Abstract
Calmodulin is a highly versatile protein that regulates intracellular calcium homeostasis and is involved in a variety of cellular functions including cardiac excitability, synaptic plasticity and signaling transduction. During osteoclastic bone resorption, calmodulin has been reported to concentrate at the ruffled border membrane of osteoclasts where it is thought to modulate bone resorption activity in response to calcium. Here we report an interaction between calmodulin and Rab3D, a small exocytic GTPase and established regulator osteoclastic bone resorption. Using yeast two-hybrid screening together with a series of protein-protein interaction studies, we show that calmodulin interacts with Rab3D in a calcium dependent manner. Consistently, expression of a calcium insensitive form of calmodulin (i.e. CaM1234) perturbs calmodulin-Rab3D interaction as monitored by bioluminescence resonance energy transfer (BRET) assays. In osteoclasts, calmodulin and Rab3D are constitutively co-expressed during RANKL-induced osteoclast differentiation, co-occupy plasma membrane fractions by differential gradient sedimentation assay and colocalise in the ruffled border as revealed by confocal microscopy. Further, functional blockade of calmodulin-Rab3D interaction by calmidazolium chloride coincides with an attenuation of osteoclastic bone resorption. Our data imply that calmodulin- Rab3D interaction is required for efficient bone resorption by osteoclasts in vitro.
Similar content being viewed by others
Introduction
Calmodulin is a versatile protein that regulates Ca(2+) homeostasis1, synaptic plasticity2, and cardiac excitability3. It has been implicated in osteoclast differentiation, function, and survival. Calmodulin regulates Ca2+/calmodulin-dependent kinase II (CaMKII) and Ca2+/calmodulin-dependent protein phosphatase, calcineurin4; both are critical for osteoclast differentiation5,6,7. It also mediates osteoclast survival through a mechanism involving the binding of calmodulin with the death receptor Fas8,9. In addition, calmodulin modulates acid transport and bone resorption10,11,12. However, the underlining molecular events by which calmodulin regulates bone resorption remain to be elucidated.
Bone resorption is a highly regulated process that requires intense vesicular trafficking to sustain the structural and functional polarity of osteoclasts and enable efficient delivery of osteolytic cargo contents (i.e. Cathepsin K) and membrane exchange between the ruffled border and the opposing basolateral surface. Vesicular trafficking is modulated by sets of genetically conserved proteins among which members of the small Rab GTPase family are now firmly established regulators13. To date, several Rab proteins have been implicated in bone resorption14, although only Rab715 and Rab3D16 and been functionally characterized in osteoclasts. Rab7 is involved in the late endocytic pathway in the osteoclast polarization and bone resorption15, whereas Rab3D modulates a post-TGN trafficking step that is required for maintenance of the osteoclastic ruffled border membrane16 and utilizes the dynein motor complex and microtubules via its direct interaction with Tctex-1 to facilitate vesicle delivery and/or retrieval during bone resorption17.
Here, we identify calmodulin as a specific Rab3D interacting molecule by yeast two hybrid screening. We show that inhibition of calmodulin calcium binding perturbs this association in vivo by bioluminescence resonance energy transfer (BRET). Disruption of calmodulin-Rab3D interaction attenuated osteoclastic bone resorption in vitro. We propose that calmodulin, via modulating calcium, imparts an additional layer of regulation on Rab3D trafficking during osteoclastic bone resorption.
Results
Calmodulin interacts with Rab3D
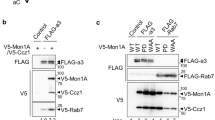
We have previously established a yeast two-hybrid approach to successfully uncover novel Rab3D interacting partners such as Tctex-117. Here we identify calmodulin as an additional binding partner of Rab3D. The interaction of calmodulin with Rab3D was verified by a yeast two hybrid assay, using a histidine-deficient plate (Fig. 1A). To further examine the interaction of calmodulin and Rab3D, we generated Rluc-camodulin and EYFP-Rab3D fusion protein constructs and performed BRET protein-protein interaction assays. As shown in Fig. 1B, co-expression of Rluc-calmodulin and EYFP-Rab3D resulted in a significant BRET signal when compared to the co-expression of Rluc and EYFP. To further confirm the interaction, an in vitro calmodulin sepharose-pull down assay was performed. Rab3D was cloned into a mammalian expression vector with an N terminal Flag-tagged (Fig. 1C). Flag-Rab3D proteins were prepared from COS cells transfected with pcDNA3.1-Flag-Rab3D expressing plasmids. COS cell lysates were harvested and subjected to immobilized calmodulin sepharose in the presence or absence of 2 mM calcium. As shown in Fig. 1C, Flag-Rab3D proteins bound immobilized calmodulin saphorose in the presence (but not in the absence) of calcium, indicative of a calcium dependent binding dependency.
