating expression of midgut-expressed osmoregulatory genes, aquaporin and alpha glucosidase of whitefly32, for which greater than 70% mortality was shown, six days post-feeding on transgenic tobacco plants, compared to negative controls.
Distinct genetic and physiological differences are expected to occur among agriculturally important insect species, making important the careful selection of target gene(s) for each particular insect species, while also minimizing negative effects on beneficial or non-target insects that may be exposed to transgenic plants. Numerous considerations are thereby important for the achievement of safe and successful transgenic plant-mediated insect management. This study demonstrated the potential for dsRNA biopesticide technology using combination of genes in a single dsRNA construct, and opens up new avenues for gene pyramiding. Perhaps RNAi could also complement other approaches, such as Bt technology, to achieve broad-spectrum resistance against different groups of sucking and chewing insects.
Materials and Methods
Whitefly B. tabaci haplotype or species identification
Approximately twenty-five adult whiteflies were collected from cotton/agricultural fields in 14 districts/different locations of Pakistan where previous analyses have shown the predominance of B. tabaci species that belong to either with the Asia II major clade, and/or identified as the exotic B biotype49, or the greenhouse whitefly Trialeurodes vaporariorum (West).
Five adults were selected from each group of whitefly samples collected and were individually subjected to PCR amplification of the mitochondrial cytochrome oxidase I (mtCOI) gene was carried out using Phire Animal Tissue Direct PCR Kit (cat no. F140WH), according to the manufacturer’s instructions. In brief, adult whiteflies were homogenized in 20 μL dilution buffer, containing 0.5 μL DNA Release provided in the kit. The mixture was heated at 98 °C for 2 minutes, and centrifuged for 2 minutes at 13,000 rpm at room temperature. The DNA was stored at −20 °C after using 1 μL of supernatant in a 20 μL PCR reaction. The primers used for mtCOI amplification were: COI-3 F5′-TTGATTTTTTGGTCATCCAGAAGT-3′ and COI-3 R5′-TCCAATGCACTAATCTGCCATATTA-3′50.
The reaction contained 10 μL 2X Phire Animal Tissue PCR Buffer, 0.4 μL Phire Hot Start II DNA Polymerase, 0.25 μL forward and reverse primer each (10 μmol) and final volume made up with ddH2O. The cycling parameters were: 1 cycle for denaturation at 95 °C for 5 minutes, 34 cycles for 30 seconds at 95 °C, 30 seconds at 45 °C, 45 seconds at 72 °C and a final extension for 1 cycle at 72 °C for 10 minutes The PCR products of 900 bp in size were subjected to DNA sequencing (Genomics and Bioinformatics Research Unit, USDA-ARS, MS, USA).
Sequence aligned was carried out and a Maximum Likelihood (ML) phylogenetic tree was reconstructed using Kimura 2-parameter model with 1000 bootstrap value using MEGA 6.0. The sequence from the greenhouse T. vaporariorum was used as an outgroup (AF418672).
The raw sequences were assembled with SeqMan software of the DNASTAR package v 5.0, and were BLAST against the NCBI nr database. The B. tabaci colony used in bioassay experiments was established by placing 2 separate pairs of Asia II adults on tobacco plants and allowing them to feed and mate. The colony was thereafter maintained by periodic serial transfer to cotton plants in a greenhouse at NIBGE, Faisalabad, Pakistan.
