Abstract
The prion protein (PRNP) gene is associated with prion diseases, whereas variants of the PRNP gene may also explain some cases of Alzheimer disease (AD) and frontotemporal dementia (FTD) in Caucasian populations. To determine the prevalence of the PRNP gene in patients with AD and FTD in China, we screened all exons of the PRNP gene in a cohort of 683 cases (606 AD and 77 FTD) in the Chinese Han population and we detected a novel missense mutation p.S17G in a late-onset AD (LOAD) patient. Furthermore, we analyzed the PRNP M/V polymorphism at codon 129, which was previously reported as a risk factor. However, there were no significant differences in genotype and allele frequency either in AD (OR = 0.75[0.378–1.49], P = 0.492), or FTD patients (OR = 2.046[0.265–15.783], P = 0.707). To our knowledge, this is the first study to reveal a correlation between the PRNP gene and Chinese AD and FTD patients in a large cohort. This study reports a novel p.S17G mutation in a clinically diagnosed LOAD patient, suggesting that the PRNP mutation is present in Chinese AD patients, whereas, M129V polymorphism is not a risk factor for AD or FTD in the Chinese Han population.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the most common form of dementia, characterized by progressive episodic memory loss, decline in learning and daily living activities, and finally, comprehensive cognitive domain damage. Over 95% of patients develop the disease after the age of 65 years, named late-onset AD (LOAD), the pathogenesis of which is complex and associated with aging, environmental factors, genetics and other unknown causes1. Approximately 1–5% of AD cases are diagnosed before 65 years old, named early-onset AD (EOAD). Genetics is the main contributor to the pathogenesis of EOAD. Family aggregation is more obvious among EOAD patients. Approximately 10% of EOAD patients have a positive family history, and most inherit in an autosomal dominant manner2. In general, the heritability of AD is estimated to be up to 79% based on twin and family studies3. Three genes involved in amyloidogenic processing have been identified as causative genes of early-onset familial AD (EOFAD): presenilin 1 (PSEN1), presenilin 2 (PSEN2), and amyloid precursor protein (APP) genes4. The ε4-allele of the apolipoprotein E gene is a major genetic risk factor for AD5,6.
Frontotemporal dementia (FTD) is the second most common form of presenile dementia after AD, comprising 10–20% of all forms of dementia7. Clinically, FTD exhibits as three forms: behavioral variant FTD (bvFTD), semantic dementia (SD), and progressive non-fluent aphasia (PNFA)8. Approximately 40% of FTD patients have a positive family history. A total of 8 genes have been identified as causative genes of both familial and sporadic FTD patients, including microtubule-associated protein tau (MAPT)9, progranulin (GRN)10, chromosome 9 open-reading frame 72 (C9orf72)11, chromatin modifying protein 2B (CHMP2B)12, TAR DNA-binding protein (TARDBP)13, valosin-containing protein (VCP)14, fused in sarcoma (FUS)15, and coiled-coil-helix-coiled-coil-helix domain containing 10 (CHCHD10)16.
Rare high-penetrant mutations in causal genes explain only a small fraction of AD and FTD cases, whereas the hereditary factors for most patients, particularly the sporadic form, remain unknown. A hypothesis suggests that a certain portion of patients diagnosed as AD or FTD may have mutations in genes involved in other neurodegenerative brain diseases (NBDs)17,18.
Prion protein (PRNP) is a causal gene of familial Creutzfeldt–Jakob disease (CJD), Gerstmann–Straussler–Scheinker disease (GSS), and fatal familial insomnia (FFI), accounting for 5–10% of inherited forms of prion diseases. Prion diseases share some overlaps with AD and FTD. Clinically, both may exhibit a dementia phenotype during progress of the disease and the symptoms usually progress gradually. They all can be divided into two forms: the sporadic and the familial. The sporadic form is the majority and the familial form is fully penetrant. Pathologically, AD, FTD and CJD are all neurodegenerative diseases, or conformational disorders caused by a common pathogenesis of the excessive accumulation of abnormal, insoluble proteins, including the accumulation of Aβ in AD, tau in FTD and prion protein (PrPc) in CJD. Recent studies on the neurotoxic effect of Aβ revealed that PrPc was required for Aβ oligomer-induced neuronal cell death, indicating that PrPc may be involved in the pathogenesis of AD19. Genetically, mutations of PRNP were found in clinically diagnosed AD and FTD patients, and the homozygosity of codon 129 (M/M alleles) was associated with higher risk for CJD, whereas the heterozygosity of codon 129 (M/V alleles) is a possible risk factor for AD and may modify the age at onset of FTD in some Caucasian populations20,21.
