Abstract
The entomopathogenic nematode Steinernema carpocapsae has been widely used for the biological control of insect pests. It shares a symbiotic relationship with the bacterium Xenorhabdus nematophila, and is emerging as a genetic model to study symbiosis and pathogenesis. We obtained a high-quality draft of the nematode’s genome comprising 84,613,633 bp in 347 scaffolds, with an N50 of 1.24 Mb. To improve annotation, we sequenced both short and long RNA and conducted shotgun proteomic analyses. S. carpocapsae shares orthologous genes with other parasitic nematodes that are absent in the free-living nematode C. elegans, it has ncRNA families that are enriched in parasites, and expresses proteins putatively associated with parasitism and pathogenesis, suggesting an active role for the nematode during the pathogenic process. Host and parasites might engage in a co-evolutionary arms-race dynamic with genes participating in their interaction showing signatures of positive selection. Our analyses indicate that the consequence of this arms race is better characterized by positive selection altering specific functions instead of just increasing the number of positively selected genes, adding a new perspective to these co-evolutionary theories. We identified a protein, ATAD-3, that suggests a relevant role for mitochondrial function in the evolution and mechanisms of nematode parasitism.
Similar content being viewed by others
Introduction
Global losses due to pests can vary from about 26 to 80% depending on the type of crop1. Chemical pesticides are commonly used to fight this problem, however, they pose threats to humans, wildlife, and might have an adverse impact on soil fertility by killing beneficial microorganisms2. Other strategies rely on biological control agents, but their use is not generalized because of their limited efficiency when compared to pesticides. Genetic improvements are possible, especially when genomic information of the biological agent is available3,4. Entomopathogenic nematodes (EPNs) from the family of Steinernematidae have been commercialized in many countries as a biological insecticide for agricultural and horticultural crops and have attracted considerable attention because they are also potential models for symbiosis and pathogenesis5. One of the most well-known is Steinernema carpocapsae that shares a symbiotic relationship with the bacterium Xenorhabdus nematophila. Since it was thought that the bacteria were the main contributor to insect death, most research has focused on the pathogenic effect of the bacteria rather than the nematode6,7,8. Nevertheless, growing evidence suggests a more active role of the nematode in the pathogenic process9,10. In fact, a set of expanded gene families that are likely involved in parasitism were predicted in a recent genome analysis of Steinernema species11. Further genomic characterization will help to better understand the evolution and the function of these genomes in the symbiotic and pathogenic contexts. Parasitism is a common way of life among nematodes that has independently arisen at least 15 times during their evolution12. Particularly interesting are the phylogenetic associations between non-vertebrate and vertebrate parasites. The entomopathogenic Steinernematidae are phylogenetically related to Strongyloidoidae (Tylenchina; Panagrolaimomorpha), which infect mammals, suggesting a transition to vertebrate parasitism through host shifting12. The study of parasitism in S. carpocapsae should help to understand the origin and mechanisms of Strongyloidoids parasites, with implications for human health. For this study we produced a high-quality draft of the genome of S. carpocapsae strain Breton, and compared it with a recently published genome from a different strain of this species11. We further assessed the genetic signatures of its adaptation to a pathogenic lifestyle, and characterized the transcriptome by RNA-Seq, including both messenger RNA (mRNA), and small RNA (sRNA). We also present the most complete characterization to date of the proteome, generated by shotgun proteomics, two-dimensional gel electrophoresis (2DE) and SDS-PAGE. Additionally we conducted genome-wide scans for signatures of natural selection. We found several distinctive features related to pathogenesis through a comparison with both pathogenic and free-living nematodes.