Calmodulin interacts with Rab3D.
(A) A yeast two hybrid assay showing that Calmodulin interacts with Rab3D, by using histidine-deficient plate. (B) BRET assays showing that co-transfection of Rluc-Camodulin and EYFP-Rab3D fusion protein constructs resulted in a significant BRET signal. Co-expression of Rluc and EYFP is shown as a negative control. (C) Flag-Rab3D proteins expressed in COS cells interact with calmodulin saphorose in the presence of 2 mM calcium. *Indicates p Value < 0.001 when compared with EYFP and Rluc. (D) Calmodulin calcium-insensitive mutant perturbs its interaction with Rab3D. Generation of a Rluc-calmodulin construct in which four aspartic acid residues at position 23, 59, 96, 132 were substituted with alanine, mimicking a calcium insensitive form of calmodulin. (E) BRET assays showing that the calcium insensitive form of camodulin failed to interact with Rab3D. 1:1, 1:2 and 1:3 indicate that transfected plasmid ratio of EYFP-Rab3D/ Rluc-camodulin or EYFP-Rab3D/ Rluc-calmodulin mutant 1234. Symbol *indicates p Value < 0.001 when compared with EYFP and Rluc-camodulin control. Symbol # indicates p Value < 0.001 when compared Rluc-camodulin with Rluc-calmodulin mutant 1234.
Calmodulin calcium insensitive mutant perturbs its interaction with Rab3D
Considering that calmodulin has four calcium binding sites via four aspartic acid residues18 and acts as a calcium modulator in the calcium sensitive regulation of many cellular processes, we next examined if calcium binding site of calmodulin is required for the interaction of calmodulin with Rab3D. For this, we generated a Rluc-calmodulin construct in which four aspartic acid residues at position 23, 59, 96, 132 were substituted with alanine, mimicking a calcium insensitive form of calmodulin18 (Fig. 1D). BRET assay results showed that the calcium insensitive form of camodulin attenuated the interaction with Rab3D (Fig. 1E).
The preferential interaction between Calmodulin and Rab3D in its GTP-bound conformation
Rab GTPases embed in organelle membranes via C-terminal prenylation moties where they function as molecular switches that oscillate between GTP “active” and GDP “inactive” conformations. In their active state, Rabs recruit GTP-dependent effector proteins through which they elicit their biological function at various stages of vesicular transport. Therefore, we next asked whether the interaction between Calmodulin and Rab3D was dependent on the nucleotide and/or prenylation status of Rab3D. To access this, we employed several well characterised Rab3D variants16, which selectively disrupt the GDP/GTP exchange i.e. GTP-bound Rab3D (Rab3DQ81L), nucleotide empty RAB3D (Rab3DN135I) and prenylation motif deletion of Rab3D (Rab3D CXC) compared to wildtype Rab3D (Fig. 2A,B). These constructs were successfully expressed as EYFP fusion proteins in transfected COS cells as confirmed by western blot analyses (Fig. 2C). As with other bona fide Rab effector protein, calmodulin exhibited a preferential association with Rab3D when locked in is GTP-bound form (Rab3DQ81L) when compared to wild-type Rab3D, nucleotide-empty (Rab3DN135I) and prenylation motif deletion of Rab3D (Rab3D CXC) in BRET assays (Fig. 2D). These data imply that the interaction of calmodulin with Rab3D is largely influenced by its active GTP-bound state.
The interaction of Calmodulin with Rab3D has a closer proximity when Rab3D is GTP-bound.
(A) Predicted molecular structures of wild-type Rab3D, GTP-bound Rab3D (Rab3DQ81L), nucleotide empty RAB3D (Rab3DN135I) and prenylation motif deletion of Rab3D (Rab3DΔCXC). (B) EYFP fusion protein constructs of EYFP-Rab3D, EYFP-Rab3DQ81L, EYFP-Rab3DN135I and EYFP-Rab3DΔCXC that were used for BRET assays. (C) Western blot analysis showing the expression of EYFP-Rab3D, EYFP-Rab3DQ81L, EYFP-Rab3DN135I and EYFP-Rab3DΔCXC proteins by anti-GFP. (D) BRET assays showing that calmodulin exhibited an enhanced association with a GTP-bound Rab3D (Rab3DQ81L) when compared to wild-type Rab3D, nucleotide empty RAB3D (Rab3DN135I) and prenylation motif deletion of Rab3D (Rab3DΔCXC) in BRET assays. *Indicates p Value < 0.001 when compared with EYFP and Rluc. # indicates p Value < 0.05 when compared to wild-type Rab3D, nucleotide-empty (Rab3DN135I) and prenylation motif deletion of Rab3D (Rab3DΔCXC).
