Abstract
Natural Cordyceps collected in Bhutan has been widely used as natural Cordyceps sinensis, an official species of Cordyceps used as Chinese medicines, around the world in recent years. However, whether Cordyceps from Bhutan could be really used as natural C. sinensis remains unknown. Therefore, DNA sequence, bioactive components including nucleosides and polysaccharides in twelve batches of Cordyceps from Bhutan were firstly investigated, and compared with natural C. sinensis. Results showed that the fungus of Cordyceps from Bhutan was C. sinensis and the host insect belonged to Hepialidae sp. In addition, nucleosides and their bases such as guanine, guanosine, hypoxanthine, uridine, inosine, thymidine, adenine, and adenosine, as well as compositional monosaccharides, partial acid or enzymatic hydrolysates, molecular weights and contents of polysaccharides in Cordyceps from Bhutan were all similar to those of natural C. sinensis. All data suggest that Cordyceps from Bhutan is a rational alternative of natural C. sinensis, which is beneficial for the improvement of their performance in health and medicinal food areas.
Similar content being viewed by others
Introduction
Cordyceps sinensis, one of the well-known tonic and traditional Chinese medicines, is a composite consisting of the stromata of the fungus, parasitized on the larva of some species of insects (Family: Hepialidae), and the dead caterpillar1,2. It is distributed on the Tibetan Plateau and its surrounding regions at an altitude above 3,000 m, including Tibet, Gansu, Qinghai, Sichuan, and Yunnan provinces in China and in certain areas such as the countries of Bhutan, India and Nepal on the southern flank of the Himalayas1,3,4. Usually, C. sinensis has been used for the prevention and treatment of a variety of diseases such as asthma, bronchitis, lung inflammation, nocturnal emissions, and night sweats5,6,7. Indeed, nucleosides and polysaccharides were considered as the mainly bioactive components in C. sinensis8,9,10,11,12. Currently, due to its various beneficial effects and limited supply, the price of C. sinensis has increased dramatically and is much more expensive, even 4 times, than gold by weight13,14. Therefore, natural Cordyceps collected from Bhutan (Bhutanese Cordyceps) has attracted much attention of the Royal Government of Bhutan, which has been considered as an economically important fungus as natural C. sinensis15. However, whether Bhutanese Cordyceps could be really used as natural C. sinensis remains unknown. Indeed, to the best of our knowledge, few chemical characters of Cordyceps from Bhutan have been investigated, and never been compared with those of natural C. sinensis. Therefore, DNA sequence, bioactive components including nucleosides and polysaccharides in twelve batches of Bhutanese Cordyceps were firstly investigated, and compared with natural C. sinensis, which are beneficial for better understanding the rational use of Cordyceps from Bhutan.
Results and Discussion
Identification of Bhutanese Cordyceps based on DNA barcoding
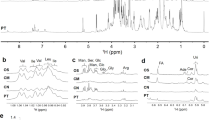
Figure 1 showed the typical morphological characteristics of Bhutanese Cordyceps. Compared to natural C. sinensis, morphological characteristics of Bhutanese Cordyceps were similar to those of the former16, but obvious difference was found in their eyes (Fig. 1A). Therefore, Bhutanese Cordyceps was further identified using DNA barcoding method, which has been widely applied for species identification of animals, plants and fungi17,18. Indeed, the nuclear ribosomal internal transcribed spacer (ITS) region has been considered as a universal DNA barcode marker for fungi identification19. Supplementary Fig. S1A showed the gel electrophoresis of DNA fragments of ITS, Cytb and COI from Bhutanese Cordyceps, and their sequences of stroma and host insect were, respectively, shown in Fig. S1B,C. ITS region of genomic DNA in stroma of Bhutanese Cordyceps was 562 bp, and GC content was 62.13%. According to GenBank NCBI nucleotide database, ITS sequence of Bhutanese Cordyceps was 99% homologous to that of C. sinensis. Therefore, the fungus in Bhutanese Cordyceps could be confirmed as C. sinensis1. In addition, mitochondrial COI and Cytb sequences in host insect of Bhutanese Cordyceps were 658 bp and 433 bp, and GC contents were 29.18% and 22.86%, respectively. Both mitochondrial COI and Cytb sequences suggested that species of host insect of Bhutanese Cordyceps belonged to Hepialidae sp (99% homologous)20, which was in accordance with those of C. sinensis3,21,22.
