Abstract
Programmed cell death (PCD) is critical for development and responses to environmental stimuli in many organisms. FUZZY ONIONS (FZO) proteins in yeast, flies, and mammals are known to affect mitochondrial fusion and function. Arabidopsis FZO-LIKE (FZL) was shown as a chloroplast protein that regulates chloroplast morphology and cell death. We cloned the FZL gene based on the lesion mimic phenotype conferred by an fzl mutation. Here we provide evidence to support that FZL has evolved new function different from its homologs from other organisms. We found that fzl mutants showed enhanced disease resistance to the bacterial pathogen Pseudomonas syringae and the oomycete pathogen Hyaloperonospora arabidopsidis. Besides altered chloroplast morphology and cell death, fzl showed the activation of reactive oxygen species (ROS) and autophagy pathways. FZL and the defense signaling molecule salicylic acid form a negative feedback loop in defense and cell death control. FZL did not complement the yeast strain lacking the FZO1 gene. Together these data suggest that the Arabidopsis FZL gene is a negative regulator of cell death and disease resistance, possibly through regulating ROS and autophagy pathways in the chloroplast.
Similar content being viewed by others
Introduction
Programmed cell death (PCD) is an integral part of proper development and responses to environmental stimuli of many organisms1,2. Apoptosis and autophagy are the two main forms of PCD in animal cells. Although not exhibiting the classic apoptosis, plant cells can undergo autophagy, involving the formation of membrane-bound vesicles to sequestrate cellular content for degradation and recycling and to determine the survival of cells3,4,5. Many core autophagy components, the autophagy-related (ATG) genes, are identified in plants and they share similar sequence and function with their homologs in other organisms6. Autophagy plays a critical role in plant development and nutrient recycling. Recent studies have also implicated a role of autophagy in plant defense against pathogens, in particular determining PCD in the infected plants7,8. However, many questions still remain regarding how autophagy affects PCD under defense conditions.
Besides pathogen-induced cell death, some mutant plants exhibit constitutive cell death in the absence of pathogen infection. Such mutants are collectively called lesion mimic mutants. These plants often have enhanced disease resistance, strengthened cell walls, express more defense-related genes, and accumulate faster and/or more defense-related molecules, such as salicylic acid (SA) and reactive oxygen species (ROS)9,10. Functional analyses of some corresponding genes of the lesion mimic mutants have revealed multiple pathways leading to PCD, among which the chloroplast appears to be an important source for pro-death signaling10. Although important for pathogen induced PCD in plants, the role of autophagy has not been well understood in plants with autoimmune defects. Mutations in some ATG genes that were associated with early senescence and cell death phenotypes indeed suggest a critical role of autophagy in cell death control observed in some lesion mimic mutants11.
The FUZZY ONIONS (FZO)-LIKE (FZL) gene of Arabidopsis is a single copy gene encoding a GTPase-domain containing protein in the dynamin superfamily. FZO genes in yeast, flies, and mammals were shown to affect mitochondrial fusion. Disruption of these FZO genes could lead to mitochondrial fragmentation and dysfunction12,13,14,15,16,17,18. An Arabidopsis fzl mutation was previously shown to confer cell death and altered chloroplast morphology, associated with higher accumulation of some defense related molecules19. However, it is unknown how the Arabidopsis FZL gene affects cell death and whether it regulates mitochondrial function and plant resistance to pathogens.
We identified a lesion mimic mutant in the background of a transposon insertional mutant for the phosphate transporter gene PHT4;120. Positional cloning revealed that the lesion mimic phenotype was not associated with the pht4;1 mutation, but was due to a mutation in the FZL gene. Besides its known role in affecting cell death and chloroplast morphology, we report here new function of the FZL gene in regulating plant defense and autophagy. We found that fzl mutations conferred enhanced disease resistance to the bacterial pathogen Pseudomonas syringae and the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa). fzl-conferred cell death was associated with the activation of the autophagy and ROS pathways and was dependent on the key defense signaling mediated by SA. Our data also support a reciprocal negative regulation between FZL and SA signaling. Although being the only close homolog of Arabidopsis to the mitochondria-localized FZO proteins from yeast, flies, and mammals, a functional FZL-YFP chimeric protein was localized the chloroplast outer membrane but not the mitochondria of Arabidopsis cells. Thus, Arabidopsis FZL gene may have evolved new function different from its other homologs. Consistent with this idea, we found that neither FZL nor an FZL variant rescued the yeast strain lacking the FZO1 gene. Together, our study suggests that the FZL gene is a negative regulator of PCD and disease resistance to biotrophic pathogens in Arabidopsis, possibly acting through the autophagy pathway.
Results
FZL is a negative regulator of disease resistance to biotrophic pathogens in Arabidopsis
We isolated a lesion mimic mutant in the background of the phosphate transporter mutant called pht4;1-2 (GT_5_110509; a Landsberg erecta allele)20. The lesion mimic phenotype progressed more severely as leaves became more senescent (Fig. S1A). This phenotype was not co-segregated with the transposon insertion in PHT4;1 and kanamycin resistance. To identify the responsible gene, we crossed this mutant to Columbia-0 (Col-0) and conducted map-based cloning with 1108 recombinant F2 lines to narrow down the mutation to a 50 kb region on chromosome 1. Further sequencing of the coding fragments in the region identified a one base pair substitution (G > A) in the FZL gene (At1g03160), which encodes a protein in the dynamin superfamily with demonstrated roles in regulating chloroplast morphology and cell death19,21. The mutation, previously called fzl-Ler, disrupts the 5′ end exon-intron junction of intron IV (Fig. 1A and ref. 19).
fzl mutations confer enhanced disease resistance to bacterial and oomycete pathogens.