Calmodulin and Rab3D are co-expressed during osteoclast formation and co-sediment in membrane fractions of osteoclasts
To begin to probe the potential relevance of the calmodulin-Rab3D association in osteoclasts, we first compared the gene expression profile of calmodulin and Rab3D in osteoclasts and their precursor cells. Bone marrow macrophages (BMM) were cultured in the present of macrophage colony stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL) for a period of 0, 1, 3, 5 days and then fixed and stained for tartrate resistant acid phosphatase (TRACP) activity, showing the presence of multinucleated TRACP positive osteoclast like cells (Fig. 3A). In a parallel experiment, semi-quantitative RT-PCR was performed. Calmodulin gene expression appears to be constitutive during osteoclastogenesis by RANKL at an expression kinetic similar to that of Rab3D (Fig. 3B). Osteoclast marker gene expression of TRACP, Cathepsin K, V-ATPase d2, and calcitonin receptor (CTR) were induced by RANKL as compared to 36B4 gene expression as an internal control (Fig. 3B). Further, Western blot analysis showed that Calmodulin protein is constitutively expressed during osteoclastogenesis similar to Rab3D protein expression (Fig. 3C).
Calmodulin and Rab3D are co-expressed during osteoclast formation and co-fractionated in the membrane fraction of osteoclasts.
(A) Representative images from BMMs cultured in the present of M-CSF and RANKL for a period of 0, 1, 3, 5 days and then fixed and stained for TRACP activity. (B) Semi-quantitative RT-PCR showing the co-expression of calmodulin and Rab3D along with osteoclast marker gene expression of TRACP, NFATc1, DC-STAMP, V-ATPase d2, and calcitonin receptor. (C) Western blot analysis showing that calmodulin and Rab3D proteins are expressed at similar kinetics during osteoclastogenesis. (D) Sucrose gradient sedimentation assays showing that Rab3D and calmodulin co-fractionate in the large membrane faction (P = Pellet). Note that all Western blot results with cropped blots are displayed from well established sizes of proteins; calmodulin, Rab3D and VATPase (d2).
Next, sucrose gradient sedimentation assays were performed to examine calmodulin and Rab3D co-fractionation. Rab3D is present in small vesicles (F9-10) and large plasma membrane fractions (P) (Fig. 3D). Interestingly, calmodulin co-fractionated with Rab3D but only in the large membrane faction (P) (Fig. 3D). By comparison, V-ATPase (d2) was also present in both vesicle and membrane fractions (Fig. 3D). Moreover, an association between Rab3D and calmodulin was further confirmed in bone-resorbing osteoclasts by immunofluorescence confocal microscopy (Fig. 4). In this instance colocalisation (yellow colour) was observed upon overlay of individual fluorescent channels for Rab3D (green) and Calmodulin (red), which were detected using antibodies specific to Rab3D and calmodulin, as validated in the Western blot analysis above (i.e. Fig. 3C). Interestingly, a subset of Rab3D-calmodulin colocalisation was observed within the F-actin ring/sealing zone (blue) that typically denotes the ruffled border membrane (Fig. 4A, region circumscribed by red line). This colocalisation was confirmed by correlative linescan analysis (Fig. 4B) which revealed overlap between the fluorescent peaks of Rab3D and calmodulin within the ruffled border region (Fig. 4A, white dashed line).
Colocalisation between Rab3D and calmodulin in bone-resorbing BMM-derived osteoclasts.
(A) Representative confocal microscopy images of individual and overlay fluorescent channels of Rab3D (green), calmodulin (red), F-actin (blue) and nuclei (magenta). Colocalisation between Rab3D and calmodulin appears as yellow in overlay. Red line demarcates the ruffled border region within the sealing zone. White line corresponds to the correlative linescan analysis in (B). Bar = 10 μm.
Functional blockade of the calmodulin-Rab3D interaction by calmidazolium chloride attenuates osteoclastic bone resorption
Finally, we set out to define the impact of the interaction of calmodulin with Rab3D on osteoclast function. To this end, we first tested the effect of calmidazolium chloride on the interaction of calmodulin and Rab3D using BRET assays. Interestingly, calmidazolium chloride perturbs the association of calmodulin and Rab3D, Rab3DQ81L, Rab3DN135I and Rab3DΔCXC (Fig. 5A). Further, to examine the effect of calmidazolium on osteoclastic bone resorption, BMM derived osteoclasts were seeded into bone slices in the presence and absence of calmidazolium chloride for 24 hours. Treatment of osteoclasts with calmidazolium chloride inhibited osteoclastic bone resorption (Fig. 5B) but did not affect the total number (Fig. 5C) and morphology of TRACP positive osteoclastic like cells at 1 μM, and 5 μM (Fig. 5D). Taken together, these data suggest that treatment of calmidazolium chloride perturbs the interaction of calmodulin with Rab3D, an effect that coincides with the attenuation of osteoclastic bone resorption in vitro.