Total RNA isolation and cDNA synthesis from adult whitefly
Total RNA isolation from the collected samples of adult whiteflies was carried out using TRIzol reagent (Life Technologies cat no. 15596018), per the manufacturer’s recommendation. The homogenate of whitefly tissue samples was made with 500 μL of TRIzol reagent and incubated at room temperature (RT) for 5 minutes followed by centrifugation at 13000 rpm (at 4 °C) for 5 minutes. The supernatant was collected in separate nuclease free 1.5 mL eppendorf tubes and 250 μL of chloroform was added. The contents were mixed briefly and were incubated at room temperature for 5 minutes followed by centrifugation at 13,000 rpm (at 4 °C) for 5 minutes. The aqueous phase was transferred to another nuclease free 1.5 mL eppendorf tube and 500 μL of pre-chilled isopropanol was added and incubated at room temperature for 5 minutes followed by centrifugation at 13000 rpm (at 4 °C) for 5 minutes. The RNA pellet was washed with 75% ethanol, and dissolved in 20 μL nuclease free water. The cDNA was synthesized using 0.2 μg of RNA template from each whitefly samples using SuperScript™ IV First-Strand cDNA synthesis kit (Thermo Fisher Scientific, cat no. 18091200) following the manufacturer’s recommended protocol.
Amplification and sequencing of whitefly AChE and EcR fragments
The cDNA from each sample was diluted to 1:9 and 1 μL of diluted cDNA was used in the PCR reaction using DreamTaq Green PCR Master mix 2x (Thermo Fisher Scientific, cat no. K1081). The PCR reaction contained 12.5 μL of Green PCR Master mix and 1 μL template cDNA with 0.5 μL each forward and reverse primers (10 μmol) added to a final volume of 25 μL using nuclease free H2O.
The PCR primers used were: AChEF5′-CGGAATTCCGCACAATATCACGCTCTTC-3′ and AChER5′-GGCTCGAGTCGATAGCCTCAGGGATCTG-3′ and EcRF5′-AAGAATTCATGAACATCCATCCCCAGAG-3′ and EcRR5′-AACTCGAGTCAGCATCATACCTCCTTGC-3′ for all the species and T. vaporariorum to amplify acetylcholinesterase and ecdysone receptor, respectively.
The PCR products were subjected to DNA sequencing (Scheffler lab, Genomics and Bioinformatics Research Unit, USDA-ARS, MS USA). The DNA sequence fragments corresponding to the B. tabaci and T. vaporariorum whitefly AChE and EcR gene fragments, respectively, were aligned using MegAlign software (DNASTAR package v 5.0) and the ClustalW algorithm. A pairwise distance matrix was computed for each gene in Sequence Demarcation Tool 1.2 (SDT) using the ClustalW method. The alignment and matrix were used to identify a 60 bp conserved region around which dsRNA could be designed.
Cloning of AChE and EcR gene fragments in TRV-VIGS plasmid vector
The primer pairs, F5′-GTGAATTCCGCACAATATCACGCTCTTCG-3′ and R5′-TGGGATCCCGTGTGAGGGCAGCCGACAGCC-3′ and F5′-AAGAATTCATGAACATCCATCCCCAGAGG-3′ and R5′-ACGGATCCACTACATGCCTTTAATAAAGC-3′ were designed to amplify a 200 bp product corresponding to AChE and EcR gene fragments, respectively, with the addition of restriction sites for EcoRI and BamHI to facilitate cloning. The PCR product was purified with 3M sodium acetate and double digested with EcoRI and BamHI using 2x Tango buffer (Thermo Fisher Scientific). The TRV2 component of the TRV-VIGS vector was also digested using the same enzymes. The restricted plasmid vector and purified restricted PCR products were then ligated. The plants were infiltrated with TRV1+ [TRV2 + ACHE](AChE was cloned in TRV2 component) and TRV1 + [TRV2 + EcR](EcR was cloned in TRV2 component). The control plants were infiltrated with wild type TRV1 + TRV2.