In our previous study, we screened the PSEN1, PSEN2, and APP genes in probands of EOFAD families, and the MAPT, GRN, and C9orf72 in FTD patients of the Chinese Han population. We found that the frequency of AD causative genes was 17.1% for PSEN1, 5.7% for APP and no mutation for PSEN222. Mutations in MAPT, GRN, and C9orf72 accounted for 4.3%, 1.4%, and 2.9%, respectively, of FTD cases in the Chinese population23. Our previous study revealed that the remaining dementia patients were genetically unexplained and comprised a large group. In this study, we screened the PRNP gene in the remaining genetically unexplained AD and FTD patients to elucidate their missing genetics. To our knowledge, this is the first study of the distribution of the PRNP gene in a large cohort of Chinese AD and FTD patients.
Results
The demographic features of 606 AD cases, 77 FTD cases and 523 controls are shown in Table 1. A total of 73 AD patients (12.04%) and 11 FTD patients (13.58%) had a positive family history. No statistically significant differences in sex distribution or age at onset were found between cases and controls. The polymorphic codon 129 of the PRNP gene was in Hardy-Weinberg equilibrium in the healthy control (P = 0.76), AD (P = 0.66) and FTD (P = 0.95) population.
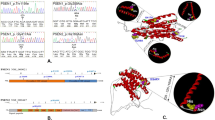
A novel rare variant p.S17G in the PRNP gene was identified in a sporadic LOAD patient
The PRNP p.S17G variant was identified in one of 606 AD cases and in none of 534 controls and public databases, including 1000 G, dbSNP, and ExAC (Fig. 1). The carrier had the M129M genotype and the ApoE genotype was ε2/3. The patient was a 70-year-old female. She first developed episodic memory loss 5 years ago, particularly short-term memory. She easily forgot tasks and words she had spoken. Over the next 2 years, her symptoms worsened with a gradual loss of calculation abilities, orientation in time and space, and perceptivity. She had difficulty taking care of herself as well as dealing with daily tasks. She also presented with personality changes characterized by irritability, depression and apathy, and barely talked to others. Neurological examination revealed no myoclonic jerks, seizures, extrapyramidal or upper motor neuron signs apart from the memory impairment and cognitive deficits. Her Mini-Mental State Examination (MMSE) and Montreal Cognitive Assessment (MoCA) scores were 2/30 and 0/30, respectively, the Activity of Daily Living Scale (ADL) score was 54, and the Clinical Dementia Rating (CDR) score was 3. The MRI revealed diffuse cortical atrophy, enlargement of the cerebral ventricle and cistern of the whole brain, especially in the frontotemporal lobe, and hippocampus (Fig. 2). She was diagnosed with probable AD according to the NINCDS-ADRDA criteria. No family members suffered from dementia, and the available DNA sequencing of her family members revealed no PRNP p.S17G mutation.
MRI scan of the patient and an age-matched control.
(A1–A3) The cross sectional, coronal, and sagittal MRI of the patient revealed diffuse atrophy of the frontal lobe, temporal lobe and parietal lobe, especially the temporal lobe and the hippocampal. (B1–B3) The cross sectional, coronal, and sagittal MRI of the age-matched control appeared much normal.
The M129V genotype may not be a risk factor for AD and FTD in the Chinese Han population
To assess the correlation between the PRNP M/V polymorphism at codon 129 and the susceptibility to AD in mainland China, we examined the genotype and allele frequencies of this polymorphism in 606 Chinese Han AD patients and in 534 healthy controls. The genotype distribution of codon 129 polymorphisms in the AD group was 96.5% MM and 3.5% MV, 98.7% MM and 1.3% MV in the FTD, and 97.4% MM and 2.6% VV among controls. No significant difference between AD patients (including EOAD and LOAD) and the controls were found for genotype or allele frequency of the PRNP 129 polymorphism (OR = 0.714[0.285–1.792], P = 0.316) (Table 2). We also investigated the genotype and allele frequencies of PRNP 129 in 77 Chinese FTD patients to determine whether this polymorphism correlated with FTD. There were no significant differences in the genotype and allele frequencies between FTD patients and controls (OR = 1.949[0.231–16.444], P = 0.461) (Table 2). This result suggests that the PRNP M/V polymorphism at codon 129 may not increase the susceptibility to AD or FTD.