Functional blockade of the interaction of Rab3D and calmodulin by calmidazolium chloride attenuates osteoclastic bone resorption.
(A) BRET assays showing the effect of calmidazolium chloride on the interaction of calmodulin and Rab3D, Rab3DQ81L, Rab3DN135I and Rab3DΔCXC; respectively. (B) Treatment of osteoclasts with calmidazolium chloride inhibits osteoclastic bone resorption with quantitative analysis of bone resorption areas. BMM derived osteoclasts were seeded into bone slices in the presence and absence of calmidazolium chloride for 24 hours. (C) Total number of TRACP positive osteoclastic like cells. (D) Representative images of bone resorption assays showing the effect of calmidazolium on TRACP positive osteoclast morphology (upper panel), and osteoclastic bone resorption SEM images (lower panel). Scale bars are shown. * and #Indicate p Value < 0.001, and < 0.05 respectively when compared to untreated control.
Discussion
Calmodulin has versatile roles in regulating intracellular calcium homeostasis and diverse cellular processes including osteoclastic bone resorption. In this study we document that calmodulin interacts with Rab3D in a calcium-dependent manner. Both calmodulin and Rab3D proteins co-occupy membranes factions by a sucrose gradient ultracentrifugation sedimentation assay and colocalise in the ruffled border by confocal microscopy. Functional blockade of calmodulin and Rab3D interaction by calmidazolium chloride resulted in an attenuation of osteoclastic bone resorption. Considering that calmodulin is concentrated on the ruffled border membrane in osteoclasts8, and that Rab3D is a functional requirement for ruffled border maintenance17, it is plausible that calmodulin, through its interaction with Rab3D facilitates the delivery and/or calcium sensitively of Rab3D-bearing vesicles at the ruffled border membrane during bone resorption (Fig. 6).
A working model illustrating that calmodulin is concentrated on the ruffled border membrane in osteoclasts and mediates bone resorption process8.
Rab3D interaction with calmodulin is implicated for a role in bone resorption when Rab3D-bearing vesicles reach the resorbing ruffled border compartment of an osteoclast.
Bone resorption by osteoclasts is a multi-step process which culminates in the removal of an inorganic mineral layer primarily composed of cystralline hydroxylapatite and subsequent degradation of the underlying organic phases. This process involves continual content delivery and membrane recycling through vesicle trafficking, governed largely small Rab GTPases. Rab proteins have been implicated in the regulation of distinct events in vesicle transport on the exocytotic, endocytotic and transcytotic pathways19,20. Because of this, in recent years, there has been mounting interest surrounding the role of Rab3D in intracellular transport. Rab3D was shown to mediate exocytosis in mast cells21, adipocytes22, and acini cells23. GTPase- deficient Rab3D decreased nutrient induced insulin release24. Moreover, actin coating of secretory granules during regulated exocytosis has been shown to correlate with the release of Rab3D25. In addition, Rab3D has been suggested to play a role in the regulation of apically directed transcytosis in rat hepatocytes26 and appears to be essential for apical transport in polarized epithelia27. Our previous data indicates that Rab3D modulates a post-TGN trafficking step that is required for osteoclastic bone resorption16. Furthermore, we have shown that Rab3D interacts with Tctex-1 which regulates microtubule-directed trafficking of Rab3D vesicles via cytoplasmic dynein17. The present study further extends the role of Rab3D in bone resorption by binding to calmodulin; a molecule previously implicated in acid secretion and bone resorption10,11,12.
During the formation of ruffled border membrane domains, the osteoclastic plasmalemma can be further divided into several specialised functional domains. These include the basolateral domain and a functional secretory domain28,29. In fact more recent evidence suggests that the ruffled border is not a continuous domain but rather segregated into an “uptake” and “secretory” domain reflecting its dynamic endo-exocytic intracellular trafficking routes30. Therefore it is likely that Rab3D mediates bone resorption along the exocytic pathway through its direct interaction with calmodulin that is located in the ruffled border membranes. In other systems, calmodulin has been shown to stimulate GTP binding to Rab3A that is complexed with GDI, which leads to the formation of an active GTP-bound form of the Rab3A-Ca (2+)/Calmodulin complex in synaptic membranes of activated nerve termini31. Similarly, interaction between Rab3B and calmodulin was Ca (2+)-dependent32. These findings provide evidence that Rab3B is primarily localized with the particulate fraction and that Ca (2+)/calmodulin could regulate function of this GTPase in platelets32. It remains to be elucidated whether other Rab proteins might also interact with calmodulin and facilitate these molecular pathways in osteoclasts and other cells.