Results and Discussion
Genome sequencing
Total DNA was extracted from isolated nuclei from a near isogenic line (~96% of estimated homozygosity) of Steinernema carpocapsae strain Breton. The use of isolated nuclei reduces the amount of symbiont and mitochondrial DNA, and the isogenic line was generated to avoid the acknowledged problems posed by heterozygosity for accurate genome assembly12. From one 454 shotgun library sequenced in three 454 FLX runs, we obtained 3,340,915 total reads with an average length of 357 bp. From one 454 paired-end library, with an insert size of 8 Kb, sequenced in two 454 FLX runs, we obtained 2,784,713 total reads with an average read length of 334 bp at each fragment end. From a SOLiD shotgun library sequenced in half a lane of SOLiD 5500xl, we obtained 24,942,584 reads of 75 bp (Table 1). By combining these long, paired-end, and short reads, we obtained a coverage of 32-fold, considering a genome size of ~110 Mb estimated by both flow cytometry and genome assembly. The final draft consists of 84,613,633 base pairs in 347 scaffolds, with an N50 of 1.24 Mega bases and with the largest scaffold of 8.7 Mb. This represents a notable improvement over a recently published genome that is more fragmented, with a much lower N50 (~0.3 Mb) and with the largest scaffold of only 1.7 Mb (Table 2). The average GC-content was of 45.67%, with 6.99% of repetitive sequences (Supplementary Table S1).
We assessed the completeness of the genome by analysing 248 ultra-conserved core eukaryotic genes13, obtaining 99.6% completeness considering partial genes and 99.2% for complete genes. These parameters indicated that our draft genome is of high quality, which gives us confidence in the genome annotation described below.
Genome annotation
From the repetitive elements, we identified 1,702 distinct retrotransposon sequences representing at least eight families. Four were long interspersed element (LINE) groups, Cr1 being the most abundant, and 588 were short interspersed elements (SINEs), of which 432 belong to the tRNA-RTE family. We identified only two long terminal repeats (LTRs): Gypsy and Pao. We also identified eight families of DNA transposons, comprising 1,202 sequences, of which hAT-Ac was the most abundant with 388 elements, followed by TcMar-Tc1, Merlin, and the rolling-circle Helitron (327, 106, and 105 elements, respectively).
We collected RNA from pooled nematodes taken from all life cycle stages and subjected to various conditions (growing in larvae of two different insect species and on two different in vitro media, as described in Materials and Methods) in order to maximize the inclusion of condition-specific genes. We obtained 15,180,085 reads with an average length of 201 bp from an Illumina paired-end library on a MiSeq, and 92,231 reads with an average length of 288 bp from a 454 library on a partial 454 FLX + plate. After quality filtering, 94.93% of the reads mapped to the masked genome, suggesting a good reliability of the genome assembly. We performed genome-guided de novo assembly of the transcriptome that resulted in 21,457,711 bp of assembled transcripts (without introns). In order to identify protein-coding genes in the assembled genome, we assigned specific weights to different types of evidence to generate consensus gene calls (see Material and Methods). The current genome sequence and annotation is available at www.genomevolution.org (ID 33774), and at the NCBI GenBank (BioProject ID# 39853).
We identified 16,333 protein-coding genes with an average length of 1,257 bp, an average exon length of 222.37 bp, and an average of six exons per gene. We also identified 6,708 alternative transcripts and 5,725 truncated genes (defined as predicted protein-coding genes missing a start codon). We verified the protein expression of 3,773 predicted genes through mass spectrometry analysis (see below and Supplementary Table S2). The total number of predicted genes in this study is much lower than the previously predicted number of genes (28,313) for the strain “All” of the same species11. In the previous study, the heterozygosity was not reduced through the generation of an isogenic line, potentially negatively impacting on their genome assembly12. In addition, they performed gene prediction using Augustus14 with parameters optimized only for Caenorhabditis elegans. However, these species diverged ~280 million years ago15, making it difficult to accurately predict genes in Steinernema by solely using C. elegans gene models. To overcome this bias, we combined predictions using Hidden Markov Models (HMM) trained on S. carpocapsae gene structures with ab initio predictions, along with HMM homology-based predictions using C. elegans genes and Brugia malayi gene predictions (all the predictions were obtained with Augustus14). Although B. malayi has the same estimated time of divergence from S. capocapsae as C. elegans15, it is a parasite and therefore might share some homologous genes with S. carpocapsae that are not represented in C. elegans. However, the full strength of our approach is given by combining predictions using gene models from these species with ab initio predictions. When we used the same annotation strategy as in Dillman et al.11 using C. elegans models, we only obtained 14,188 protein coding genes. This is lower than the previous study, and also lower to the one we obtained with our combined strategy, in which the number of predicted genes probably increased due to the inclusion of B. malayi models, along with the ab initio predictions.