Agro-infiltration with TRV-VIGS containing dsRNA
The vector component the TRV2, containing gene of interest, and TRV1 were transformed in Agrobacterium tumefaciens strain GV3101. Positive colonies were confirmed by a modified method involving colony PCR51, and inoculated into 50 mL Luria Bertani medium with addition of 50 mg ml−1 kanamycin and 20 mg ml−1 rifampicin. The cultures were centrifuged after 48 hours of incubation at 28 °C and were resuspended in induction medium (10 mM 2-N-morpholino ethane sulfonic acid with pH = 5.5, 10 mM MgCl2 and 200 μM acetosyringone) and were held overnight at room temperature. The optical density (O.D.) was maintained at 2.0, and cells were suspended in 10 mM MES buffer, pH 5.5. The VIGS vector components, TRV1 and TRV2, were mixed 1:1 and used to infiltrate N. tabacum plant leaves 25-days after plants were transplanted. The tobacco plants were held in the dark for two days, and then placed in an environmentally controlled greenhouse, maintained as described above. Similarly, agro-inoculation of tobacco plants with the plasmid vector minus the dsRNA hairpins, were established separately using the approach described above.
Construction of dsRNA expression cassette
The sequence of the whitefly B. tabaci, MEAM I haplotype (B biotype) genes used to design the dsRNA hairpin cassettes for AChE (GenBank accession number EF675187.1) and EcR (GenBank accession number EF174329.1) were retrieved from the NCBI database. These sequences were analyzed for homology with other available sequences in the nr database using the NCBI BLAST search tool, and specifically against the human genome. Sequence regions with a 0% BLAST hit to the human genome sequence were used for commercial synthesis (Life Technologies, Thermo Fisher Scientific, USA) of the fragments, joined in the sense (200 bp each) and antisense (200 bp each) orientation, using an intron of A. thaliana as a spacer between the sense and anti-sense fragments to yield a hairpin structure upon expression. Suitable restriction sites were introduced into the fragments to allow cloning into the plasmid vector, pJIT163. The resultant 931 bp fragment was cloned into the plasmid vector for expression under the control of a Figwort mosaic virus (FMV) promoter and CaMV35S terminator. The SacI and HindIII restriction sites were introduced and used to replace the 2xCaMV35S promoter of pJIT163 vector with the FMV promoter. The complete cassette [SacI-acetylcholinesterase (sense)-ecdysone receptor (sense)-Intron-acetylcholinesterase (anti sense)-ecdysone receptor (anti sense)-EcoRV] was cloned into the binary vector pCambia2300 (GSL Biotech LLC Chicago, USA) for tobacco plant transformation using restriction sites SacI and EcoRV. The EcoRV restriction site produced a blunt end cut and was used because of the limited number of options available for further cloning. The pCambia2300 vector was restricted using SacI and SmaI sites for compatible ligation because SmaI produced a blunt end compatible with the EcoRV restriction site.
Transgenic tobacco plants
The cassette developed in pJIT163 vector initially, was restricted with restriction enzymes SacI and EcoRV and ligated into the plant transformation binary vector pCambia2300. Using the binary plasmid vector pCambia2300 containing the cloned dsRNA cassette, the plasmid was transformed in the A. tumefaciens strain LBA4404 using electroporation, at 1.8 kV. Positive transformants were inoculated in 50 mL LB media for 48 h at 28 °C. The N. tabacum explants were agro-transformed52, with the gene constructs, and selected by direct organogenesis using established procedure and the transformants were selected on 500 mg/L of kanamycin. Transgenic plants were grown in a tissue culture room at 25 ± 2 °C. Plants were transplanted to plastic pots, and transferred to a greenhouse maintained under controlled conditions, at 25 ± 2 °C with 60–70% relative humidity. The insertion of T-DNA was confirmed by PCR (Fig. 3) using Dream Taq Green PCR Master Mix (2x) (Thermo Fisher scientific, USA). The cycling parameters of PCR were 94 °C for 3 minutes (1 cycle), 94 °C for 30 seconds, 54 °C for 30 seconds and 72 °C for 45 seconds (35 cycles) and a final extension step at 72 °C for 10 minutes. The primer pairs used for the confirmation were: G2F5′-CGCACAATATCACGCTCTTCGGC-3′ and G2R5′-ACTACATGCCTTTAATAAAGCTA-3′ for transgene confirmation while nptIIF5′-CTCACCTTGCTCCTGCCGAGA-3′ and nptIIR5′-CGCCTTGAGCCTGGCGAACAG-3′ for kanamycin resistance marker gene nptII.