Discussion
Located on chromosome 20p13, the PRNP gene encodes the prion protein containing 253 amino acids. Thus far, more than 50 mutations in PRNP have been reported to cause autosomal dominant inherited CJD, and a large number of polymorphisms in PRNP (including codon 129, 219, 178, 200, and 232, etc.) have been identified as susceptible factors in sporadic CJD (sCJD) cases. For example, the M/M genotype of codon 129 is associated with a higher portion of sCJD, early-onset and a shorter incubation time of kuru. Due to the overlap of clinical phenotypes and pathological characteristics between CJD and other neurodegenerative disorders, a series of studies supported the hypothesis that mutations and polymorphisms of PRNP also accounted for other neurodegenerative diseases, including AD, FTD, Parkinson’s disease (PD) and primary progressive aphasia24,25,26. For example, the nonsense mutation p.Q160* and p.Y145* in PRNP were reported in clinically diagnosed AD cases27,28. A missense mutation p.P39L was identified as causative in an FTD family29, and the M/V polymorphism at codon 129 was demonstrated to confer a higher risk to AD in several European studies, but yielded controversial results in populations in Japan and Korea30,31. Given the clinical heterogeneity of PRNP, we propose that the PRNP gene may also account for Chinese AD and FTD patients. In this study, we screened 606 AD patients, 77 FTD patients and 534 age-matched controls to determine whether the PRNP gene is causative in the Chinese AD and FTD population. We also examined the correlation between the PRNP M/V polymorphism at codon 129 and the susceptibility of AD and FTD in China.
As a result, we identified a novel missense mutation p.S17G of the PRNP gene in a LOAD patient. To our knowledge, this is the first reported in the Chinese Han AD population. The mutation was absent in other family members of the carrier and 534 cognitively normal, age-matched controls, suggesting that the p.S17G maybe a de novo, likely pathogenic mutation in this patient. The carrier presented this variation in a heterozygosis status. She started episodic memory loss at the age of 65, and gradually exhibited depression, calculation, orientation and perceptivity disabilities. Neuropsychological assessments were consistent with her clinical manifestations. Cranial MRI screening revealed diffuse cortical atrophy, most seriously in the frontotemporal lobe and hippocampus. Moreover, the clinical course in this patient was not rapidly progressive as the duration of the disease was over 5 years. Unfortunately, we didn’t receive permission from her relative to conduct a brain biopsy; however, based on the clinical course and MRI findings, the patient seems more likely to have AD than CJD. Previous reports of PRNP mutations in AD and other dementia phenotype are summarized in Table 3. Thus far, a total of 8 point mutations of the PRNP gene have been reported to be associated with the clinical diagnosis of AD, FTD, DLB, and AD/FTD phenotype of GSS and CJD. The reason for the phenotypic heterogeneity of PRNP mutant remains unclear; therefore, we propose that AD, FTD and other neurodegenerative dementia maybe a low penetrance phenotype of PRNP mutation. The clinically diagnosed AD patients with a PRNP mutation appear to share some common features, such as initial symptom of short-memory loss, followed by depression, spatial and temporal orientation, absence of myoclonus and extrapyramidal signs, prolonged clinical courses and a negative family history in a few patients. The phenotype of our patient is consistent with previous reports. The Ser-to-Gly substitution is located in the signal peptide near the N-terminal of the human prion protein and little is known about the exact function of the (1–22) peptide. Studies on the N-terminal (1–28) part of the mouse prion protein revealed that it is a cell penetrating peptide, capable of transporting large hydrophilic cargoes through a cell membrane with a strong tendency for aggregation and beta-structure formation32. It will thus be of considerable interest to determine whether this processing peptide influences the transformation and aggregation of the prion protein.