Previously, structural analysis revealed four Ca (2+)-binding domains in calmodulin that are important for the function of calmodulin, together with hydrophobic regions represent the sites of interaction with pharmacological agents33. The three-dimensional structure of calmodulin has been determined crystallographically at 3.0 A resolution. The molecule consists of two globular lobes connected by a long exposed alpha-helix, and each lobe is able to bind two calcium ions through helix-loop-helix domains34. The flexibility of the protein may explain the fact that calmodulin is able to bind many different targets35. We have found that the interaction of calmodulin with Rab3D is calcium dependent. During osteoclastic bone resorption, free calcium is to be released in the resorption compartment, which could in turn further facilitate the interaction of calmodulin with Rab3D. It has been suggested that a Rab3-calmodulin complex generated by elevated Ca (2+) concentrations mediated at least some of the effects of the GTPase and limited the number of exocytic events that occurred in response to secretory stimuli36. We propose that the recruitment of calmodulin by Rab3D is an important requirement for osteoclast-mediated bone resorption.
Identification of interacting partners to Rab proteins will be important for drug design, for instance, Plekhm1 was found to co-localize with Rab7 to late endosomal/lysosomal vesicles, a putative function in vesicular transport in the osteoclast. This has been implicated in the development of osteopetrosis37. Interestingly, alendronate inactivates osteoclasts by mechanisms that impair their intracellular vesicle transport, with apoptosis being only a secondary phenomenon to this38. The anti-resorptive activity of NE10790 is thus likely due to disruption of Rab-dependent intracellular membrane trafficking in osteoclasts39. Given that Rab-dependent intracellular membrane trafficking in osteoclastic bone resorption has been proposed to be a target of nitrogen-containing bisphosphonate drug NE1079039, defining Rab3D interacting partners might facilitate the design of the next generation of bisphosphonate drugs.
Materials and Methods
Two-hybrid screening
Mouse Rab3D cDNA was inserted into pBTM116 and used to screen a pVP16-based yeast two-hybrid cDNA library as previously described17. Briefly, for cDNA library screening, the yeast reporter strain L40 was first transfected with baits pLexA-Rab3D and subsequently transfected with the pVP16 mouse embryo cDNA library using lithium acetate and polyethylene glycol. Library plasmids were grown in the presence or absence of histidine. Positive clones isolated from the cDNA library were further analysed by co-transfection with pLexA-Rab3D and DNA sequencing.
RT-PCR
To determine the calmodulin and Rab3D gene transcripts in osteoclasts, total RNA was isolated from bone marrow cells treated with RANKL at various time points according to the manufacturer’s instructions (Qiagen, Sydney). For RT-PCR, single-stranded cDNA was prepared from 2 μg of total RNA using reverse transcriptase with an oligo-dT primer. Two μl of each cDNA was subjected to 30 cycles of PCR (94 °C, 40 sec; 55 °C, 40 sec; and 72 °C, 40 sec) using specific primers to mouse rab3d gene (forward: 5′-ATG GCA TCC GCT AGT GAG-3′; reverse:5′-CTA ACA GCT GCA GCT GCT-3′) and calmodulin (forward: 5′-CCA TGG CTG ACC AGC TGA-3′; reverse: 5′-CCT TCA CTT TGC AGT CAT-3′). Osteoclast markers were also used; including cathepsin K (forward: 5′-CCA GTG GGA GCT ATG GAA GA-3′; reverse: 5′-AAG TGG TTC ATG GCC AGT TC-3′); calcitonin receptor (forward: 5′-CGG ACT TTG ACA CAG CAG AA-3′, reverse: 5′-CAG CAA TCG ACA AGG AGT GA-3′); V-ATPase-d2 (forward: 5′-GTG AGA CCT TGG AAG ACC TGA A-3′; reverse:5′-GAG AAA TGT GCT CAG GGG CT-3′). As an internal control, the single stranded cDNA was PCR-amplified for 25 cycles using 36 B4 primers (forward: 5′-TCA TTG TGG GAG CAG ACA-3′; reverse: 5′-TCC TCC GAC TCT TCC TTT-3′).