In summary, the use of an isogenic line (see Material and Methods), the combination of different sequencing platforms (Table 1), and an improved annotation strategy, resulted in a higher quality genome compared to a recent publication (Table 2). In any case, the studies used different strains of the nematode, and their goals were different, with Dillman et al.10 focusing on comparing their genome to that of other species of Steinernema. In our study, we obtained a higher quality genome, included analyses of the proteome and small RNAs, and performed a genome-wide scan of positive selection.
The most abundant GO terms in predicted genes are shown in Fig. 1. Our analysis revealed 135 enriched GO terms in S. carpocapsae when compared to those in C. elegans (Fisher’s exact test, FDR ≤ 0.05) (Supplementary Table S3). Many of these GO terms are involved in degradation, protein modification, binding and transport, and could be associated to parasitism (reviewed in ref. 16). At least 22 GO terms are also enriched in at least two other pathogenic worms, but not in the free-living nematode Pristionchus pacificus (Table 3). Supplementary Table S4 shows the abundance of the different protein families in 10 different nematode genomes compared with the 30 most abundant families in S. carpocapsae (Supplementary Fig. S1). Most of the expanded families (16 out of 20) identified previously in the “All” strain of S. carpocapsae11 are also overrepresented in our strain (Breton). In addition, Peptidase S1 is also overrepresented in S. carpocapsae and other parasites (Bursaphelenchus xylophilus, Meloidogyne hapla, and M. incognita) compared to C. elegans. Integrase, enoyl-acyl-carrier-protein reductase (ENR) (IPR014358), retrotransposon pao, and pimelyl-acyl-carrier protein methyl ester esterase PFAM domains, are also overrepresented in some parasites, including S. carpocapsae.
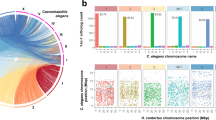
Since C. elegans and S. carpocapsae are phylogenetically distant from one another, we found no evident macro-synteny. However, there are genes located in single chromosomes of C. elegans that match genes located in single scaffolds of S. carpocapsae. A similar result was obtained in a comparison with the Brugia malayi genome (version WS253) (Fig. 2 and Table 4), even though this genome is not of the same quality as that of C. elegans. This reinforces the idea that the use of B. malayi gene models in the annotation strategy is at least as good as the use of C. elegans models.
Beyond the protein-coding potential of the genome, we predicted non-coding RNA (ncRNA) using a variety of tools (see Materials and Methods), identifying 1,097 tRNAs, 40 rRNAs (15 5S rRNA, 1 5.8S rRNA and 24 8S rRNA), 38 micro-RNA hairpins and 146 other ncRNAs. Using the same annotation pipeline, we compared the abundance of each ncRNA family in parasitic (Ascaris suum, Bursaphelenchus xylophilus, Brugia malayi, S. carpocasae, M. incognita, M. hapla and Heterorhabditis bacteriophora) and free-living (Panagrellus redivivus, Pristionchus pacificus, C. remanei and C. elegans) nematodes (Supplementary Table S5). By comparing the average number of elements in each family between parasitic and free-living nematodes, we derived a simple metric to decide if a family had a tendency to be enriched in one of the two lifestyles (see Materials and Methods). The families enriched in parasitic nematodes are, ACEA_U3 (a snoRNA), SeC (a tRNA), mir-100/mir-10, mir-227, mir-2b, mir-2444, and mir-4455 (microRNAs), all of which have at least twice the number of elements on average in the parasites (Supplementary Fig. S2). Although the correlation between these families and parasitism needs to be further investigated, this is a first indication that these ncRNA families might have a functional role in the pathogenic lifestyle.