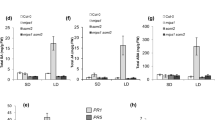
(A) The FZL gene structure and fzl mutant alleles. Exons are indicated in black and introns in grey. (B) SA quantification. Total SA (glucosylated form) was extracted from the indicated plants and quantified with an HPLC instrument. (C) Expression of PR1 analyzed by qRT-PCR. (D) Bacterial growth assay. Plant leaves were infiltrated with virulent P. syringae pv. tomato strain DC3000 (OD600 = 0.0004). Leaf discs were taken 3-day post infection for bacterial growth measurement. (E) Hpa Noco2 infection assay. Seven-day-old seedlings were spray-infected with Hpa Noco 2. Sporangiophore production in cotyledons of each genotype was counted at 7 dpi. Data represent the average number of sporangiophores from 50 seedlings with SEM. Statistical analysis was performed with Student’s t-test (StatView 5.0.1). The asterisk indicates significant difference between the labeled sample and other samples (P < 0.01). These experiments were repeated at least two times and similar results were obtained.
Using a chimeric construct FZL-YFP, created by fusing the FZL cDNA translationally at the 3′ end with the YFP reporter under the control of the CAMV 35S promoter, we were able to complement phenotypes conferred by fzl-Ler, including cell death, high SA accumulation, and high expression of SA marker gene PR1 (Fig. 1B,C and S1B-S1D). To test if FZL regulates disease resistance, we infected fzl-Ler with Pseudomonas syringae pv. tomato strain DC3000 (DC3000). The mutant showed more resistance than Ler, which was rescued by the FZL-YFP transgene (Fig. 1D).
We obtained two additional mutant alleles, fzl-2 (SALK_118335) and fzl-3 (SALK_152584) in Columbia-0 (Col-0) background (Fig. 1A). Compared with Col-0 leaves, leaves of fzl-2 and fzl-3 mutants appeared to be paler but did not show obvious cell death (Fig. 2). The fzl-2 and fzl-3 alleles showed either higher SA accumulation nor enhanced resistance to P. syringae infection, compared with Col-0 (data not shown). However, when infected with the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) isolate Noco2, these mutants demonstrated higher resistance than Col-0 (Fig. 1E). Ler and fzl-Ler plants cannot be tested in this experiment because the Ler background confers hyper-resistance to Hpa Noco2. Thus these data suggest that FZL is a negative regulator of disease resistance to P. syringae and Hpa strains and cell death.
fzl-conferred cell death phenotype can be enhanced in the Ler background.
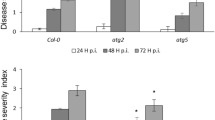
The fzl alleles in Col-0 were crossed to Ler. The resulting homozygous plants were isolated for cell death analysis. (A) Picture of leaves. (B) Cell death staining with trypan blue dye. These experiments were repeated two times with similar results.
Consistent with the defense role of FZL, we found that expression of FZL was suppressed by P. syringae infection (Fig. 3A). To further test if FZL expression can be affected by SA, we treated plants with an SA analog, benzo (1, 2, 3) thiadiazol-7-carothioic acid (BTH), which was shown to activate similar defense responses in plants as SA but without the toxicity caused by SA22,23,24. Indeed, expression of FZL was lower in BTH treated plants in a dosage-dependent manner (Fig. 3B). We also observed a lower expression of FZL in the accelerated cell death 6-1 (acd6-1) mutant that shows constitutive defense and high SA levels (Fig. 3B and refs 25,26). Thus these results suggest a negative feedback regulation of FZL by SA.
Expression of FZL is suppressed by P. syringae infection and by SA activation.
(A) P. syringae infection suppresses FZL expression. Plants were infected by the virulent strain P. syringae pv. maculicola ES4326 DG3 (PmaDG3) or the avirulent strain PmaDG34 (expressing the avirulence effector avrRpm1)66. (B) Activation of SA signaling suppresses FZL expression. Col-0 plants were treated with 100 μM BTH for 24 hrs. qRT-PCR was performed with RNA extracted from the treated and control plants. These experiments were repeated two times with similar results. Different letters indicate significant difference among the samples (P < 0.01).
The cell death phenotype of fzl mutants can be enhanced by the Ler background
We were intrigued by the effects of different genetic backgrounds on cell death formation in the fzl mutants. We confirmed that the two Col-0 fzl mutants were allelic to fzl-Ler by crossing fzl-Ler with fzl-2 or fzl-3. The resulting F1 plants demonstrated strong cell death in leaves as fzl-Ler (data not shown). Such phenotypic discrepancy in the fzl alleles suggests genetic background could affect cell death formation mediated by FZL in Arabidopsis. To test this possibility, we crossed fzl-2 and fzl-3 with Ler and found that fzl-2-Ler and fzl-3-Ler homozygous plants showed more severe cell death than the original mutants in Col-0 background (Fig. 2).
fzl-conferred cell death phenotype is regulated by SA
Some lesion mimic mutants showed SA-dependent cell death9. To test if fzl-conferred cell death phenotype is also SA-dependent, we conducted a mutant analysis. Given the complexity of genetic background on the manifestation of fzl-conferred cell death (Fig. 2), it is important to use the same genetic background in the mutant analysis. While most SA mutants are in Col-0 background20,25,27,28, the eds1-2 mutant is a Ler allele and is impaired in major SA accumulation29. Thus we crossed fzl-Ler with eds1-2 and found that the double mutant fzl-Ler eds1-2 had much suppressed but not completely abolished cell death and restored wt-levels of SA, compared with fzl-Ler (Fig. S2). Our data are largely consistent with the results from a previous study19, supporting a major role of SA in regulating cell death in fzl-Ler.