In vitro protein interaction
The full length of mouse Rab3D cDNA was subcloned into Flag-tagged expression vector with CMV promoter to generate a Flag-Rab3D plasmid. To express Rab3D protein, 5 × 105 COS-7 cells was transfected with 10 μg of Flag-Rab3D plasmid using effectant reagents (Qiagen, Sydney). After incubation for 48 hours, transfected cells were washed twice with PBS and lysed for 30 mins on ice with 750 μl of lysis buffer (50 mM Tris.Cl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 1 M EDTA, 1 μg/ml pepstatin A). Following centrifugation at 12,000 RPM for 5 mins at 4 °C, supernatant was collected and 750 μl of binding buffer (20 mM Tris. Cl, pH 7.5, 50 mM KCl, 100 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM DTT) was added to the supernatant and mixed. One half of the sample was added to 50 μl of calmodulin immobilized in sepharose beads (Sigma, Sydney) in the presence and absence of 2 mM of calcium. The mixture was incubated overnight at 4 °C with a shaker. The beads were washed three times with 300 μl of buffer containing equal volumes of lysis buffer and binding buffer. The bound proteins were boiled with 1 x SDS-PAGE sample buffer and analysed by Western blot analysis using an anti-FLAG monoclonal antibody (Sigma, Sydney).
Construction of EYFP-Rab3D and Rluc-calmodulin
EYFP-Rab3DWT, EYFP-Rab3DQ81L, EYFP-Rab3DN135I, and EYFP-Rab3DΔCXC fusion constructs were reported previously17. To generate Rluc-calmodulin construct, calmodulin mRNA was PCR-amplified with primers (forward: 5′-AAG AAT TCG GAC CAT GGC TGA CCA GCT GA-3′; reverse: 5′-GGA TCC TCT AGA CCT TCA CTT TGC AGT CAT-3′). The amplified fragment was cloned into a pCR 2.1 T/A-cloning vector as outlined by the manufacturers’ instructions. The BamHI and EcoRI fragment was then cloned into the BamHI and EcoRI sites of pcDNA3.1-Rluc (Invitrogen) to make pcDNA-Rluc as previously described40. To generate a Rluc-calmodulin calcium insensitive mutant, pcDNA3.1 calcium insensitive calmodulin mutant, in which the first Asp of each EF-hand is changed to Ala, was obtained from Dr. Blaise Z. Peterson18 and then subcloned into the pcDNA3.1-Rluc. These constructs were sequenced to confirm. Bioluminescence resonance energy transfer (BRET) assay and cell transfections were performed in COS cells as previously described using the Mithras LB940 BRET plate reader (Berthold Technologies, Inc., Germany)40.
Generation and isolation of osteoclastic cells and bone resorption assay
OCs were generated from mouse BMMs treated with RANKL (100 ng/ml) and M-CSF (10 ng/ml) in vitro as previously described41. Bone resorption assay was carried out using mouse BMM-derived mature osteoclasts as previously described17.
Sucrose gradient ultracentrifugation
Sucrose gradient centrifugation was performed as described previously with minor modifications. Briefly, ~3 × 107 OCs were pelleted, 500 μl of the clarified supernatant was layered on top of 12 ml of 5–20% linear sucrose density gradient prepared in the lysis buffer without Triton X-100. After centrifugation at 150,000 g for 18 h in a SW40 rotor (Beckman Coulter), 1 ml fractions were collected and analyzed by immunoblotting17 using antibodies to GFP (Santa Cruz, CA, USA), Rab3D (SynapicSystems, Goettingen), V-ATPase d242 and calmodulin (Sigma, Sydney).
Immunofluorescence confocal microscopy
Immunofluorescence detection of Rab3D in osteoclasts by confocal microscopy was performed as previously described16. Briefly BMM-derived osteoclasts cultured on bone for 7-days where fixed with 4% paraformaldehyde, permeablised with Triton X-100, blocked in 0.2% BSA-PBS, immunostained with primary antibodies against Rab3D (Rabbit polyclonal; SynapicSystems, Goettingen) or Calmodulin (mouse monoclonal, Sigma, Sydney) and then corresponding secondary Alexa-Fluor conjugated antibodies against rabbit (Alexafluor-488) and mouse Alexafluor-555). F-actin rings where detected by staining with Alexfluor647-conjugated Phalloidin (Molecular Probes, USA) and nuclei were visualised by staining with DAPI (Sigma, USA).
Statistics
The results are representative of at least three independent experiments. Single comparison tests were performed by using paired Student’s t-test in Microsoft Excel. All data are presented as the mean ± standard error of the mean (SEM). Statistical significance was determined at P values < 0.05.
Additional Information
How to cite this article: Zhu, S. et al. Calmodulin interacts with Rab3D and modulates osteoclastic bone resorption. Sci. Rep. 6, 37963; doi: 10.1038/srep37963 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Liu, X., Yang, P. S., Yang, W. & Yue, D. T. Enzyme-inhibitor-like tuning of Ca(2+) channel connectivity with calmodulin. Nature 463, 968–972 (2010).