RNA dot blot analysis of dsRNA expression in transgenic tobacco
RNA was isolated from PCR positive transgenic lines according to the standard protocol by TRIzol reagent (Thermo Fisher scientific, USA). RNA concentrations of the samples were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher scientific, USA). Dilutions of the RNA samples were made by dissolving 15 μg of total RNA in 66% formamide, 21% formaldehyde and 13% MOPS buffer and incubating at 65 °C for 15 minutes for denaturation. RNA samples dissolved in dilution buffer were spotted onto positively charged nylon membrane (Hybond-N+, GE-Amersham, UK) soaked with 6x SSC solution (pH = 7) and dried on sterilized Whatman filter paper. Spots of RNA were then fixed under UV, cross linked with the energy output 1200 μjoules/cm2 per second for 30 seconds (twice) at 300 nm. DIG (Dioxigenin-Labelled UTP) labelled RNA probe of AChE and EcR were made using DIG RNA labelling kit, according to the manufacturer’s protocol (Sigma-Aldrich). The probe was denatured at 65 °C for 5 minutes before hybridization onto a nylon membrane. After washing and blocking, RNA spots were detected by the chromogenic method, with a 6-hr incubation in NBT/BCIP at room temperature.
Whitefly mortality on transgenic tobacco
Adults of the B. tabaci (Asia II I) were collected from cotton plants grown in the greenhouse, at 60–70% relative humidity, 26 ± 2 °C, and 16:8 h day/night photoperiod. Adults were transferred to and allowed to feed and oviposit on 70-day old tobacco plants grown maintained under the conditions described above.
Newly emerged adult whiteflies were released either on TRV-VIGS infiltrated, or transgenic tobacco plants, and the respective negative control tobacco plants, and allowed feeding exposure to the plants. Plants were monitored for ten continuous days in two replicated experiments for TRV-VIGS. Mortality was also recorded on daily basis for three consecutive days in case of transgenic tobacco plant bioassays (as insects were only able to survive for three days on transgenic tobacco plants). The mean percent mortality was calculated, and subjected to an analysis of variance (ANOVA) using the statistical software Graph Pad Prism 5 and SPSS version 20 for transient and transgenic experiments respectively. Tukey’s post-test of significance was performed for both bioassays to assess pairwise differences, and a p-value < 0.05 was considered significant.
Adult whiteflies were collected from transgenic plants at 1, 2 and 3 days after their initial release on transgenic tobacco plants. Total RNA was isolated from whiteflies using TRIzol reagent, and then treated with DNase I, according to the manufacturer’s instructions (Thermo Fisher Scientific, USA). A Nanodrop 2000 spectrophotometer (Thermo Fisher scientific, USA) was used to quantify whitefly total RNA. From each RNA sample, 0.5 μg was used for cDNA synthesis, according to manufacturer’s protocol (Thermo Fisher Scientific, USA).
Real time quantitative PCR analysis
Expression of the target genes, AChE and EcR, in adult B. tabaci was quantified by RT-qPCR. The 25 μl reaction mixture contained 12.5 μl SYBR® Green, 0.25 μl each of the forward and reverse primers (0.1 pmole), 2.5 μl cDNA (~25 ng) and 9.5 μl water. Primer sequences: qAChEF5′-GGAGGAGGGCAACTACTGGAT-3′ and qAChER5′-CACCGCCTGGATGAAACTG-3′ for AChE gene, qEcRF5′-GTTTGCAACTAACCAGCCGTATA-3′ and qEcRR5′-GGAGCAGATCCTCCACAGTATC-3′ for EcR gene while q18SF5′-GACCGGAGCTTGCAATTGTTC-3′ and q18SR5′-ATCGCCGCGAGGTTATGAC-3′ were used for 18S ribosomal RNA gene as internal control. The PCR reaction parameters for both primer pair combinations were: 1 cycle at 94 °C for 10 minutes followed by 40 cycles, at 94 °C (for 30 seconds), 53 °C (for 30 seconds) and 72 °C (for 30 seconds). The RT-qPCR reactions were run in triplicate, in a 96 well microtiter plate using Bio-Rad iQ5 thermal cycler (Bio-Rad, USA).