To further test the role of SA in regulating cell death in fzl mutants, we used a single-cell system. We treated protoplasts of Ler, fzl-Ler and a complementation line (FZL-YFP + fzl-Ler #1) with BTH. Protoplast survival rate was recorded by using fluorescein diacetate (FDA) staining. We found that mock treatment did not induce much difference in cell death in the protoplasts. Upon BTH treatment, fzl-Ler protoplasts showed much faster and more cell death (Fig. 4). By 1 hr of BTH treatment, all fzl-Ler cells were dead while most Ler and FZL-YFP/fzl-Ler cells remained alive. Similarly, protoplasts of the two Col-0 FZL alleles, fzl-2 and fzl-3, also showed enhanced cell death with BTH treatment, compared with Col-0 control (Fig. S3). Since BTH treatment of wild type (wt) protoplasts did not activate cell death, these results suggest that SA signaling works together with FZL to control cell death.
Cell death conferred by fzl-Ler can be enhanced by SA treatment.
Protoplasts were prepared from 21-day old plants and treated with 100 μM BTH or water. Cell survival at the indicated times was assessed with fluorescein diacetate staining and recorded by fluorescence microscopy. The survival rate was calculated as the follow: No. of living protoplasts/no. of total protoplasts *100. Different letters indicate significant difference among the samples at the same time point (P < 0.01; Mann-Whitney test). This experiment was repeated three times and similar results were obtained.
fzl-Ler shows increased autophagosomes and ROS accumulation besides altered chloroplast morphology
Previous studies showed that fzl mutations caused abnormal chloroplast morphology19,21. We confirmed this phenotype with transmission electron microscopy (TEM), using the fourth to sixth leaves of 21-day old plants. The chloroplast of fzl-Ler cells showed reduced thylakoid stacks but more elongated thylakoid grana, compared with that of Ler cells (Fig. 5 and S4). The average number of chloroplasts in fzl-Ler cells is 3.35 ± 0.16 (standard error of the mean; n = 78), which is significantly lower than that in Ler cells (4.76 ± 0.40; n = 60). No major difference was observed in the mitochondrial morphology.
TEM analyses of chloroplast morphology, autophagosome, and ROS deposition.
The fourth to sixth leaves of each genotype were collected and cut into 1 × 2 mm sections, which were incubated with freshly prepared 5 mM CeCl3 in 50 mM MOPS at pH 7.2 or MOPS only for 1 hr. The samples were then fixed and processed for TEM imaging. At least three different leaf samples of each genotype were used in each experiment. Images represent typical observations in two independent experiments. (A,B) Images of Ler cells. Note that all organelles are cerium-free. (C) Image of fzl-Ler cells to show electron-dense cerium deposits mainly on the cell wall. (D–F) Images of fzl-Ler cells to show autophagosome accumulation. Arrows in panels (C,F) indicate cerium deposits. The size bar in panels (A,E,F) is 1 μm, in panel (B,D) is 5 μm, and in panel (C) is 500 nm. *autophagosome; Ch, chloroplast; CW, cell wall; and M, mitochondrion.
Interestingly, we noticed the accumulation of vesicles in the cytoplasm of fzl-Ler cells, some of which were adjacent to the chloroplast in a cell (Fig. 5D–F). These vesicles have a double membrane surrounding the vesicle content, resembling autophagosomes that represent the ancient vesicle mechanism to engulf and deliver cytoplasm content for degradation3,30. Leaf cells of Ler and fzl-Ler/FZL-YFP plants did not have obvious autophagosome accumulation (Fig. 5A,B and data not shown). To further confirm the activation of the autophagy pathway in fzl-Ler, we measured expression of several ATG genes (ATG5, 6, 7, 8c, 8f, 8i, 9, and 10) that are important for the formation of autophagosomes. We found that expression of ATG7, 8C, 8i, and 9 in fzl-Ler was at least two-fold higher than that in Ler and fzl-Ler/FZL-YFP plants (Fig. 6). Thus cell death formation in fzl-Ler likely involves the activation of autophagy.
Oxidative bursts lead to the production of reactive oxygen species (ROS), which are important for activation of defense and cell death31,32,33. Autophagy is tightly connected with ROS production by removing damaged proteins due to oxidation and thus affects cell survival34,35. To see how FZL could affect ROS accumulation and localization, we performed histochemical staining using cerium chloride (CeCl3), which reacts with H2O2 to produce electron-dense precipitates of ceriumperhydroxide that can be visualized using TEM36,37. The fourth to sixth leaves of fzl-Ler and Ler plants were fixed in the presence of cerium chloride followed by embedding and sectioning. Analysis of ultra-thin sections of the embedded samples showed abundant electron-dense cerium deposits, an indicator of H2O2 accumulation, mainly on the cell wall of the fzl-Ler mutant (Fig. 5C and S4). Minor cerium deposits were seen in the chloroplast, cytoplasm, and autophagosome membrane adjacent to the chloroplast (Fig. 5C and F, arrows). However, Ler and fzl-Ler/FZL-YFP plants generally lack such dark deposits (Fig. 5A,B and data not shown). These results indicate that fzl-Ler-conferred cell death is associated with increased ROS production and autophagosome formation.