To complement these bioinformatic predictions we performed small RNA-seq of S. carpocapsae with and without induction with insect hemolymph. We obtained a total of 42.8 million reads from 6 libraries. Less than 10% of the cleaned reads failed to map to the genome (see Materials and Methods), another indication that the genome assembly is very complete. We used two of the most popular tools to annotate known and novel microRNAs using small RNA sequencing data: miRDeep17 and ShortStack18. Both tools coincided in predicting 100 miRNAs, while miRDeep predicted an additional 162, and ShortStack 25 more, giving a total of 287 miRNA hairpins, each with a potential 5′ and 3′ mature product (Supplementary Table S6). These predictions followed an expected length distribution, with a dominant peak centred at 22 nucleotides. Of the sRNA sequencing reads of 20–24 nucleotides that mapped to the genome, 83% overlapped with the 287 miRNA hairpins (Supplementary Fig. S3). Interestingly, we detected a large number of novel miRNA genes, since only 25 out of the 287 predicted miRNA hairpins correspond to known miRNAs according to homology searches. Although the majority of the conserved miRNAs tend to have high expression in our experiments, half of the 20 most highly expressed miRNA predictions correspond to novel sequences (Supplementary Table S7). This confirms the great diversity of ncRNA genes that are species or lineage specific, and highlights the importance of using experimental data when annotating genomes, particularly for species that are distant to well annotated model organisms.
We were also interested to see if any of the microRNAs that we detected changed their expression in response to insect haemolymph. None of the miRNAs showed a significant decrease in expression, but five increased their expression after hemolymph induction (Supplementary Table S7 and Supplementary Fig. S4). These miRNAs were miR-84–3p, miR-84-5p, miR-31-3p, let-7-5p and Cluster_21397_3p (a new prediction with no similarity to known miRNAs). Interestingly, the induced miRNAs included miR-84 and let-7, members of the let-7 family of miRNAs that are important players during development. In Caenorhabditis elegans, double mutants of miR-84 and miR-48 (another let-7 family member) show a delayed moulting phenotype and accumulate a double cuticle19. This is interesting because S. carpocapsae infective juveniles have a double cuticle that is lost upon entering the insect host8. The up-regulation of miR-84 and let-7 could thus be involved in the moulting process, triggered by the contact with insect hemolymph.
Differentially expressed proteins
During infection, the nematodes first invade the insect intestine and then cross the intestinal wall by expressing putative effectors that facilitate parasite penetration to the hemocoel, where they continue to counteract insect defences10,20. Therefore, we were interested in comparing the soluble proteins from Infective Juveniles (IJs) induced with either insect intestines or insect hemolymph, against non-induced controls. We opted for a detection strategy combining shotgun proteomics strategy, with two-dimensional electrophoresis (2DE), and SDS-PAGE, that resulted in the identification of 7,527 proteins. By eliminating duplicates (proteins that were detected more than once), we obtained 3,773 non-redundant proteins (Supplementary Table S2). Among the non-redundant proteins, 1,625 were expressed in the three conditions, 155 were only expressed under both hemolymph and intestine induction, and 349 were expressed in the control and one other condition. In addition, 489 proteins were exclusively expressed in nematodes induced with intestine, 510 in those induced with hemolymph, and 645 in the non-induced controls (Fig. 3). This suggests that specific activities occur at different stages during the pathogenic process. Proteins expressed specifically in the induced conditions, were associated with functional categories (GO terms) using Blast2GO (Supplementary Figs S5 and S6). We found four GO terms enriched among the 489 proteins expressed exclusively in the intestine-induced sample, 10 GO terms in the 510 proteins of the hemolymph-induced sample, and 8 GO terms in the 1,154 combined proteins from both induction conditions (Fisher’s exact test, p < 0.05); in all cases in relation to the untreated control (Supplementary Table S8). One of the differentially expressed proteins was a transthyretin-like protein (TLP). Unlike transthyretins, which are known for transporting thyroxine and related molecules, the function of TLPs is not well understood21. They seem to have a role in the uricase reaction pathway as 5-hydroxyisourate hydrolases22. Their abundance in parasitic nematodes and more specifically their expression during parasitic stages, suggests an involvement of TLPs in parasitism22,23. We also found other differentially expressed peptides in the parasitic stages of S. carpocapsae that, being found in other parasites, could be involved in the infective processes (Supplementary Table S2).