The typical samples (A), and representative HPLC-DAD chromatograms of nucleosides (B) and GC-MS profiles of compositional monosaccharides of polysaccharides (C) from Bhutanese Cordyceps (Left) and natural C. sinensis (Right). 2, uracil; 3, cytidine; 4, guanine; 5, hypoxanthin; 6, adenine; 7, uridine; 8, thymine; 10, inosine; 11, guanosine; 12, thymidine; 13, adenosine; Ara, arabinose; Fuc, fucose; Man, mannose; Glc, Glucose; Gal, Galactose; IS, internal standard.
Determination of nucleosides in Bhutanese Cordyceps
Although Bhutanese Cordyceps was identified as the same as the species of C. sinensis, their chemical characters could be different because of their different locations. Generally, nucleosides and their bases, involved in the regulation and modulation of various physiological processes in body through purinergic and/or pyrimidine receptors23,24, are considered as the main bioactive components in C. sinensis9,25. To date, more than ten nucleosides and nucleobases, as well as their analogues, including cytosine, uracil, cytidine, guanine, guanosine, hypoxanthine, adenine, adenosine, uridine, thymine, thymidine, 2′-deoxyuridine, inosine and cordycepin have been found in C. sinensis25. Indeed, adenosine also has been used as a marker for the quality control of C. sinensis in Chinese Pharmacopoeia (2015). Therefore, determination of nucleosides and their bases in Bhutanese Cordyceps is extremely important for better understanding its chemical characters and quality.
The typical HPLC-DAD chromatograms of mixed standards and water extract of Bhutanese Cordyceps were shown in Supplementary Fig. S2A and Fig. 1B, respectively. The contents of individual investigated component in Bhutanese Cordyceps were summarized in Table S1. Results showed that types26,27,28 of nucleosides and their bases in water extract, and contents of adenine, uridine, inosine, guanosine and adenosine27,28,29 in Bhutanese Cordyceps were similar to those of C. sinensis. In addition, the overall contents of nucleosides are much higher in Bhutanese Cordyceps than those of C. sinensis 26,27,29.
Determination of polysaccharides in Bhutanese Cordyceps
Compositional monosaccharides and their molar ratios
Besides nucleosides and their bases, polysaccharides are major contributors to the most of biological activities of C. sinensis, and the content of polysaccharides in C. sinensis is ranged about 3% to 8% of its total dry weight8. Generally, bioactivities of polysaccharides are closely correlated to their chemical structures such as compositional monosaccharides, types of glycosidic linkages, and molecular weight distributions, as well as their absolute content30. Therefore, determination of physicochemical properties of polysaccharides in Bhutanese Cordyceps is also extremely important for evaluation of its beneficial effects.
GC-MS analysis has been widely employed for the qualitative and quantitative analysis of compositional monosaccharides in polysaccharides from medicinal plants and fungi31. The typical GC-MS profiles of monosaccharides standards and monosaccharides released from polysaccharides in Bhutanese Cordyceps were shown in the Fig. S2B and Fig. 1C, respectively, and their molar ratios were summarized in Table S2. The data showed that compositional monosaccharides of polysaccharides from Bhutanese Cordyceps were mannose, glucose, and galactose, which were in accordance with those of C. sinensis11,32, though their molar ratios in few samples (CC1, CC2, and CC5) of Bhutanese Cordyceps were different to those of C. sinensis32.
PACE and HPTLC fingerprints of partial acid and enzymatic hydrolysates
Saccharide mapping based on HPTLC and PACE analysis has been proven to be a feasible and desirable technique for qualitative analysis of monosaccharides and oligosaccharides released from polysaccharides, which has been successfully applied for partial characterization and comparison of polysaccharides from natural and cultural C. sinensis and their related species33,34. Therefore, partial acid hydrolysates and enzymatic digestions of polysaccharides in Bhutanese Cordyceps were investigated and compared using saccharide mapping based on PACE and HPTLC analysis. Supplementary Fig. S4 showed the HPTLC and PACE fingerprints of hydrolysates of polysaccharides in Bhutanese Cordyceps. Partial acid hydrolysates of polysaccharides from Bhutanese Cordyceps (CC1-CC12), which did not exist in samples before acid hydrolysis (see in the Supplementary Fig. S3), were similar in both PACE and HPTLC fingerprints (Fig. S4A). To improve the specificity, pectinase, α-amylase, and β-D-glucanase were selected for enzymatic digestion of polysaccharides from Bhutanese Cordyceps34,35. Results showed that both PACE and HPTLC fingerprints of pectinase, α-amylase, and β-D-glucanase digested polysaccharides in Bhutanese Cordyceps were similar (Fig. S4B–D), which suggested that α-1,4-galactosidic, α-1,4-glucosidic, and β-1,4-glucosidic linkages might exist in polysaccharides from Bhutanese Cordyceps34,35.