FZL is a chloroplast protein that functions differently from its yeast homolog
FZO1 proteins in yeasts, flies, and mammals are known to localize to the mitochondria and affect mitochondrial fusion38. Although being the only close homolog of FZO1 proteins, Arabidopsis FZL was localized to the chloroplast and affected chloroplast morphology. The predicted FZL protein has a potential mitochondrial targeting sequence. Whether FZL could also localize to the mitochondria and regulate mitochondrial function has not been explicitly ruled out. To address this question, we used a single cell system, utilization protoplasts from plants expressing a functional FZL-YFP under the control of the constitutive promoter CAMV 35 S (Fig. 1 and S1B-1D). We first confirmed chloroplast-localization of FZL-YFP, which resided on the outer chloroplast membrane (Fig. S5A and B and21). To test if the FZL-YFP protein also resides in the mitochondria in Arabidopsis, we stained protoplasts expressing FZL-YFP with the mitochondria-specific dye MitoTracker Red CMXRos. However, we did not observe a co-localization of YFP and MitoTracker red signals (Fig. S5C). Similarly, co-expression of a mitochondrial marker gene tagged with the red fluorescent reporter mCherry39 in FZL-YFP protoplasts did not reveal a co-localization of mCherry with FZL-YFP (Fig. S6). Thus FZL-YFP is unlikely localized to the mitochondria.
Although not detected in the mitochondria of Arabidopsis cells, it is still possible that the Arabidopsis FZL gene shares conserved function with its yeast homolog FZO1 because the two proteins share significant similarity with 23% identity16,17,18. To test this, we expressed the full-length FZL cDNA and a truncated version with a deletion of the DNA fragment encoding the chloroplast transient peptide (FZL-ΔCTP) in yeast strains with or without the FZO1 gene. These yeast strains also expressed mitochondria-targeted GFP, mtGFP40. GFP fluorescence images showed typical wt-like tubular mitochondria in the strains expressing FZO1 (Fig. 7A left). However, the lack of FZO1 led to fragmented mitochondria (Fig. 7A right). Expression of Arabidopsis FZL or FZL-ΔCTP failed to rescue the mitochondrial defect in the FZO1-deletion yeast strain.
Expression of Arabidopsis FZL gene fails to rescue the yeast FZO1-lacking mutant.
The [FZO1] yeast strain has the chromosomal FZO1 gene deleted and expresses the FZO1 gene on a plasmid. The minus sign (−) indicates strains not expressing the FZO1 gene. Both [FZO1] and “-” yeast strains expressed mitochondria-targeted GFP (mtGFP). In a plasmid shuffling experiment yeast strains were generated to carry an empty vector, the vector with the full-length FZL cDNA (FZL) or an FZL variant with the chloroplast transit peptide-coding sequence being deleted (FZL-ΔCTP). (A) Mitochondrial morphology. Note there was no rescue of the FZO1 lacking yeast mutant by Arabidopsis FZL or FZL-ΔCTP. DIC: Differential interference contrast (DIC). (B) Yeast growth assay. Fermentable YPD medium and non-fermentable YPG medium were used to grow yeast strains. These experiments were repeated two times with similar results.
Yeast cells lacking FZO1 eventually lose mitochondrial DNA and can only grow on the fermentable YPD medium (containing glucose) but not on the non-fermentable YPG medium (containing glycerol) (Fig. 7B and ref. 17). Consistent with results shown in Fig. 7A, only the strains expressing FZO1 but not the FZO1-lacking strains that expressed Arabidopsis FZL or FZL-ΔCTP, were able to grow on YPG (Fig. 7B). Together these data further support that the Arabidopsis FZL gene has evolved new function different from its yeast homolog FZO1.
Discussion
The FZL gene was previously reported to affect chloroplast morphology and cell death in Arabidopsis19,21. We identified an fzl mutant based on its lesion mimic phenotype. Further characterization of fzl mutants showed that besides altered chloroplast morphology and cell death, disruption in the FZL gene led to more resistance to both bacterial and oomycete pathogens, the activation of autophagy and ROS pathways. Cell death conferred by fzl is SA-dependent and can be further exacerbated by SA. While the lack of FZL led to increased SA accumulation, more SA could in turn suppress expression of FZL. Protein co-localization study and yeast complementation test suggest that the Arabidopsis FZL gene may have evolved new function different from its homologs in yeast, flies, and mammals. This new function of FZL likely involves the autophagy and ROS pathways, leading to the regulation of defense and cell death in Arabidopsis.
The yeast FZO1 is the founding member of FZO-like genes and its protein product was shown to localize to the mitochondria and affect mitochondrial function16,17,18. The mitochondria are dynamic organelles with constant movement inside of the cell along with frequent fusion and fission. Such dynamic behavior of the mitochondria is critical for mitochondrial function in many biochemical reactions, energy production, and cellular respiration38. The yeast fzo1 mutant showed highly fragmented mitochondria and instable mitochondrial DNA. Later studies with disrupted FZO homologs in flies, worms and mammals showed similar mitochondrial fragmentation12,13,14,15,18. These studies indicate that FZO genes regulate mitochondrial fusion, disruption of which could lead to disease in some organisms41,42,43,44,45. FZL is the only Arabidopsis gene that shares significant homology to the FZO genes in yeast, flies, and mammals. It was originally hypothesized to function similarly to these other FZO genes in regulating mitochondrial function. However two previous studies19,21 and our data presented here support new function of the Arabidopsis FZL gene in the chloroplast. First, while mutations in FZL led to cell death in Arabidopsis, the yeast fzo1 mutant did not show a cell death phenotype (this study and refs 19,46). Second, it is the morphology of the chloroplast, not the mitochondria, that is altered in fzl-Ler when compared with Ler (Fig. 5, S4, and refs 19,21). Third, the FZL protein was localized to the chloroplast but not in the mitochondria of Arabidopsis cells (Figs S5 and S6, and ref. 21). Fourth, Arabidopsis FZL or an FZL variant did not rescue the FZO-lacking yeast strain (Fig. 7). Thus the Arabidopsis FZL gene likely functions differently from its homologs in yeast, flies, and mammals and Arabidopsis uses an FZL-independent mechanism in regulating mitochondrial fusion.