To assess the specificity of the amplification product, at the end of each run, a melt curve analysis was performed, from 60 to 95 °C, with an increment of 0.5 °C every 10 seconds Quantification results were analyzed using 2 −ΔΔCT method53. The 18 S ribosomal RNA (rRNA) gene was used to normalize the corresponding Ct values. Transcript levels were measured in whiteflies fed on transgenic lines and/or control plants and the fold change in the expression levels of AChE and EcR was determined as described, above.
Additional Information
How to cite this article: Malik, H. J. et al. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 6, 38469; doi: 10.1038/srep38469 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Onkaramurthy, S., Goud, K. B. & Udikeri, S. Season long bio-efficacy of first and second generation bt cotton genotypes against Helicoverpa armigera (hubner), Earias vittella (fabricius), Spodoptera litura (fabricius) and Pectinophora gossypiella (saunders) under rainfed conditions. Interaction 4, 11.96 (2015).
Carrière, Y., Crowder, D. W. & Tabashnik, B. E. Evolutionary ecology of insect adaptation to Bt crops. Evolutionary Applications 3, 561–573 (2010).
Duan, J. J., Marvier, M., Huesing, J., Dively, G. & Huang, Z. Y. A meta-analysis of effects of Bt crops on honey bees (Hymenoptera: Apidae). PLoS One 3, e1415 (2008).
Chougule, N. P. & Bonning, B. C. Toxins for transgenic resistance to Hemipteran pests. Toxins 4, 405–429 (2012).
Pusch, O. et al. Nucleotide sequence homology requirements of HIV‐1‐specific short hairpin RNA. Nucleic Acids Research 31, 6444–6449 (2003).
Bellés, X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annual Review of Entomology 55, 111–128 (2010).
Voinnet, O. RNA silencing as a plant immune system against viruses. Trends in Genetics 17, 449–459 (2001).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Winston, W. M., Molodowitch, C. & Hunter, C. P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002).
Scott, J. G. et al. Towards the elements of successful insect RNAi. Journal of Insect Physiology 59, 1212–1221 (2013).
Huvenne, H. & Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. Journal of Insect Physiology 56, 227–235 (2010).
Garbutt, J. S., Bellés, X., Richards, E. H. & Reynolds, S. E. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. Journal of Insect Physiology 59, 171–178 (2013).
Jaubert-Possamai, S. et al. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnology 7, 63 (2007).
Zha, W. et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One 6, e20504 (2011).
Zhang, X., Zhang, J. & Zhu, K. Chitosan/double‐stranded RNA nanoparticle‐mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Molecular Biology 19, 683–693 (2010).
Zhang, L. et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Research 22, 107–126 (2012).
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J. & Hogenhout, S. A. Silencing of aphid genes by dsRNA feeding from plants. PLoS One 6, e25709 (2011).
Mao, Y. B. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature Biotechnology 25, 1307–1313 (2007).
Bolognesi, R. et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7, e47534 (2012).
Rao, Q., Luo, C., Zhang, H., Guo, X. & Devine, G. Distribution and dynamics of Bemisia tabaci invasive biotypes in central China. Bulletin of Entomological Research 101, 81–88 (2011).
Chen, W. et al. Estimation of the whitefly Bemisia tabaci genome size based on k-mer and flow cytometric analyses. Insects 6, 704–715 (2015).