Rodriguez-Castaneda, F. et al. Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity. Embo J 29, 680–691 (2010).
Tan, H. L. et al. A calcium sensor in the sodium channel modulates cardiac excitability. Nature 415, 442–447 (2002).
Fujisawa, H. Regulation of the activities of multifunctional Ca2+/calmodulin-dependent protein kinases. J Biochem (Tokyo) 129, 193–199 (2001).
Ang, E. S. et al. Calcium/calmodulin-dependent kinase activity is required for efficient induction of osteoclast differentiation and bone resorption by receptor activator of nuclear factor kappa B ligand (RANKL). Journal of cellular physiology 212, 787–795 (2007).
Sato, K. et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med 12, 1410–1416 (2006).
Hirotani, H., Tuohy, N. A., Woo, J. T., Stern, P. H. & Clipstone, N. A. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem 279, 13984–13992 (2004).
Seales, E. C., Micoli, K. J. & McDonald, J. M. Calmodulin is a critical regulator of osteoclastic differentiation, function, and survival. J Cell Biochem 97, 45–55 (2006).
Wu, X., Ahn, E. Y., McKenna, M. A., Yeo, H. & McDonald, J. M. Fas binding to calmodulin regulates apoptosis in osteoclasts. J Biol Chem 280, 29964–29970 (2005).
Williams, J. P., Thames, A. M. 3rd, McKenna, M. A. & McDonald, J. M. Differential effects of calmodulin and protein kinase C antagonists on bone resorption and acid transport activity. Calcif Tissue Int 73, 290–296 (2003).
Williams, J. P., Dong, S. S., Whitaker, C. H., Jordan, S. E. & Blair, H. C. Effects of cell culture time and bone matrix exposure on calmodulin content and ATP-dependent cell membrane acid transport in avian osteoclasts and macrophages. J Cell Physiol 169, 411–419 (1996).
Williams, J. P., Blair, H. C., McKenna, M. A., Jordan, S. E. & McDonald, J. M. Regulation of avian osteoclastic H+-ATPase and bone resorption by tamoxifen and calmodulin antagonists. Effects independent of steroid receptors. J Biol Chem 271, 12488–12495 (1996).
Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513–525 (2009).
Coxon, F. P. & Taylor, A. Vesicular trafficking in osteoclasts. Semin Cell Dev Biol 19, 424–433 (2008).
Zhao, H., Laitala-Leinonen, T., Parikka, V. & Vaananen, H. K. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J. Biol. Chem. 276, 39295–39302 (2001).
Pavlos, N. J. et al. Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Molecular and cellular biology 25, 5253–5269, (2005).
Pavlos, N. J. et al. Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption. Mol Cell Biol 31, 1551–1564 (2011).
Peterson, B. Z., DeMaria, C. D., Adelman, J. P. & Yue, D. T. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron 22, 549–558 (1999).
Pfeffer, S. R. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11, 487–491 (2001).
Pfeffer, S. R. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol 6, 522–526 (1994).
Roa, M., Paumet, F., Le Mao, J., David, B. & Blank, U. Involvement of the ras-like GTPase rab3d in RBL-2H3 mast cell exocytosis following stimulation via high affinity IgE receptors (Fc epsilonRI). J Immunol 159, 2815–2823 (1997).
Baldini, G., Hohl, T., Lin, H. Y. & Lodish, H. F. Cloning of a Rab3 isotype predominantly expressed in adipocytes. Proc Natl Acad Sci USA 89, 5049–5052 (1992).
Ohnishi, H., Samuelson, L. C., Yule, D. I., Ernst, S. A. & Williams, J. A. Overexpression of Rab3D enhances regulated amylase secretion from pancreatic acini of transgenic mice. J Clin Invest 100, 3044–3052 (1997).
Iezzi, M. et al. Subcellular distribution and function of Rab3A, B, C, and D isoforms in insulin-secreting cells. Mol Endocrinol 13, 202–212 (1999).
Valentijn, J. A., Valentijn, K., Pastore, L. M. & Jamieson, J. D. Actin coating of secretory granules during regulated exocytosis correlates with the release of rab3D. Proc Natl Acad Sci USA 97, 1091–1095 (2000).
Larkin, J. M. et al. Rab3D, a small GTP-binding protein implicated in regulated secretion, is associated with the transcytotic pathway in rat hepatocytes. Hepatology 32, 348–356 (2000).
Tai, A. W., Chuang, J. Z. & Sung, C. H. Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. J Cell Biol 153, 1499–1509 (2001).