The function of FZL is closely related to the chloroplast. Besides its critical role in photosynthesis, the chloroplast has been shown as the primary source of many important defense molecules, such as SA biosynthesis, production of ROS and some secondary compounds47,48. Like the mitochondria, the chloroplast is also an important player in PCD49. Mutations in several genes affect chloroplast-derived metabolites and confer the lesion mimic phenotype. Examples of such mutants include acd1 (impaired in pheophorbide an oxygenase)50, acd2 (impaired in red chlorophyll catabolite reductase)44,51, flu (impaired in a protein that is a part of a complex inhibiting tetrapyrrole synthesis)52,53, and the maize mutant les22 (impaired in uroporphyrinogen decarboxylase)54. FZL is an outer-membrane chloroplast protein (Figs S5 and S6, and ref. 21) and likely functions differently from these chloroplast metabolic proteins in affecting PCD. The lack of FZL could cause the change in chloroplast morphology and subsequently affect the function of the chloroplast. Such changes in the chloroplast can activate stress signals, leading to the production of ROS, which in turn could cause the accumulation of damaged proteins and result in cell death. Consistent with the role of autophagy in removing damaged proteins and regulating PCD, we observed increased autophagosomes using high-resolution TEM images and enhanced expression of autophagic genes in the fzl mutant exhibiting cell death (Figs 5 and 6). Thus our data support the regulation of autophay and ROS pathways by FZL. Further detailed morphological analysis of autophagosomes in fzl-Ler and genetic analysis of fzl mutants and other mutants impaired in autophagy and ROS pathways could contribute to a better understanding of FZL function.
Another factor affecting FZL function is SA as demonstrated by these supporting data: (1) fzl-Ler accumulates higher SA levels, associating with more cell death and disease resistance, compared to Ler (Fig. 1 and S1); (2) The high SA level in fzl-Ler is suppressed by a normal FZL gene and by the eds1-2 mutation. Such suppression in SA accumulation is associated with reduced cell death and defense (Fig. 1, S1, and S2); (3) SA treatment activates cell death in fzl mutants but not in wt alone (Fig. 4 and S3) and (4) expression of FZL is suppressed by high SA levels and defense activation (Fig. 3). Together these observations suggest a negative feedback loop formed between FZL and SA in regulating cell death and defense. The FZL-SA signaling loop likely also involves the ROS and autophagy pathways. To support this notion, ROS bursts are known to trigger SA production and signaling and in turn SA signal activation can induce more ROS bursts33. A similar interplay was found with autophagy and SA3,11,55,56. Thus FZL may normally function be to inhibit the activation of the signaling cascade involving ROS, SA and autophagy. In the absence of FZL, the activation of this signaling event leads to degradation of the chloroplast (the source of some pro-death signals) and eventually cell death. Indeed, we observed reduced numbers of chloroplasts in addition to cell death in fzl-Ler. While we know disrupting SA could suppress fzl-conferred phenotypes, it would be interesting to further investigate how disrupting ROS and autophagy pathways could interfere with fzl-conferred phenotypes in future studies.
Besides ROS, SA, and autophagy, fzl-conferred cell death can be affected by other factors. The fzl-2 and fzl-3 alleles in Col-0 background demonstrated minor defense and no cell death while the fzl-ler allele in Ler background showed strong cell death and defense phenotypes (Figs S1 and 2). We showed that the Ler background can enhance cell death in fzl-2 and fzl-3 mutants. Consistent with the influence of genetic background on fzl-conferred cell death, we observed residual cell death when the SA mutant eds1-2 in Ler background was crossed into fzl-ler while another study showed a complete suppression of cell death in fzl-Ler by a different eds1 mutant (Fig. S2 and ref. 19). Such discrepancy could be due to genetic background and/or the growth environment in different laboratories. Developmental stages could also affect fzl-conferred cell death (Fig. S1A). Together these observations suggest that additional molecules existing under certain conditions could affect FZL function.
The roles of ROS and SA in plant defense against pathogens have been relatively well understood. Pathogen infection and activation of defense signaling are known to induce autophagy5,6,7. However how autophagy regulates plant innate immunity remains unclear. While they all had much reduced autophagosomes and increased cell death, atg mutants showed opposing effects on disease resistance. One atg mutant, atg6, was shown to be more susceptible to biotrophic pathogens57,58. Consistent with atg6-conferred disease susceptibility, studies on fzl and the mutant defective in glyceraldehyde-3-phosphate dehydrogenase (GAPDH) showed an association of constitutive autophagy with enhanced disease resistance against biotrophic pathogens. On the other hand a group of atg mutants (i.e. atg2, 5, 10, and 18a) was shown to be more resistant to biotrophic pathogens56,59,60,61. Plants usually respond to biotrophic and necrotrophic pathogens with opposing defense phenotypes62. Consistent with this idea, these atg mutants were more susceptible to necrotrophic pathogens. Interestingly another group of atg mutants (i.e. atg7 and 9) was shown to be more susceptible to both biotrophic and necrotrophic pathogens. It appears that there is a lack of consistent association between the number of autophagosome formation and plant defense. These seemly controversial data could suggest that different ATG genes are differentially required by pathogens of different lifestyles and the severity of cell death is not always coupled with the level of disease resistance in plants. Thus the mechanism of autophagy signaling in defense, in particular its connection with FZL function, still remains to be elucidated.