Nadeem, A., Xiong, Z. & Nelson, M. Cotton Leaf Curl Virus, A threat to Arizona cotton? Cotton: A College of Agriculture Report (1995).
Brown, J. K. Phylogenetic biology of the Bemisia tabaci sibling species group. Bemisia: Bionomics and Management of a Global Pest (eds. Stansly, P. A., Naranjo, S. E. ) 31–67 (Springer, 2010).
Ahmed, M. Z. et al. Genetic identity of the Bemisia tabaci species complex and association with high cotton leaf curl disease (CLCuD) incidence in Pakistan. Pest Management Science 67, 307–317 (2011).
Simón, B. et al. Genetic structure of field populations of begomoviruses and of their vector Bemisia tabaci in Pakistan. Phytopathology 93, 1422–1429 (2003).
Ashfaq, M. et al. DNA barcoding of Bemisia tabaci complex (Hemiptera: Aleyrodidae) reveals southerly expansion of the dominant whitefly species on cotton in Pakistan. PLoS One 9, e104485 (2014).
Ahmad, M., Arif, M. I., Ahmad, Z. & Denholm, I. Cotton whitefly (Bemisia tabaci) resistance to organophosphate and pyrethroid insecticides in Pakistan. Pest Management Science 58, 203–208 (2002).
Denholm, I., Cahill, M., Dennehy, T. & Horowitz, A. Challenges with managing insecticide resistance in agricultural pests, exemplisfied by the whitefly Bemisia tabaci. Philosophical Transactions of the Royal Society B: Biological Sciences 353, 1757–1767 (1998).
Elbert, A. & Nauen, R. Resistance of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides in southern Spain with special reference to neonicotinoids. Pest Management Science 56, 60–64 (2000).
Shadmany, M., Omar, D. & Muhamad, R. Biotype and insecticide resistance status of Bemisia tabaci populations from Peninsular Malaysia. Journal of Applied Entomology 139, 67–75 (2015).
Thakur, N. et al. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase a gene. PLoS One 9, e87235 (2014).
Raza, A. et al. RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): Potential technology for the control of whitefly. PLoS One 11, e0153883 (2016).
Olivera, S., Rodriguez-Ithurralde, D. & Henley, J. M. Acetylcholinesterase promotes neurite elongation, synapse formation, and surface expression of AMPA receptors in hippocampal neurones. Molecular and Cellular Neuroscience 23, 96–106 (2003).
Shapira, M., Thompson, C. K., Soreq, H. & Robinson, G. E. Changes in neuronal acetylcholinesterase gene expression and division of labor in honey bee colonies. Journal of Molecular Neuroscience 17, 1–12 (2001).
Kumar, M., Gupta, G. P. & Rajam, M. V. Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. Journal of Insect Physiology 55, 273–278 (2009).
Revuelta, L. et al. RNAi of ace1 and ace2 in Blattella germanica reveals their differential contribution to acetylcholinesterase activity and sensitivity to insecticides. Insect Biochemistry and Molecular Biology 39, 913–919 (2009).
Schwedes, C., Tulsiani, S. & Carney, G. E. Ecdysone receptor expression and activity in adult Drosophila melanogaster. Journal of Insect Physiology 57, 899–907 (2011).
Schwedes, C. C. & Carney, G. E. Ecdysone signaling in adult Drosophila melanogaster. Journal of Insect Physiology 58, 293–302 (2012).
Tricoire, H. et al. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mechanisms of Ageing and Development 130, 547–552 (2009).
Senthil-Kumar, M. & Mysore, K. S. Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nature Protocols 9, 1549–1562 (2014).
Younis, A., Siddique, M. I., Kim, C. K. & Lim, K. B. RNA interference (RNAi) induced gene silencing: a promising approach of hi-tech plant breeding. International Journal of Biological Sciences 10, 1150–1158 (2014).
Benbrook, C. M. Impacts of genetically engineered crops on pesticide use in the US–the first sixteen years. Environmental Sciences Europe 24, 2190–4715 (2012).