Molecular weight distributions and contents of polysaccharides and their fractions
HPSEC-MALLS-RID, which has been proven as the powerful and efficient technique for the determination of the molecular weight, molecular weight distribution, as well as contents of polysaccharides and their fractions from natural resources30,36, was used for the determination of molecular weight distributions and their fractions contents of polysaccharides from Bhutanese Cordyceps. In order to exclude the possible interference from the presence of proteins in the sample solutions, UV absorbance was simultaneously detected at UV 280 nm. The typical HPSEC-RID-UV chromatograms of polysaccharides in Bhutanese Cordyceps were shown in Supplementary Fig. S5, and three peaks were found in samples. Proteins were almost absent in the peaks 1 and peak 2 of all tested samples, while peak 3 had high UV (280 nm) absorbance. Therefore, the molecular weights, polydispersity (Mw/Mn) and contents of polysaccharide fractions (peak 1 and peak 2) were determined. As shown in Table 1, molecular weights of polysaccharide fractions in Bhutanese Cordyceps were ranging from 1.12 × 106 to 5.51 × 106 Da (peak 1) and 0.45 × 105 to 4.89 × 105 Da (peak 2), respectively. The Mw/Mn of polysaccharide fraction (peak 1) was ranging from 1.4 to 2.6, while peak 2 was ranging from 1.3 to 1.7. Moreover, total contents of polysaccharides in Bhutanese Cordyceps were ranging from 2.38% to 8.71%, and their average content was 4.53 ± 1.91% (n = 12), which were similar to those of C. sinensis8.
Comparison of both nucleosides and polysaccharides in Bhutanese Cordyceps and C. sinensis
Hierarchical clustering analysis (HCA) could calculate the distance matrices of data objects, and organize objects with great similarities into clusters. The applicability of this method has been recognized in many studies37,38. In order to further investigate the difference and similarity between Bhutanese Cordyceps and C. sinensis, HCA was performed based on their nucleosides and polysaccharides analysis, including the contents of the investigated nucleosides and bases, molar ratios of compositional monosaccharides, molecular weights, and contents of polysaccharides. As shown in Fig. 2, almost all samples, except CS1 and CC5, were grouped into one cluster, which suggested that Bhutanese Cordyceps was very similar to C. sinensis in active components. However, CS1 and CC5 were grouped into other clusters due to their significant difference in the molar ratio of compositional monosaccharides. The molar ratios of glucose in CS1 and galactose in CC5 were, at least about two folds, much higher than others, respectively, which might attribute to their different locations32.
The dendrogram of HCA analysis for all tested samples.
HCA analysis of all tested samples was based on their contents of nucleosides and nucleobases, and molar ratios of compositional monosaccharides, molecular weights, and contents of polysaccharides; Sample codes were the same as in Table 1.
In addition, the professional software named “Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine” (Matlab version, Ver1.315) was used for the evaluation of the similarity of Bhutanese Cordyceps and C. sinensis based on their PACE fingerprints of partial acid and enzymatic hydrolysates. The correlation coefficients of individual chromatogram to their simulative mean chromatograms of partial acid, pectinase, α-amylase, and β-D-glucanase hydrolysates of polysaccharides from Bhutanese Cordyceps and C. sinensis were summarized in Table 2. The average correlation coefficients of partial acid (SMC-C), pectinase (SMC-P), α-amylase (SMC-A), and β-D-glucanase (SMC-B) hydrolysates of polysaccharides were 0.958 ± 0.021 (n = 18), 0.979 ± 0.011 (n = 18), 0.934 ± 0.024 (n = 18), and 0.965 ± 0.022 (n = 18), respectively. The data further supported that chemical structures of polysaccharides in Bhutanese Cordyceps and C. sinensis were similar.
In summary, this study suggests that fungus of Cordyceps from Bhutan is C. sinensis and the host insect belongs to Hepialidae sp. Their bioactive components, including nucleosides and their bases, and polysaccharides, in Bhutanese Cordyceps are greatly similar to those of C. sinensis, which is beneficial for the rational usage of Bhutanese Cordyceps.
Materials and Methods
Materials and chemicals
Twelve batches of natural Cordyceps were collected from Himalayas of Bhutan (CC1-CC12,), and six batches of natural C. sinensis (CS1-CS6) were collected from Tibet and Qinghai of China (Table 1). Identity of natural C. sinensis was confirmed by Professor Shao-Ping Li, University of Macau, Macau SAR, China. The voucher specimens were deposited at the Institute of Chinese Medical Sciences, University of Macau, Macao, China.