Taken together the fzl-ler mutant exhibits hallmarks of lesion mimic mutant phenotypes, including cell death, increased accumulation of defense-related molecules (i.e. ROS, SA and defense gene transcripts). We report that the activation of the autophagy pathway in this mutant is a possible mechanism leading to cell death in the plant. Data from this and other studies clearly support that the FZL gene of Arabidopsis has evolved new function different from its homologs in yeast, flies, and mammals. This new function lies in the regulation of chloroplast morphology and function, activation the signaling cascade involving ROS, SA and autophagy. The FZL-regulated processes ultimately affect plant growth, development, and response to pathogen attacks. Although we are still far from a complete understanding of the molecular mechanism by which FZL regulates innate immunity, FZL and its related genes can be used as excellent resources to uncover mechanisms of PCD and disease resistance. These genes can also be used as powerful tools to manipulate plant defense response in order to achieve a broad spectrum of disease resistance in plants.
Methods
Plant Materials
Most Arabidopsis plants used in this research were grown in growth chambers with a 12 hr light/12 h dark cycle, light intensity at 200 μmol m−2 s−1, 60% humidity, and 22 °C. For protoplast isolation, plants were grown under lower light intensity (100 μmol m−2 s−1) with other conditions the same. The fzl-Ler and eds1-2 mutants were previously described19,29. The fzl-2 (SALK_118335) and fzl-3 (SALK_152584) mutants were obtained from the Arabidopsis Biological Resource Center.
Protoplast Analyses
Arabidopsis protoplasts were isolated from leaves of 21-day old plants following the tape sandwich technique described previously63 with few modifications. Briefly, the lower epidermal surface cell layer was peeled away from leaves using plastic tape. Fifteen peeled leaves were transferred to a Petri dish containing 10 ml of enzyme solution (20 mM MES (pH 5.7), 1% (w/v) cellulase R10, 0.25% (w/v) macerozyme R10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, 0.1% (w/v) BSA). Leaves were gently agitated in the dark for 60 to 120 min till the protoplasts were completely released into the solution. Protoplasts were centrifuged three minutes at 100 × g, washed twice with 10 ml of pre-chilled modified W5 solution (2 mM MES (pH 5.7)), 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and incubated on ice for 30 min. Protoplasts were then centrifuged and resuspended to a final concentration of 5 × 105 cells/ml in modified MMG solution (4 mM MES (pH 5.7), 0.4 M mannitol, 15 mM MgCl2) for further experiments.
For co-localization studies, protoplasts expressing FZL-YFP were stained with the mitochondria-specific dye MitoTracker Red CMXRos (Molecular Probes) or transfected with a mitochondrial marker gene tagged with the red fluorescent reporter gene mCherry39. Images were captured using a confocal laser scanning microscope (Leica TCS SP2 AOBS) and analyzed using the Imaris software (version 7.0.0).
Complementation Tests
The complementation construct for fzl-Ler was made by fusing FZL cDNA translationally at the 3′ end with the YPF reporter and the chimeric gene was expressed under the control of the CAMV 35 S promoter. This construct, named FZL-YFP, was used to transform fzl-Ler. At least 15 independently transformed lines were obtained and they all showed abolished cell death phenotype. Primers used for making the construct were listed in Table S1.
In order to complement the yeast FZO1-lacking mutant, we cloned the full-length FZL cDNA and a truncated version with a deletion of the sequence encoding the chloroplast transient peptide (FZL-ΔCTP) in the pYX122 vector under the control of the constitutive TPI promoter. These constructs and the empty vector were introduced into a yeast strain that has the chromosomal copy of the FZO1 gene deleted and carries a copy of the FZO1 gene on a plasmid with a URA3 marker. Upon growth on the 5-FOA medium, the URA3 plasmid was counter-selected against and thereby the FZO1 gene was eliminated in yeast cells, thus creating FZO1-lacking yeast mutant strains. Primers used for making yeast-complementation constructs were listed in Table S1.
Pathogen Infection
Bacterial culture and preparation of P. syringae strains were conducted as described64. The fourth to sixth leaves of 21-day old plants were infiltrated with P. syringae-containing solution, using a 1 ml needleless syringe. Infected leaf discs were collected three days later for bacterial growth assay. The infection with Hyaloperonospora arabidopsidis isolate Noco 2 was conducted with 7-d old seedlings as previously described64.
Cell Death Analyses
The fourth to sixth leaves from each genotype were stained with trypan blue for cell death visualization, according to a published protocol27. Photographs of the stained leaves were taken with a CCD camera (cool Snap HQ2, Photometrics, USA) connected to a dissecting microscope (Leica M205 FA, Leica Microsystems, Germany). At least three leaves from different plants of each genotype were stained and examined for cell death.
To assess cell death in protoplasts, 80 μl of protoplasts at 5 × 105/ml for each genotype were treated with 100 μM of benzo (1,2,3)-thiadiazole-7-carbothioic acid (BTH, a kind gift from Robert Dietrich, Syngenta) or water (mock). Three replicates for each genotype were used in each experiment. Protoplasts were collected at the indicated times and were stained with fluorescein diacetate (FDA) (Sigma-Aldrich Co. LLC, St. Louis MO) for cell viability test, using 2 μl of 5 mg/ml FDA per sample for 2 min in darkness. Living cells gave out green fluorescence that was detected with fluorescence microscopy. The cell survival rate was calculated based on the ratio between living cells and the total number of protoplasts in a sample.
RNA Analysis
The fourth to sixth leaves of 21-day old plants were harvested for RNA extraction followed by qRT-PCR analysis as previously described65. Primers used for qRT-PCR were listed in Table S1.
SA Measurement
Free and total SA (glucosylated SA) were extracted from 21-day old plants and quantified with a high-performance liquid chromatography (HPLC) instrument as previously described25,28.