Nesbitt, S. A. & Horton, M. A. Trafficking of matrix collagens through bone-resorbing osteoclasts. Science 276, 266–269 (1997).
Salo, J., Lehenkari, P., Mulari, M., Metsikko, K. & Vaananen, H. K. Removal of osteoclast bone resorption products by transcytosis. Science 276, 270 (1997).
Mulari, M. T. et al. The architecture of microtubular network and Golgi orientation in osteoclasts–major differences between avian and mammalian species. Exp Cell Res 285, 221–235 (2003).
Park, J. B., Farnsworth, C. C. & Glomset, J. A. Ca2+/calmodulin causes Rab3A to dissociate from synaptic membranes. J Biol Chem 272, 20857–20865 (1997).
Sidhu, R. S. & Bhullar, R. P. Rab3B in human platelet is membrane bound and interacts with Ca(2+)/calmodulin. Biochem Biophys Res Commun 289, 1039–1043 (2001).
Babu, Y. S., Bugg, C. E. & Cook, W. J. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol 204, 191–204 (1988).
Babu, Y. S. et al. Three-dimensional structure of calmodulin. Nature 315, 37–40 (1985).
Yang, C., Jas, G. S. & Kuczera, K. Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin. Biochim Biophys Acta 1697, 289–300 (2004).
Coppola, T. et al. Disruption of Rab3-calmodulin interaction, but not other effector interactions, prevents Rab3 inhibition of exocytosis. Embo J 18, 5885–5891 (1999).
Van Wesenbeeck, L. et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest 117, 919–930 (2007).
Alakangas, A. et al. Alendronate disturbs vesicular trafficking in osteoclasts. Calcif Tissue Int 70, 40–47 (2002).
Coxon, F. P. et al. Identification of a novel phosphonocarboxylate inhibitor of Rab geranylgeranyl transferase that specifically prevents Rab prenylation in osteoclasts and macrophages. J Biol Chem 276, 48213–48222 (2001).
Feng, H. et al. Cytoplasmic terminus of vacuolar type proton pump accessory subunit Ac45 is required for proper interaction with V(0) domain subunits and efficient osteoclastic bone resorption. J Biol Chem 283, 13194–13204 (2008).
Xu, J. et al. Cloning, sequencing, and functional characterization of the rat homologue of receptor activator of NF-kappaB ligand. J Bone Miner Res 15, 2178–2186 (2000).
Feng, H. et al. Myocyte enhancer factor 2 and microphthalmia-associated transcription factor cooperate with NFATc1 to transactivate the V-ATPase d2 promoter during RANKL-induced osteoclastogenesis. J Biol Chem 284, 14667–14676 (2009).
Acknowledgements
This study was also supported in part by the Australian National Health and Medical Research Council (NHMRC, No. 1107828). Arthritis Foundation of Australia (The H J & G J Mckenzie grant), University of Western Australia Research Collaboration Awards. The authors also acknowledge technical suggestions from Jay Steer, Vincent Keuk, Dian Teguh, Gaurav Jadhav, Mingli Yang, Lin Zhou, Jasreen Kular, Eecheng Khor, Jacob Kenny, Prof. Ming-H. Zheng, and Prof. Karin Eidne, and equipment operation assistance from the Centre for Microscopy, Characterization and Analysis (CMCA) of The University of Western Australia, a facility funded by the University, State and Commonwealth Governments. This work is supported by a grant from the National Natural Science Foundation of China (NSFC, no. 81572227), Zhejiang Provincial Natural Science Foundation of China (LY14H170002), the Opening Project of Zhejiang Provincial Top Key Discipline of Clinical Medicine (no. LKFJ017), which in part support Dr. Sipin Zhu as visiting scholar to UWA from Wenzhou Medical University. Dr. Sipin Zhu and Jiake Xu made mutual collaborative visits.
Author information
Authors and Affiliations
Contributions
Sipin Zhu, Shek Man Chim, Taksum Cheng, Estabelle Ang, Kai Chen, Heng Qiu carried out experiments and data analysist. Benjamin Ng, Baysie Lim, Jennifer Tickner provided technical assistance. Sipin Zhu and Jiake Xu participated in draft manuscript. Jennifer Tickner, and Nathan Pavlos revised manuscript. Hua-Zi Xu, Nathan Pavlos and Jiake Xu supervised the project, experimental designs and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhu, S., Chim, S., Cheng, T. et al. Calmodulin interacts with Rab3D and modulates osteoclastic bone resorption. Sci Rep 6, 37963 (2016). https://doi.org/10.1038/srep37963
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37963
This article is cited by
-
Pattern of protein expression in the epididymis of Oligoryzomys nigripes (Cricetidae, Sigmodontinae)
Cell and Tissue Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.