Cytosine, uracil, cytidine, guanine, hypoxanthine, adenine, uridine, thymine, 2′-deoxyuridine, inosine, guanosine, thymidine, adenosine, and cordycepin were purchased from Sigma (purity ≥ 99.0%, St. Louis, MO, USA). QIAGEN DNeasy plant mini kit, Promega Wizard SV genomic DNA purification system, PrimeSTAR HS DNA Polymerase, and Ex Taq DNA Polymerase were purchased from Takara Biotech Inc. Glucose, mannose, galactose, fucose, arabinose, starch (STN), α-amylase (EC 3.2.1.1), pectinase (EC 3.2.1.15), β-D-glucanase (EC 3.2.1.6), polygalacturonic acid (PGN), dextran (DEN) and acetic anhydride were purchased from Sigma (St. Louis, MO, USA). Laminaribiose (DP2), laminaritriose (DP3), laminaritetraose (DP4), laminaripentaose (DP5), laminarihexaose (DP6) and guar galactomannan (GGN) were purchased from Megazyme (Wicklow, Ireland). Polyacrylamide containing a ratio of acrylamide/N,N-methylenebisacrylamide (19:1, w/w) was obtained from Bio-Rad (Hercules, CA, USA). Silica gel 60 TLC plates were obtained from Merck (Merck, Darmstadt, Germany). Deionized water was prepared by a Millipore Milli-Q Plus system (Millipore, Bedford, MA, USA). All the other reagents were of analytical grade.
DNA barcoding analysis
DNA extraction, amplification and sequencing were performed according to our previously reported methods with modification39,40. In brief, specimens of Bhutanese Cordyceps were divided into stroma and host insect. Genomic DNA in stroma and host insect was then isolated using a QIAGEN DNeasy plant mini kit and a Promega Wizard SV genomic DNA purification system, respectively. The ITS regions of genomic DNA in stroma were amplified with a forward primer of ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and a reverse primer of ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The samples were amplified using an ABI Veriti PCR (Applied Biosystems, USA) under the following conditions, initial denaturation at 95 °C for 4 min, followed by 30 cycles of denaturation at 95 °C for 0.5 min, annealing at 52 °C for 0.5 min, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 7 min. In addition, both COI and Cytb sequences of genomic DNA in host insect were also amplified. The COI sequence was amplified with two primers including COI-F (5′-GGTCAACAAATCATAAAGATATTG-3′) and COI-R (5′-TAAACTTCAGGGTGACCAAAAAAT-3′), and the samples were amplified under the following conditions, initial denaturation at 95 °C for 4 min, followed by 35 cycles of denaturation at 95 °C for 0.5 min, annealing at 50 °C for 0.5 min, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 7 min. Moreover, the Cytb sequence was amplified with two primers including Cytb1 (5′-TATGTACTACCATGAGGACAAATATC-3′) and Cytb2 (5′-ATTACACCTCCTAATTTATTAGGAAT-3′), and the samples were amplified under the following conditions: initial denaturation at 95 °C for 4 min, followed by 40 cycles of denaturation at 95 °C for 0.5 min, annealing at 48 °C for 1.0 min, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 7 min. After all PCR products were confirmed by 1.5% agarose gel electrophoresis, the fragments were purified, and then sequenced with the help of INVITROGEN TRADING (SHANGHAI) CO., LTD. Finally, the sequences of ITS, COI and Cytb were blasted against the GenBank NCBI nucleotide database online (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), respectively.
Sample preparation
Samples were carefully cleaned without water by a small brush, dried at 40 °C for 24 h, and pulverized via grinding. Nucleosides and their bases in sample materials were extracted using boiling water extraction according to our previously reported method with minor modification28. In brief, the powder of Cordyceps (0.5 g) was mixed with 20 mL Milli-Q water in a glass tube, accurately weighted, and placed onto a Syncore Reactor (BUCHI-Syncore, Flawil, Switzerland) and heat reflux (100 °C) for 60 min. After extraction, the extract was cooled down to the room temperature, and made up the lost weight with water, then centrifuged (4000 × g for 5 min). The supernatant was filtered through a 0.45 μm Econofilter before HPLC-DAD analysis.