We further examined the M/V polymorphism at codon 129 in our cohort, and failed to find an association in the AD or FTD cohort, even when the AD cohort was divided into EOAD and LOAD subgroups. The frequency of the genotypes of our cohort was MM 96.5% and MV 3.5% in AD, MM 98.7% and MV 1.3% in FTD and 97.4% MM and 2.6% VV among controls. The VV genotype was absent in our cohort. Our results are consistent with the previous reports in Asian and part of Caucasian populations30. It is possible that the PRNP 129 polymorphism does not affect the risk of AD or FTD in Asians because the V allele is thoroughly rare in both cases and healthy controls. Another possibility is that a false negative result was obtained due to the limited sample size, particularly the FTD cases. However, our study is fundamental for identifying the mutations and polymorphisms of the PRNP gene in China. These data will help advance our understanding of PRNP gene in the Chinese and East Asian populations.
In conclusion, we found a novel p.S17G mutation in a clinically diagnosed LOAD patient, suggesting that the PRNP mutation is present in Chinese AD patients. Further functional studies on the PRNP p.S17G and replication in other cohorts are necessary to confirm the pathogenicity of this PRNP mutation. Genetic screening of the PRNP gene is warranted in neurodegenerative dementia, particularly in suspected AD cases. The V allele of codon 129 is rare in our cohort and the M/V polymorphism at codon 129 was not significantly associated with the incidence of AD and FTD in China.
Method
Subjects
This study included 606 AD cases (including 533 sporadic AD and 73 probands from FAD families), 77 FTD patients (including 66 sporadic FTD and 11 probands from FTD families), and 523 sex, age-matched normal controls. All subjects were enrolled from the outpatient neurology clinics of Xiangya Hospital and Jiangsu Geriatric Hospital. The AD cohort met the NINCDS-ADRDA criteria for probable AD, and the FTD cohort met the consensus criteria for bvFTD, SD or PNFA. All patients underwent standard neuropsychological assessments including MMSE, MoCA, CDR, ADL, verbal fluency test, voice fluency test, and Alzheimer’s Disease Assessment Scale-cognitive subscale (ADAS-Cog). The study was approved by the Ethics Committee of Xiangya Hospital, Central South University in China (equivalent to an Institutional Review Board) and performed in accordance with the approved guidelines and regulations. Written informed consent was obtained from each subject. For all patients, careful clinical, neurological examination and blood tests for vitamin status, thyroid function, HIV and Treponema pallidum infection were conducted to avoid the possibility of reversible dementia.
DNA isolation and genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a QIAGEN kit following the supplier’s instructions. All DNA samples were normalized to 50–100 ng/μl. Polymerase chain reaction (PCR) was performed on the exonic regions of PRNP (NM_001080123), as well as their corresponding flanking intronic sequences. All PCR products were sequenced using identical forward and reverse primers with BigDye terminator v3.1 sequencing chemistry on an ABI 3730xl DNA analyzer (Applied Biosystems). DNA sequences were analyzed using the Sequencher software. All related primer information and protocols are presented in the Supplementary materials.
Patients with mutations of APP, PSEN1, PSEN2, MAPT, GRN and C9orf72 were excluded from the study. The prevalence of all novel mutations and polymorphisms at codons were further tested in 534 healthy individuals of matched geographical ancestry and age to provide further evidence of pathogenicity. The control group comprised volunteers from communities and the health examination center. All control individuals were older than 65 years and had scores indicative of no cognitive impairment (>26/30) on the MMSE.
Statistics analysis
According to the polymorphism at codon 129, patients and controls were classified into three groups as follows: MM, MV and VV genotype. Differences in the distribution of polymorphisms at codon 129 between the patients and controls were tested using Pearson χ2 test or Fisher’s exact test when the sample size was small, and significance was set at P = 0.05. Descriptive statistics were expressed as the mean ± the standard deviation. The associations between polymorphisms at codon 129 and disease risk were determined in logistic regression models adjusted for the age at onset and gender. Statistical analysis was performed using SPSS Statistics software (version 21.0).
Additional Information
How to cite this article: Zhang, W. et al. Mutational analysis of PRNP in Alzheimer’s disease and frontotemporal dementia in China. Sci. Rep. 6, 38435; doi: 10.1038/srep38435 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Borenstein, A. R., Copenhaver, C. I. & Mortimer, J. A. Early-life risk factors for Alzheimer disease. Alzheimer Dis Assoc Disord 20, 63–72, doi: 10.1097/01.wad.0000201854.62116.d7 (2006).
Campion, D. et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 65, 664–670, doi: 10.1086/302553 (1999).
Gatz, M. et al. Role of genes and environments for explaining Alzheimer disease. Archives of general psychiatry 63, 168–174, doi: 10.1001/archpsyc.63.2.168 (2006).