Proteases and excretory/secretory proteins
We found all types of proteases in the genome of S. carpocapsae. Proportions of most of them (Aspartic, Cysteine, and Threonine) are similar to the proportions found in free-living (C. elegans), necromeric (P. pacificus), and parasitic (B. malayi, Strongyloides ratti) nematodes. However, the amount of serine proteases in S. carpocapsae and P. pacificus genomes is higher than in other nematodes (39% and 33.2% of all proteases, respectively); while in S. carpocapsae the percentage of metalloproteases, although high (31%), is the lowest in the comparison (the highest is in the Strongyloides ratti genome with 52%).
Excretory/secretory (ES) products are complex mixtures of hundreds of different proteins that are thought to have important roles in the life cycle of a parasite and during host-parasite interactions24. A total of 1,421 putative excreted proteins were predicted in the genome (see Methods), including various families of proteases, protease inhibitors, cuticular collagens, and C-type lectins, as well as putative signalling molecules such as warthog, ground and ground-like proteins (Supplementary Table S9). Some ES proteins are predicted to be involved in immuno-evasion; such as collagen25, whereas others play crucial roles in the suppression of host immune responses by mimicking host molecules, such as C-lectin26,27. Furthermore, C-lectins have been found to be upregulated in Ancylostoma ceylanicum at the onset of heavy blood feeding from the host28. We also found three putative copies of parasitic stage specific protein 1, a protein without known domains, which is present in several parasitic nematodes and is expressed during the transition to the parasitic lifestyle in Haemonchus contortus29. Other interesting putative secreted peptides in the S. carpocapsae genome were lipases, saposins, and transthyretin-like protein, all of which are expressed during early parasitic stages in H. contortus29. Serine proteases, including Sc-SP-1 and Sc-SP-3, can mediate the invasion or apoptosis of host cells10,20. Astacin metalloprotease is one of the effector molecules involved in tissue invasion of parasitic nematodes30. The family of papain-type aspartic and cysteine proteases are thought to have the same role in invertebrate digestion as trypsin in vertebrates31. Therefore, it is possible that the dependence of S. carpocapsae on aspartic protease activities is related to the digestion of nutrients. Cysteine proteases are involved in digestive processes or moulting and cuticle renewal in free-living and parasitic nematodes32,33.
Orthologous proteins
We compared 7,724 orthologous groups of proteins among several species with different lifestyles. We found 318 orthologous groups that are absent in non-pathogenic species (Caenorhabditis angaria, C. remanei, C. briggsae, C. japonica, C. elegans, and Pristionchus pacificus) but present in S. carpocapsae and in at least another pathogenic nematode (Supplementary Table S10). The annotations of these orthologues revealed enrichment of protein functions with possible associations with parasitism, such as serine proteases and other terms related to degradation and binding (Table 5). We also found 134 additional groups from the orthoMCL-DB (version 5) database that are present in the genome of S. carpocapsae but not in the other tested species (Supplementary Table S11).
Positive selection
Because of the co-evolutionary arms-race relationship between hosts and their pathogens, genes involved in their interaction are expected to evolve under positive selection34, potentially resulting in specific genomic signatures associated with their lifestyles35. We used the branch-sites test of positive selection36,37 to analyse 2,034 orthologous genes in three species of Clade IV nematodes (as defined in ref. 38). This test is based on a maximum likelihood estimation of the nucleotide nonsynonymous and synonymous substitutions rates. The ratio of nonsynonymous to synonymous rates (ω) can be used to identify purifying selection (ω < 1), neutral evolution (ω = 1), or positive selection (ω > 1), assessing the significance with a Likelihood Ratio Test (LRT)36. We found 83 genes with sites evolving under positive selection (ω > 1, LRT, p < 0.05; 14 of which had an FDR < 0.1) in S. carpocapsae (Table 6). Among the 83 genes, 23 GO terms were significantly enriched (Supplementary Table S12) when compared to the genes with no sites under positive selection (1,951 genes) (Fisher’s exact test, p < 0.01). Although we found more genes (95) with sites evolving under positive selection in the free-living nematode Panagrellus redivivus, there were no enriched GO terms among them (even with an alpha value of 0.05), indicating that the consequence of an arms-race relationship is better characterized by positive selection preferentially altering genes of specific functions than just increasing the number of positively selected genes. Although we would need to increase the number of analysed genes to increase the power of these analyses, the initial results suggest a new perspective to the co-evolutionary arms-race theories.