Water soluble polysaccharides were extracted with microwave assisted extraction according to a previously reported method with minor modification41. In brief, the powder of sample materials was immersed in water (20 mL), and extracted with microwave assisted extraction (Multiwave 3000, Anton paar GmbH, Graz, Austria). The microwave irradiation program was performed at 600 W and 90 °C for 20.0 min. Then the extract solution was centrifuged at 4000 × g for 10 min (Allegre X-15 centrifuge; Beckman Coulter, Fullerton, CA, USA). Subsequently, ethanol was added to the final concentration of 80% (v/v) for precipitation of crude polysaccharides. After standing for 12 h at 4 °C, centrifugation (4000 × g for 10 min) was performed. The precipitate was redissolved in 10 mL of hot water (60 °C). After centrifugation (4000 × g for 15 min), the supernatant was collected and the powder of the supernatant was obtained by freeze-drying.
HPLC-DAD analysis
Qualitative and quantitative analysis of nucleosides and their bases was performed on an Agilent Series 1200 (Agilent Technologies, USA) liquid chromatography system according to a previously reported method with minor modification29. In brief, a grace prevail select C18 column (4.6 × 150 mm, 3 μm) was used. The column temperature was maintained at 25 °C. The standards and samples were separated using a gradient mobile phase consisting of water (A) and acetonitrile (B). The gradient condition is: 0–6 min, 0% acetonitrile; 6–20 min: 0–5% acetonitrile; 20–30 min 5–25% acetonitrile. The flow rate was 1.0 mL/min and the injection volume was 10.0 μL. Peaks were detected at 260 nm of UV detection.
GC-MS analysis
Compositional monosaccharides of polysaccharides were investigated using GC-MS analysis according to a previously reported method with minor modification11. Briefly, the sample (~4.0 mg/mL, 0.5 mL) was hydrolyzed with 2.0 M trifluoroacetic acid under microwave irradiation (Multiwave 3000, Anton paar GmbH, Graz, Austria). The microwave irradiation program was performed at 300 W for 6 min. After hydrolysis, the hydrolysates were evaporated to dryness by using nitrogen and washed with methanol for three times to remove the residue of trifluoroacetic acid. Subsequently, 0.5 mL of pyridine and 10.0 mg of hydroxylamine hydrochloride were added and incubated at 90 °C for 30 min, 0.5 mL of acetic anhydride was then added and incubated at 90 °C for 30 min. Furthermore, the derivatives of mixed monosaccharide standards (Ara, Fuc, Gal, Glc, and Man, respectively) were prepared as described above. The derivatives were analyzed by using an Agilent 6890 gas chromatography instrument coupled to an Agilent 5973 mass spectrometer (Agilent Technologies, Palo Alto, CA) according to our previously reported method11. In brief, a capillary column (30 m × 0.25 mm, i.d.) coated with 0.25 μm film 5% phenyl methyl siloxane was used for separation. The column temperature was set at 165 °C and held for 7 min for injection, then programmed at 5 °C/min to 185 °C and held for 5 min, then at 4 °C/min to 200 °C, and finally at 20 °C/min to 280 °C, and held for 2 min.
Saccharide mapping analysis
Partial acid and enzymatic hydrolysis of refined polysaccharides
The crude polysaccharides of each sample (40.0 mg) were redissolved in 10.0 mL of hot water (60 °C). Then the low molecular weight compounds were removed by centrifugation (3500 × g, 25 min) with an ultra centrifugal filter (molecular weight cutoff: 3 kDa, Millipore, Billerica, MA, USA) for seven times. Finally, the concentration of crude polysaccharides in each sample was adjusted to the same concentration for further partial acid and enzymatic hydrolysis.
Polysaccharide solutions (~2.0 mg/500 μL) were treated with trifluoroacetic acid at a final concentration of 0.5 mol/L in a total volume of 1000 μL, and incubated at 80 °C for 5 h according to our previously reported method with minor modification34. After hydrolysis, the hydrolysates were washed with methanol and evaporated to dryness with a nitrogen evaporator at 35 °C for three times to remove the residue trifluoroacetic acid. The dried products were stored in 4 °C before derivatization, and redissolved in 100 μL of ethanol (70%, v/v) for HPTLC analysis, respectively.
In addition, polysaccharide solutions (~2.0 mg/ 500 μL) were mixed with selected enzyme (the final concentration of β-D-glucanase, α-amylase and pectinase was 2, 20 and 20 U/mL respectively) in a total volume of 1000 μL and digested overnight (16 h) at 40 °C. Then the mixtures were heated at 80 °C for 30 min to denature the enzymes. The supernatants were evaporated to dryness with a nitrogen evaporator and then were used for derivatization, and redissolved in 100 μL of ethanol (70%, v/v) for HPTLC analysis, respectively. Polysaccharide solution without enzymes, treated as described above, was used as blank control. Subsequently, the partial acid and enzymatic digestions were derivatized with ANTS at 37 °C for 17 h according to a previously reported method41.