Bertram, L., Lill, C. M. & Tanzi, R. E. The genetics of Alzheimer disease: back to the future. Neuron 68, 270–281, doi: 10.1016/j.neuron.2010.10.013 (2010).
Corder, E. H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993).
Bekris, L. M., Yu, C. E., Bird, T. D. & Tsuang, D. W. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol 23, 213–227, doi: 10.1177/0891988710383571 (2010).
Neary, D. et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 51, 1546–1554 (1998).
Rohrer, J. D. et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology 73, 1451–1456, doi: 10.1212/WNL.0b013e3181bf997a (2009).
Hutton, M. et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705, doi: 10.1038/31508 (1998).
Baker, M. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442, 916–919, doi: 10.1038/nature05016 (2006).
Daoud, H. et al. C9orf72 hexanucleotide repeat expansions as the causative mutation for chromosome 9p21-linked amyotrophic lateral sclerosis and frontotemporal dementia. Archives of neurology 69, 1159–1163, doi: 10.1001/archneurol.2012.377 (2012).
Isaacs, A. M., Johannsen, P., Holm, I., Nielsen, J. E. & consortium, F. R. Frontotemporal dementia caused by CHMP2B mutations. Current Alzheimer research 8, 246–251 (2011).
Synofzik, M. et al. Targeted high-throughput sequencing identifies a TARDBP mutation as a cause of early-onset FTD without motor neuron disease. Neurobiology of aging 35, 1212 e1211–1215, doi: 10.1016/j.neurobiolaging.2013.10.092 (2014).
Watts, G. D. et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nature genetics 36, 377–381, doi: 10.1038/ng1332 (2004).
Van Langenhove, T. et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology 74, 366–371, doi: 10.1212/WNL.0b013e3181ccc732 (2010).
Bannwarth, S. et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain: a journal of neurology 137, 2329–2345, doi: 10.1093/brain/awu138 (2014).
Jarmolowicz, A. I., Chen, H. Y. & Panegyres, P. K. The patterns of inheritance in early-onset dementia: Alzheimer’s disease and frontotemporal dementia. American journal of Alzheimer’s disease and other dementias 30, 299–306, doi: 10.1177/1533317514545825 (2015).
Cacace, R., Sleegers, K. & Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 12, 733–748, doi: 10.1016/j.jalz.2016.01.012 (2016).
Kudo, W. et al. Cellular prion protein is essential for oligomeric amyloid-beta-induced neuronal cell death. Human molecular genetics 21, 1138–1144, doi: 10.1093/hmg/ddr542 (2012).
Dermaut, B. et al. PRNP Val129 homozygosity increases risk for early-onset Alzheimer’s disease. Annals of neurology 53, 409–412, doi: 10.1002/ana.10507 (2003).
Moreno, F. et al. Prion protein codon 129 polymorphism modifies age at onset of frontotemporal dementia with the C.709-1G > A progranulin mutation. Alzheimer disease and associated disorders 25, 93–95, doi: 10.1097/WAD.0b013e3181eff695 (2011).
Jiao, B. et al. Mutational analysis in early-onset familial Alzheimer’s disease in Mainland China. Neurobiology of aging 35, 1957 e1951-1956, doi: 10.1016/j.neurobiolaging.2014.02.014 (2014).
Tang, M. et al. Analyses MAPT, GRN, and C9orf72 mutations in Chinese patients with frontotemporal dementia. Neurobiology of aging, doi: 10.1016/j.neurobiolaging.2016.05.013 (2016).
Li, X. et al. Prion protein codon 129 genotype prevalence is altered in primary progressive aphasia. Annals of neurology 58, 858–864, doi: 10.1002/ana.20646 (2005).
Poleggi, A. et al. Codon 129 polymorphism of prion protein gene in sporadic Alzheimer’s disease. European journal of neurology 15, 173–178, doi: 10.1111/j.1468-1331.2007.02021.x (2008).
Gossrau, G. et al. Analysis of the polymorphic prion protein gene codon 129 in idiopathic Parkinson’s disease. Journal of neural transmission 113, 331–337, doi: 10.1007/s00702-005-0329-x (2006).
Guerreiro, R. et al. Nonsense mutation in PRNP associated with clinical Alzheimer’s disease. Neurobiology of aging 35, 2656 e2613-2656, doi: 10.1016/j.neurobiolaging.2014.05.013 (2014).