Phylogenetic analysis
To explore the phylogenetic relationships of S. carpocapsae, we reconstructed a phylogeny using 245 proteins from strictly 1-1 orthologous genes from nine nematode species. According to Blaxter et al.38, Steinernema is phylogenetically closer to Strongyloides than to Caenorhabditis, as inferred from a tree reconstructed using the small subunit ribosomal DNA (18S) sequences from 53 nematode species. A similar result was obtained in a more extensive analysis using 339 18S sequences39. However, Montiel et al.40 found Steinernema to be closer to Caenorhabditis than to Strongyloides using complete mtDNA sequences. Although this discrepancy may result from differential reproductive strategies and/or differential selective pressures acting on nuclear and mitochondrial genes40, an analysis of large subunit ribosomal DNA sequences (28S) also showed Steinernema to be closer to Caenorhabditis41. Defining these relationships is relevant because if Steinernema is phylogenetically closer to Strongyloides, it could be used as a more general model for parasitism, with implications for human health. Steinernema is more tractable than Strongyloides because it does not require a vertebrate host to reproduce in the laboratory. Our new phylogenetic analysis supports Steinernema being closer to Strongyloides than to Caenorhabditis (Fig. 4). In addition, its basal position in relation to Strongyloides, gives support to the hypothesis that this vertebrate parasite originated by host shifting from an entomopathogenic ancestor12, in this case Steinernema.
Gene functions enriched at several levels
To assess how the pathogenic lifestyle is affecting specific gene functions at different levels, we compared the GO terms enriched in the genome of S. carpocapsae when compared to C. elegans (Supplementary Table S3), with those in differentially expressed proteins due to hemolymph or intestine induction (Supplementary Table S8), and with those in genes with putative sites evolving under positive selection (Supplementary Table S12). One GO term was enriched in the genome and in differentially expressed proteins (transcription factor activity – sequence-specific DNA binding); and two GO terms in both the genome and genes under positive selection (macromolecular complex and ribonucleoprotein complex). No GO terms were shared between the three analyses. However, one protein, ATAD-3 (ATPase family AAA domain-containing protein 3), is associated with 33 enriched GO terms in the annotated sequences, and two additional enriched GO terms from genes with evidence of positive selection (Supplementary Table S13). This protein is differentially expressed in nematodes induced with insect tissues (intestines) and presents amino acid sites evolving under positive selection. Functions or proteins shared between these analyses might reveal relevant effects of the pathogenic lifestyle in the genome. A deeper analysis of positive selection (i.e. including more orthologous genes or conducting population genetic analyses) could expand the number of shared genes, which should be good candidates for further studies. In this case we have identified a putative homolog of C. elegans’ ATAD-3. Its deficiency in C. elegans causes early larval arrest, gonadal dysfunction, and embryonic lethality. It is also associated with defects in organellar structure and mtDNA depletion42,43, suggesting that ATAD-3 is important for increased mitochondrial activity during the transition to later larval stages42. S. carpocapsae needs to go through developmental changes to establish itself in the insect body during the pathogenic process, which might explain the relevance of this, and probably other mitochondria-related genes, in nematode parasitism. For example, the defective mitochondrial respiration family member protein 1, with functions in regulation of growth rate, was differentially expressed in nematodes induced with insect tissues (hemolymph) and presented evidence of positive selection. Mitochondria have been identified as important contributors to the virulence of fungal pathogens44, and it has been previously hypothesised that differential selective constraints in mitochondrial genes might explain discrepancies between nuclear and mitochondrial gene phylogenies in nematodes40. In addition, depletion of ATAD-3 in C. elegans resulted in reduced intestinal fat storage42, and it would be interesting to explore if fat metabolism might also be relevant in nematode parasitism.