Saccharide mapping based on PACE analysis
All samples (1–8 μL depending of the sugar concentration) were separated using a vertical slab gel electrophoresis apparatus, Mini-Protean Tetra System (Bio-Rad, Hercules, CA, USA) according to a previously reported method34. In brief, electrophoresis of 30% (w/v) polyacrylamide in the resolving gel with a stacking gel of 8% (w/v) polyacrylamide was used for the separation of partial acid and enzymatic hydrolysates, respectively. The samples were electrophoresed first at 200 V for 15 min and then at 700 V for 45 min, to move bromophenol blue (migration indicator) to the desired level. Gels were imaged using an InGenius LHR CCD camera system (Syngene, Cambridge, UK) under UV 365 nm.
Saccharide mapping based on HPTLC analysis
All the samples (4–10 μL depending of the sugar concentration) were separated on a silica gel 60 plate with an AS30 HPTLC Applicator (Desaga GmbH, Germany) according to a previously reported method34. In brief, the bands were 8 mm wide, 13 mm distance, and 10 mm from the bottom edge. Then the plate was firstly developed to a distance of 95 mm with 1-butanol/isopropanol/acetic acid/water, 7:5:1:2 (v/v/v/v) as mobile phase at room temperature. Then the plate was dried and placed in the same chamber to develop a distance of 95 mm with the same mobile phase as described above. Finally, the developed plates were dried and colorized with aniline-diphenylamine-phosphoric acid solution, then heated at 105 °C for 10 min on a YOKO-XR plate heater (Wuhan YOKO technology Ltd., China) and photographed under white night.
HPSEC-MALLS-RID analysis
The molecular weights (Mw), molecular weight distributions and contents of polysaccharides and their fractions in Bhutanese Cordyceps and C. sinensis were measured using HPSEC-MALLS-RID according to a previously reported method with minor modification36. In brief, HPSEC-MALLS-RID measurements were carried out on a multi-angle laser light scattering (DAWN HELEOS, Wyatt Technology Co., Santa Barbara, CA, USA) with an with an Agilent 1100 series LC/DAD system (Agilent Technologies, Palo Alto, CA, USA) equipped with a column of TSK-Gel G5000PWXL (300 mm × 7.8 mm, i.d.) and TSK-Gel G3000pwXL (300 mm × 7.8 mm, i.d., Tosoh Bioscience, Tokyo, Japan) in series at 35 °C. A refractive index detector (RID, Optilab rEX refractometer, DAWN EOS, Wyatt Technology Co., Santa Barbara, CA, USA) was simultaneously connected. The Mw was calculated by the Zimm method of static light scattering based on the basic light scattering equation according to our previously reported method11. The content of polysaccharides was calculated based on the refractive index difference with dn/dc value according to the following equation42,

where Ci is the concentration of polymers; α is the RID calibration constant (in RI units per volt), which is determined as 3.4756 × 10−5 RIU/pixel using the aqueous solutions of reference standard (sodium chloride); Vi and Vi, baseline are the RID voltages of sample and baseline, respectively; dn/dc is the specific refractive index increment of polysaccharides, which is defined as 0.15 mL/g according to our previously reported method36.
The mobile phase was 0.9% NaCl aqueous solution at a flow rate of 0.5 mL/min. All of polysaccharide solutions were filtered by a 0.22 μm membrane before use. The injection volume was 50 μL for each sample. The Astra software (Version 6.0.2, Wyatt Tech. Corp. Santa Barbara, CA, USA) was utilized for data acquisition and analysis.
Data analysis
Hierarchical cluster analysis (HCA) was performed by using Origin86 software, the nearest neighbor cluster method with euclidean distance type was selected as measurement for hierarchical clustering analysis. In addition, the optical densities of bands in electronic images and digital scanning profiles of PACE analysis were generated and analyzed using Quantity-One software (version 4.6.2, Bio-Rad, Hercules, USA). The similarities of the tested samples, as well as the simulative mean chromatogram were calculated and generated using the professional software named “Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine” (Matlab version, Ver1.315, developed by the Research Center of Modernization of Chinese Herbal Medicine, Central South University, and the Hong Kong Polytechnic University).
Additional Information
How to cite this article: Wu, D.-T. et al. Cordyceps collected from Bhutan, an appropriate alternative of Cordyceps sinensis. Sci. Rep. 6, 37668; doi: 10.1038/srep37668 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zhang, Y., Li, E., Wang, C., Li, Y. & Liu, X. Ophiocordyceps sinensis, the flagship fungus of China: terminology, life strategy and ecology. Mycology 3, 2–10 (2012).