Fan, Y. & Petitt, F. L. Dispersal of the broad mite, Polyphagotarsonemus latus (Acari: Tarsonemidae) on Bemisia argentifolii (Homoptera: Aleyrodidae). Experimental and Applied Acarology 22, 411–415 (1998).
Greenberg, S., Legaspi, B., Jones, W. & Enkegaard, A. Temperature-dependent life history of Eretmocerus eremicus (Hymenoptera: Aphelinidae) on two whitefly hosts (Homoptera: Aleyrodidae). Environmental Entomology 29, 851–860 (2000).
Bedford, I. D., Briddon, R. W., Brown, J. K., Rosell, R. & Markham, P. G. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Annals of Applied Biology 125, 311–325 (1994).
Matsuda, K. et al. Neonicotinoids: insecticides acting on insect nicotinic acetylcholine receptors. Trends in Pharmacological Sciences 22, 573–580 (2001).
Bengochea, P. et al. Insect growth regulators as potential insecticides to control olive fruit fly (Bactrocera oleae Rossi): insect toxicity bioassays and molecular docking approach. Pest Management Science 69, 27–34 (2013).
Knight, A. L., Dunley, J. E. & Jansson, R. K. Baseline monitoring of codling moth (Lepidoptera: Tortricidae) larval response to benzoylhydrazine insecticides. Journal of Economic Entomology 94, 264–270 (2001).
Attique, M., Rafiq, M., Ghaffar, A., Ahmad, Z. & Mohyuddin, A. Hosts of Bemisia tabaci (Genn.)(Homoptera: Aleyrodidae) in cotton areas of Punjab, Pakistan. Crop Protection 22, 715–720 (2003).
Frohlich, D., Torres‐Jerez, I., Bedford, I., Markham, P. & Brown, J. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Molecular Ecology 8, 1683–1691 (1999).
Zhao, Y., Dong, Z., Li, T. & Huang, G. A simple negative selection method to identify adenovirus recombinants using colony PCR. Electronic Journal of Biotechnology 17, 8–8 (2014).
Burow, M. D., Chlan, C. A., Sen, P., Lisca, A. & Murai, N. High-frequency generation of transgenic tobacco plants after modified leaf disk cocultivation with Agrobacterium tumefaciens. Plant Molecular Biology Reporter 8, 124–139 (1990).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
This work was funded by a fellowship to AR by the Pakistan Higher Education Commission, Pakistan and USDA-ICARDA to carry out research at NIBGE, Pakistan. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Author information
Authors and Affiliations
Contributions
S.M. and I.A. conceived the idea. H.J.M. and A.R. conducted all the experiments and wrote first draft of the paper. J.K.B. hosted and facilitated A.R. during his stay at The University of Arizona. I.A., J.A.S., B.E.S., J.K.B., and S.M. edited and/or reviewed the paper and coordinated the research work. All the authors have read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Malik, H., Raza, A., Amin, I. et al. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep 6, 38469 (2016). https://doi.org/10.1038/srep38469
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep38469
This article is cited by
-
Host-Delivered RNA Interference for Durable Pest Resistance in Plants: Advanced Methods, Challenges, and Applications
Molecular Biotechnology (2023)
-
Foliar application of clay-delivered RNA interference for whitefly control
Nature Plants (2022)
-
Silencing of multiple target genes via ingestion of dsRNA and PMRi affects development and survival in Helicoverpa armigera
Scientific Reports (2022)
-
RNA interference-mediated tolerance to whitefly (Bemisia tabaci) in genetically engineered tomato
Plant Cell, Tissue and Organ Culture (PCTOC) (2022)
-
Predicting the insecticide-driven mutations in a crop pest insect: Evidence for multiple polymorphisms of acetylcholinesterase gene with potential relevance for resistance to chemicals
Environmental Science and Pollution Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.