Ghetti, B. et al. Vascular variant of prion protein cerebral amyloidosis with tau-positive neurofibrillary tangles: the phenotype of the stop codon 145 mutation in PRNP. Proceedings of the National Academy of Sciences of the United States of America 93, 744–748 (1996).
Oldoni, E. et al. PRNP P39L Variant is a Rare Cause of Frontotemporal Dementia in Italian Population. Journal of Alzheimer’s disease: JAD 50, 353–357, doi: 10.3233/JAD-150863 (2015).
Ohkubo, T. et al. Absence of association between codon 129/219 polymorphisms of the prion protein gene and Alzheimer’s disease in Japan. Annals of neurology 54, 553–554, author reply 555, doi: 10.1002/ana.10748 (2003).
Jeong, B. H. et al. Polymorphisms at codons 129 and 219 of the prion protein gene (PRNP) are not associated with sporadic Alzheimer’s disease in the Korean population. European journal of neurology 14, 621–626, doi: 10.1111/j.1468-1331.2007.01786.x (2007).
Lundberg, P. et al. Cell membrane translocation of the N-terminal (1–28) part of the prion protein. Biochemical and biophysical research communications 299, 85–90 (2002).
Munoz-Nieto, M. et al. A novel mutation I215V in the PRNP gene associated with Creutzfeldt-Jakob and Alzheimer’s diseases in three patients with divergent clinical phenotypes. Journal of neurology 260, 77–84, doi: 10.1007/s00415-012-6588-1 (2013).
Jayadev, S. et al. Familial prion disease with Alzheimer disease-like tau pathology and clinical phenotype. Annals of neurology 69, 712–720, doi: 10.1002/ana.22264 (2011).
Guerreiro, R. J., Vaskov, T., Crews, C., Singleton, A. & Hardy, J. A case of dementia with PRNP D178Ncis-129M and no insomnia. Alzheimer disease and associated disorders 23, 415–417, doi: 10.1097/WAD.0b013e3181ae3a76 (2009).
Bernardi, L. et al. Novel N-terminal domain mutation in prion protein detected in 2 patients diagnosed with frontotemporal lobar degeneration syndrome. Neurobiology of aging 35, 2657 e2657-2611, doi: 10.1016/j.neurobiolaging.2014.06.006 (2014).
Koide, T. et al. A patient with dementia with Lewy bodies and codon 232 mutation of PRNP. Neurology 59, 1619–1621 (2002).
Giovagnoli, A. R. et al. Atypical frontotemporal dementia as a new clinical phenotype of Gerstmann-Straussler-Scheinker disease with the PrP-P102L mutation. Description of a previously unreported Italian family. Neurological sciences: official journal of the Italian Neurological Society and of the Italian Society of Clinical Neurophysiology 29, 405–410, doi: 10.1007/s10072-008-1025-z (2008).
Heinemann, U. et al. Novel PRNP mutation in a patient with a slow progressive dementia syndrome. Medical science monitor: international medical journal of experimental and clinical research 14, CS41-43 (2008).
Acknowledgements
The authors are grateful to patients and families for the interest and generous participation in our research effort.
Author information
Authors and Affiliations
Contributions
B.S.T., and L.S. designed the study. W.W.Z., B.J., T.T.X., C.Z.P., X.X.L. conducted the experiments, analyzed and interpreted the data. W.W.Z. wrote the manuscript. L.Z., B.S.T., and L.S. supervised the study.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, W., Jiao, B., Xiao, T. et al. Mutational analysis of PRNP in Alzheimer’s disease and frontotemporal dementia in China. Sci Rep 6, 38435 (2016). https://doi.org/10.1038/srep38435
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep38435
This article is cited by
-
Mapping the genetic landscape of early-onset Alzheimer’s disease in a cohort of 36 families
Alzheimer's Research & Therapy (2022)
-
Ensemble disease gene prediction by clinical sample-based networks
BMC Bioinformatics (2020)
-
The characterization of AD/PART co-pathology in CJD suggests independent pathogenic mechanisms and no cross-seeding between misfolded Aβ and prion proteins
Acta Neuropathologica Communications (2019)
-
Clinical and neuropathological phenotype associated with the novel V189I mutation in the prion protein gene
Acta Neuropathologica Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.