Hu, X. et al. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 58, 2846–2854 (2013).
Li, Y. et al. Complete mitochondrial genome of the medicinal fungus Ophiocordyceps sinensis. Sci. Rep. 5 (2015).
Li, Y. et al. A survey of the geographic distribution of Ophiocordyceps sinensis. J. Microbiol. 49, 913–919 (2011).
Zhu, J. S., Halpern, G. M. & Jones, K. The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis Part I. J. Altern. Complem Med. 4, 289–303 (1998).
Zhu, J. S., Halpern, G. M. & Jones, K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis–Part II. J. Altern. Complem Med. 4, 429–457 (1998).
Mi, J. N., Wang, J. R. & Jiang, Z. H. Quantitative profiling of sphingolipids in wild Cordyceps and its mycelia by using UHPLC-MS. Sci. Rep. 6 (2016).
Li, S. P., Yang, F. Q. & Tsim, K. W. K. Quality control of Cordyceps sinensis, a valued traditional Chinese medicine. J. Pharm. Biomed. Anal. 41, 1571–1584 (2006).
Shashidhar, M. G., Giridhar, P., Sankar, K. U. & Manohar, B. Bioactive principles from Cordyceps sinensis: A potent food supplement–A review. J. Funct. Food. 5, 1013–1030 (2013).
Paterson, R. R. M. Cordyceps–A traditional Chinese medicine and another fungal therapeutic biofactory? Phytochemistry 69, 1469–1495 (2008).
Wu, D. T. et al. Chain conformation and immunomodulatory activity of a hyperbranched polysaccharide from Cordyceps sinensis. Carbohydr. Polym. 110, 405–414 (2014).
Ng, T. B. & Wang, H. X. Pharmacological actions of Cordyceps, a prized folk medicine. J. Pharm. Pharmacol. 57, 1509–1519 (2005).
Winkler, D. Caterpillar fungus (Ophiocordyceps sinensis) production and sustainability on the Tibetan Plateau and in the Himalayas. Asian Medicine 5, 291–316 (2009).
Winkler, D. Yartsa Gunbu (Cordyceps sinensis) and the fungal commodification of Tibet’s rural economy. Econ. Bot. 62, 291–305 (2008).
Cannon, P. F. et al. Steps towards sustainable harvest of Ophiocordyceps sinensis in Bhutan. Biodivers. Conserv. 18, 2263–2281 (2009).
Liu, H.-j., Hu, H.-b., Chu, C., Li, Q. & Li, P. Morphological and microscopic identification studies of Cordyceps and its counterfeits. Acta pharm. Sinica B 1, 189–195 (2011).
Seifert, K. A. Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 9, 83–89 (2009).
Xiang, L. et al. DNA barcoding the commercial Chinese caterpillar fungus. FEMS Microbiol. Lett. 347, 156–162 (2013).
Schoch, C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246 (2012).
Maczey, N., Dhendup, K., Cannon, P., Hywel-Jones, N. & Rai, T. B. Thitarodes namnai sp nov and T. caligophilus sp nov (Lepidoptera: Hepialidae), hosts of the economically important entomopathogenic fungus Ophiocordyceps sinensis in Bhutan. Zootaxa, 42–52 (2010).
Quan, Q. M. et al. Comparative phylogenetic relationships and genetic structure of the caterpillar fungus Ophiocordyceps sinensis and its host insects inferred from multiple gene sequences. J. Microbiol. 52, 99–105 (2014).
Wang, X. L. & Yao, Y. J. Host insect species of Ophiocordyceps sinensis: a review. Zookeys, 43–59 (2011).
Ralevic, V. & Burnstock, G. Receptors for purines and pyrimidines. Pharmacol. Rev. 50, 413–492 (1998).
Jacobson, K. A., Jarvis, M. F. & Williams, M. Purine and pyrimidine (P2) receptors as drug targets. J. Med. Chem. 45, 4057–4093 (2002).
Zhao, J., Xie, J., Wang, L. Y. & Li, S. P. Advanced development in chemical analysis of Cordyceps. J. Pharm. Biomed. Anal. 87, 271–289 (2014).
Yang, F. Q. & Li, S. P. Effects of sample preparation methods on the quantification of nucleosides in natural and cultured Cordyceps. J. Pharm. Biomed. Anal. 48, 231–235 (2008).