Conclusion
Our genomic analyses of S. carpocapsae confirm a role in pathogenicity beyond simply vectoring the symbiotic bacteria. S. carpocapsae shares orthologous genes with other parasitic nematodes that are absent in the free-living nematode C. elegans, it encodes ncRNA families that are enriched in parasites, and presents putative proteins associated with functions related to parasitism and pathogenesis. Until now, the best examples of positive selection in genes related to host-pathogen interactions were pathogen effectors and genes of the host immune and defence systems34. Our analyses indicate that positive selection can also alter genes belonging to other functional categories, such as metabolism and development, adding a new aspect to the arms-race co-evolutionary theories. Through a comprehensive analysis, we identified a protein, ATAD-3, suggesting a relevant role for mitochondria during the evolution of nematode parasitism that warrants further investigation. We provide additional evidence for the phylogenetically relatedness of S. carpocapsae to Strongyloides, making this high-quality genome valuable for comparative studies with potential implications for human health. Our genome also represents a useful resource to aid ongoing efforts towards the genetic improvement of entomopathogens as biological control agents as well as to better understand host-parasite interactions in nematodes.
Materials and Methods
Organisms, maintenance and storage
Steinernema carpocapsae strain Breton was obtained from Nelson Simões, and cultured using in vitro methods. Nematodes were grown using a modified protocol for mass production in artificial medium according to ref. 45, as well as in small-scale, on plates containing Fortified Lipid Agar (FLA) prepared with 1.6% TSB (nutrient broth), 1% vegetable oil, 1.2% bacteriological agar, and 5% yeast extract (modified from ref. 46). A near isogenic line of S. carpocapsae strain Breton was generated by reproduction of single couples of brother and sister for 12 generations (F12). This produces ~96% homozygosis47.
DNA isolation, sequencing and quality control
Total genomic DNA was isolated from the nuclei48 using the phenol/chloroform extraction protocol described by Sambrook et al.49. Total DNA yield and integrity was measured with a 2100 Bioanalyzer (Agilent) using an Expert High Sensitivity DNA chip. Three high-quality libraries for Next Generation Sequencing (NGS) were prepared following manufacturer’s instructions. One shotgun 454 library was sequenced in three 454 FLX runs. One 454 paired-end library with 8-kb inserts was sequenced in two 454 FLX runs. Finally, a SOLiD shotgun library was tagged and sequenced, along with a different library (from other organisms), in a lane of SOLiD 5500xl, equivalent to half a lane of SOLiD sequencing. Low-quality sequences, base-calling duplicates and adapters were removed from all the sequence data (see below).
Genomic assembly and filtering
All DNA-sequence reads were filtered to remove contamination of the endosymbiotic bacteria Xenorhabdus nematophila (Xn). A genomic dataset was created by adding the published genome of Xn strain ATCC 19061 to the unpublished genome of the Xn strain isolated from the S. carpocapsae strain Breton nematodes, produced in our laboratory. The dataset was used for contamination screening and filtering using GS Assembler 2.7.
Raw standard flowgram format (sff) files coming from 454 platforms were assembled using GS Assembler 2.7 with the default trimming parameters. Basespace reads coming from the SOLiD platform using the Exact Call Chemistry Module (that allows conversion from colour to basespace) were filtered to remove PCR clonal repeats as well as reads with ambiguous bases. Subsequently, sequences were filtered based on Phred quality values. Bases below Phred18 were removed from 3′ ends and only reads longer than 20 bp were kept. Filtered data were assembled into contigs using GS Assembler 2.7, and joined into scaffolds using the paired-end data.