H2O2 Localization by Cerium Chloride Staining
The localization of H2O2 by cerium chloride staining was described previously66. The fourth to sixth leaves of 21-day-old plants were cut into 1 × 2 mm pieces, which were first incubated with freshly prepared 5 mM CeCl3 in 50 mM 3-(N-morpholino) propanesulfonic acid (MOPS) at pH 7.2 for 1 h. Control samples were incubated in MOPS buffer only. The treated leaf sections were further fixed in 2.5% (v/v) glutaraldehyde and 2% (v/v) paraformaldehyde in 0.1 M cacodylate buffer (pH 7.2–7.4) and embedded in Epon 812 resin (Electron Microscopy Sciences). Ultra-thin sections (90 nm) were used for observation with a transmission electron microscope (JEM-1400, JEOL, Tokyo, Japan) at an accelerating voltage of 120 kV.
Additional Information
How to cite this article: Tremblay, A. et al. A Role of the FUZZY ONIONS LIKE Gene in Regulating Cell Death and Defense in Arabidopsis. Sci. Rep. 6, 37797; doi: 10.1038/srep37797 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
van Doorn, W. G. & Woltering, E. J. Many ways to exit? Cell death categories in plants. Trends Plant Sci 10, 117–122 (2005).
Greenberg, J. T. & Yao, N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6, 201–211 (2004).
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R. & Avin-Wittenberg, T. Autophagy in Plants–What’s New on the Menu? Trends Plant Sci 21, 134–144, doi: 10.1016/j.tplants.2015.10.008 (2016).
Bassham, D. C. & Crespo, J. L. Autophagy in plants and algae. Front Plant Sci 5, 679, doi: 10.3389/fpls.2014.00679 (2014).
Lv, X., Pu, X., Qin, G., Zhu, T. & Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 19, 905–921, doi: 10.1007/s10495-014-0981-4 (2014).
Kim, S. H., Kwon, C., Lee, J. H. & Chung, T. Genes for plant autophagy: functions and interactions. Mol Cells 34, 413–423, doi: 10.1007/s10059-012-0098-y (2012).
Hayward, A. P. & Dinesh-Kumar, S. P. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol 49, 557–576, doi: 10.1146/annurev-phyto-072910-095333 (2011).
Patel, S., Caplan, J. & Dinesh-Kumar, S. P. Autophagy in the control of programmed cell death. Curr Opin Plant Biol 9, 391–396, doi: 10.1016/j.pbi.2006.05.007 (2006).
Lorrain, S., Vailleau, F., Balague, C. & Roby, D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8, 263–271 (2003).
Moeder, W. & Yoshioka, K. Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3, 764–767 (2008).
Yoshimoto, K. et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927, doi: 10.1105/tpc.109.068635 (2009).
Rolland, S. G., Lu, Y., David, C. N. & Conradt, B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. J Cell Biol 186, 525–540, doi: jcb.200905070 (2009).
Chen, H. et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289, doi: S0092-8674(10)00179-0 (2010).
Ishihara, N., Eura, Y. & Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117, 6535–6546, doi: jcs.01565 (2004).
Eura, Y., Ishihara, N., Yokota, S. & Mihara, K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem 134, 333–344 (2003).
Hermann, G. J. et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143, 359–373 (1998).
Rapaport, D., Brunner, M., Neupert, W. & Westermann, B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem 273, 20150–20155 (1998).
Hales, K. G. & Fuller, M. T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129, doi: S0092-8674(00)80319-0 (1997).
Landoni, M. et al. A mutation in the FZL gene of Arabidopsis causing alteration in chloroplast morphology results in a lesion mimic phenotype. J Exp Bot 64, 4313–4328, doi: 10.1093/jxb/ert237 (2013).
Wang, G. Y. et al. Circadian clock-regulated phosphate transporter PHT4;1 plays an important role in Arabidopsis Defense. Mol. Plant 4, 516–526, doi: ssr016 (2011).
Gao, H., Sage, T. L. & Osteryoung, K. W. FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc Natl Acad Sci USA 103, 6759–6764, doi: 0507287103 (2006).
Friedrich, L. et al. A benzothiadiazole derivative induces systemic acquired resistance. Plant J. 10, 61–70 (1996).
Lawton, K. A. et al. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82 (1996).
Kinkema, M., Fan, W. & Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. (2000).
Ng, G. et al. Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189, 851–859, doi: genetics.111.132332 (2011).
Lu, H., Rate, D. N., Song, J. T. & Greenberg, J. T. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420 (2003).
Wang, G., Zhang, C., Battle, S. L. & Lu, H. The Phosphate Transporter PHT4;1 is a Salicylic Acid Regulator Likely Controlled By the Circadian Clock Protein CCA1. Frontiers in Plant Science 5, doi: 10.3389/fpls.2014.00701 (2014).
Wang, G. F. et al. Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156, 1508–1519, doi: pp.111.176776 (2011).
Falk, A. et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297 (1999).
Mizushima,. N. Autophagy: process and function. Genes Dev 21, 2861–2873, doi: 10.1101/gad.1599207 (2007).
Torres,. M. A., Jones,. J. D. & Dangl, J. L. Reactive oxygen species signaling in response to pathogens. Plant Physiol 141, 373–378 (2006).
Lamb, C. & Dixon, R. A. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48, 251–275 (1997).
Herrera-Vásquez, A., Salinas, P. & Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Frontiers in Plant Science 6, doi: 10.3389/fpls.2015.00171 (2015).
Minibayeva, F., Dmitrieva, S., Ponomareva, A. & Ryabovol, V. Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol Biochem 59, 11–19, doi: 10.1016/j.plaphy.2012.02.013 (2012).