Yang, F.-Q., Ge, L., Yong, J. W. H., Tan, S. N. & Li, S.-P. Determination of nucleosides and nucleobases in different species of Cordyceps by capillary electrophoresis–mass spectrometry. J. Pharm. Biomed. Anal. 50, 307–314 (2009).
Yang, F. Q., Li, D. Q., Feng, K., Hu, D. J. & Li, S. P. Determination of nucleotides, nucleosides and their transformation products in Cordyceps by ion-pairing reversed-phase liquid chromatography–mass spectrometry. J. Chromatogr. A 1217, 5501–5510 (2010).
Fan, H. et al. Qualitative and quantitative determination of nucleosides, bases and their analogues in natural and cultured Cordyceps by pressurized liquid extraction and high performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC–ESI–MS/MS). Anal. Chim. Acta 567, 218–228 (2006).
Li, S. P., Wu, D. T., Lv, G. P. & Zhao, J. Carbohydrates analysis in herbal glycomics. Trac-Trends Anal. Chem. 52, 155–169 (2013).
Hu, D. J., Cheong, K. L., Zhao, J. & Li, S. P. Chromatography in characterization of polysaccharides from medicinal plants and fungi. J. Sep. Sci. 36, 1–19 (2013).
Guan, J., Yang, F. Q. & Li, S. P. Evaluation of carbohydrates in natural and cultured Cordyceps by pressurized liquid extraction and gas chromatography coupled with mass spectrometry. Molecules 15, 4227–4241 (2010).
Wu, D. T. et al. Characterization of bioactive polysaccharides from Cordyceps militaris produced in China using saccharide mapping. J. Funct. Food. 9, 315–323 (2014).
Wu, D. T. et al. Characterization and discrimination of polysaccharides from different species of Cordyceps using saccharide mapping based on PACE and HPTLC. Carbohydr. Polym. 103, 100–109 (2014).
Guan, J., Zhao, J., Feng, K., Hu, D. J. & Li, S. P. Comparison and characterization of polysaccharides from natural and cultured Cordyceps using saccharide mapping. Anal. Bioanal. Chem. 399, 3465–3474 (2011).
Cheong, K.-L., Wu, D.-T., Zhao, J. & Li, S.-P. A rapid and accurate method for the quantitative estimation of natural polysaccharides and their fractions using high performance size exclusion chromatography coupled with multi-angle laser light scattering and refractive index detector. J. Chromatogr. A 1400, 98–106 (2015).
Pattathil, S. et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525 (2010).
Lo, T. C. T., Kang, M. W., Wang, B. C. & Chang, C. A. Glycosyl linkage characteristics and classifications of exo-polysaccharides of some regionally different strains of Lentinula edodes by amplified fragment length polymorphism assay and cluster analysis. Anal. Chim. Acta 592, 146–153 (2007).
Chen, S. Y. et al. Molecular identification of Metacordyceps Liangshanensis, its adulterants and its relative species based on DNA barcode. World Science and Technology/Modernization of Traditional Chinese Medicine and Materia Medica 16, 1336–1346 (2014).
Chen, S. Y. et al. Feasibility analysis on mitochondrial COI and Cytb gene used for identification of paratitic insect of Cordyceps. World Science and Technology/Modernization of Traditional Chinese Medicine and Materia Medica 17, 182–188 (2015).
Wu, D. T., Xie, J., Hu, D. J., Zhao, J. & Li, S. P. Characterization of polysaccharides from Ganoderma spp. using saccharide mapping. Carbohydr. Polym. 97, 398–405 (2013).
Podzimek, S. In Light Scattering, Size Exclusion Chromatography and Asymmetric Flow Field Flow Fractionation 37–98 (John Wiley & Sons, Inc., 2011).
Acknowledgements
This research was partially supported by grants from the Science and Technology Development Fund of Macao (FDCT059/2011/A3), and the University of Macau (MYRG2014-00041 and MYRG2015-00202) to S.P. Li and MYRG2015-00122 to J. Zhao.
Author information
Authors and Affiliations
Contributions
S.-P.L., J.Z., and S.-C.M. conceived and designed the research. D.-T.W., G.-P.L., J.Z., and Q.L. conducted the experiments. S.-P.L., J.Z., S.-C.M., D.-T.W., and G.-P.L. analyzed the data. S.-P.L. and D.-T.W. drafted the manuscript, and S.-P.L. corrected the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wu, DT., Lv, GP., Zheng, J. et al. Cordyceps collected from Bhutan, an appropriate alternative of Cordyceps sinensis. Sci Rep 6, 37668 (2016). https://doi.org/10.1038/srep37668
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37668
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.