Perez-Perez, M. E., Lemaire, S. D. & Crespo, J. L. Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160, 156–164, doi: 10.1104/pp.112.199992 (2012).
Bestwick, C. S., Brown, I. R., Bennett, M. H. & Mansfield, J. W. Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9, 209–221 (1997).
Yao, N. et al. Mitochondrial oxidative burst involved in apoptotic response in oats. Plant J. 30, 567–579 (2002).
Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11, 872–884, doi: nrm3013 (2010).
Nelson, B. K., Cai, X. & Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51, 1126–1136, doi: TPJ3212 (2007).
Westermann, B. & Neupert, W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427, doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U (2000).
41 Vellosillo, T. et al. Defense activated by 9-lipoxygenase-derived oxylipins requires specific mitochondrial proteins. Plant Physiol 161, 617–627, doi: pp.112.207514 (2013).
Laluk, K., Abuqamar, S. & Mengiste, T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol 156, 2053–2068, doi: pp.111.177501 (2011).
Gleason, C. et al. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108, 10768–10773, doi: 1016060108 (2011).
Yao, N. & Greenberg, J. T. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18, 397–411 (2006).
Yao, N., Eisfelder, B., Marvin, J. & Greenberg, J. The mitochondrion- an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J 40, 596–610 (2004).
Braun, R. J. & Westermann, B. Mitochondrial dynamics in yeast cell death and aging. Biochem Soc Trans 39, 1520–1526, doi: 10.1042/BST0391520 (2011).
Dempsey, D. A., Vlot, A. C., Wildermuth, M. C. & Klessig, D. F. Salicylic Acid biosynthesis and metabolism. Arabidopsis Book 9, e0156, doi: 10.1199/tab.0156 (2011).
Seyfferth, C. & Tsuda, K. Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Frontiers in Plant Science 5, doi: 10.3389/fpls.2014.00697 (2014).
Kim, C. et al. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039, doi: 10.1105/tpc.112.100479 (2012).
Tanaka, R., Hirashima, M., Satoh, S. & Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44, 1266–1274 (2003).
Mach, J. M., Castillo, A. R., Hoogstraten, R. & Greenberg, J. T. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98, 771–776 (2001).
Kauss, D., Bischof, S., Steiner, S., Apel, K. & Meskauskiene, R. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett 586, 211–216, doi: 10.1016/j.febslet.2011.12.029 (2012).
Meskauskiene, R. et al. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98, 12826–12831, doi: 10.1073/pnas.221252798 (2001).
Hu, G., Yalpani, N., Briggs, S. P. & Johal, G. S. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize [see comments]. Plant Cell 10, 1095–1105 (1998).
Hofius, D. et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783, doi: 10.1016/j.cell.2009.02.036 (2009).
Lenz, H. D. et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66, 818–830, doi: 10.1111/j.1365-313X.2011.04546.x (2011).
Liu, Y. et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577, doi: 10.1016/j.cell.2005.03.007 (2005).
Patel, S. & Dinesh-Kumar, S. P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20–27 (2008).
Lai, Z., Wang, F., Zheng, Z., Fan, B. & Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 66, 953–968, doi: 10.1111/j.1365-313X.2011.04553.x (2011).
Wang, Y., Nishimura, M. T., Zhao, T. & Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68, 74–87, doi: 10.1111/j.1365-313X.2011.04669.x (2011).
Wang, Y., Wu, Y. & Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal Behav 6, 1408–1410 (2011).
Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43, 205–227 (2005).
Wu, F. H. et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16, doi: 10.1186/1746-4811-5-16 (2009).
Lu, H. et al. Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Front Plant Sci 4, 157, doi: 10.3389/fpls.2013.00157 (2013).
Hamdoun, S. et al. Differential Roles of Two Homologous Cyclin-Dependent Kinase Inhibitor Genes in Regulating Cell Cycle and Innate Immunity in Arabidopsis. Plant Physiol 170, 515–527, doi: 10.1104/pp.15.01466 (2016).
Hamdoun, S., Liu, Z., Gill, M., Yao, N. & Lu, H. Dynamics of Defense Responses and Cell Fate Change during Arabidopsis-Pseudomonas syringae Interactions. PLoS One 8, e83219, doi: 10.1371/journal.pone.0083219 (2013).
Acknowledgements
We thank members in the Lu laboratory for assistance with this work. We thank Dr. Benedikt Westermann from Universität Bayreuth for his assistance in the yeast assays and critical comments for this manuscript. This work was partially supported by a grant from National Science Foundation (NSF 1456140) to H.L and National Natural Science Foundation of China (31570255) to N.Y.
Author information
Authors and Affiliations
Contributions
A.T., H.Z., S.B., and H.L. wrote the manuscript with input from other authors. A.T. performed fzl-Ler complementation, characterization of fzl mutants in different genetic background, and protoplast experiments. S.S. and H.L. cloned the FZL gene and performed initial genetic analysis. D.N.T. and V.T.D. assisted in obtaining fzl mutants and in genetic analyses. H.Z. and N.Y. conducted the TEM and confocal microscopy experiments. S.B. performed the yeast complementation test. C.Z. did the qRT-PCR experiment for gene expression. All authors reviewed the manuscript and agreed on the content of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tremblay, A., Seabolt, S., Zeng, H. et al. A Role of the FUZZY ONIONS LIKE Gene in Regulating Cell Death and Defense in Arabidopsis. Sci Rep 6, 37797 (2016). https://doi.org/10.1038/srep37797
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37797
This article is cited by
-
FZL is primarily localized to the inner chloroplast membrane however influences thylakoid maintenance
Plant Molecular Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.