GC-content was estimated from the scaffolds using 10-kb non-overlapping sliding windows, and GC-bias was assessed based on a frequency distribution of these data. To evaluate the completeness of the genome assembly, we followed two strategies. RNA-seq sequences representing all different stages and diverse culture conditions of S. carpocapsae were mapped to the final assembly using Newbler (GS Reference Mapper v.2.7). In addition, we analysed the completeness of 248 ultra-conserved core eukaryotic genes13. We expect a complete genome will contain a higher number of complete ultra-conserved genes.
Estimation of genome size
Genome size was estimated from the genomic assembly using GS Assembler 2.7 and corroborated through flow cytometry of the isolated cellular nuclei of S. carpocapsae using the nuclei of C. elegans strain N2 (genome size approx. 100 Mb50) as a size control. Nuclei were stained with CyStain® UV Ploidy (Partec 05-5001), and fluorescence was detected at λ ≤ 420 nm and quantified using a PARTECPAII (Partec, Germany) flow cytometer with a mercury lamp (100 W UV light).
Assessment of repeat content
Following genome assembly, repeats were identified using a combination of homology-based comparisons (using RepeatMasker51) and a de novo approach (using RepeatModeler52).
Annotation of non-coding RNA
Covariance models from Rfam53 were used to scan the genomes using Infernal software54. In addition, tRNAs were predicted using tRNAscan-SE55 and rRNAs were predicted using RNAmmer56. Finally, microRNA precursor sequences (miRNA hairpins) were located using MapMi57, using all mature miRNA sequences from miRBase 2158 as input. MapMi results were filtered selecting only microRNA precursor sequences with score ≥30. Results from Rfam, tRNAscan-SE, RNAmmer and MapMi were processed within R (R: A Language and Environment for Statistical Computing; http://www.r-project.org), using ‘GenomicFeatures’ and ‘rtracklayer’ packages59,60.
To compare ncRNA families present in parasitic (Ascaris suum, Bursaphelenchus xylophilus, Brugia malayi, S. carpocasae, Meloidogyne incognita, M. hapla, and Heterorhabditis bacteriophora) and free-living (Panagrellus redivivus, Pristionchus pacificus, Caenorhabditis remanei, and C. elegans) nematodes, the average number of genes belonging to each ncRNA family, were calculated for each group. Families with at least twice the average number of genes in the parasitic compared to free-living group were selected.
RNA isolation, sequencing and assembly
Total RNA was extracted from a pool of individuals from all lifecycle stages (eggs at different stages, L1, L2, L3, IJ, L4, and adults), cultured in vivo infecting Galleria mellonella and Tenebrio molitor larvae as described61, as well as all lifecycle stages cultured in vitro using the methods described in the Organisms, maintenance and storage section45,46. The final pool consisted of approximately 3 mg of individuals from each stage/condition. RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions with an additional step using Qiagen RNAeasy Mini Elute Clean up columns and buffers to clean and concentrate the RNA.
An Illumina paired-end library and a 454 library were generated for RNA-seq, which were run on a full plate of MiSeq and a partial plate of a 454 FLX+, respectively. RNA-seq reads were quality filtered and mapped to the repeat-masked genome using Newbler gsmapper. Read alignments were provided to Trinity62 (r2013-02-25) as a coordinate-sorted bam file. Trinity was used to assemble the aligned reads. The Trinity-reconstructed transcripts were aligned and assembled using the PASA263 (r20130425 beta) pipeline.
For small RNA (sRNA) sequencing, nematodes were induced for 2 hour with hemolymph of Galleria mellonella and with buffer as control64. Nematodes were grinded under liquid nitrogen and RNA was extracted with Trizol according to the manufacturer’s instructions. Six sRNA-Seq tagged libraries were prepared from three replicates of each condition, which were run in an Illumina HiSeq lane.
Processing small RNA sequencing results
All sRNA-Seq libraries were 3′-adaptor trimmed using the reaper tool from Kraken65. After trimming, reads between 18 and 36 nucleotides were mapped to the genome using ShortStack 3.318, setting the maximum number of mismatches to 1, no stich, multi-mappers guided by unique-mappers and removing reads that mapped to more than 101 locations. The raw and processed sRNA-Seq results were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo), under accession GSE85256.
Predicting expressed microRNA loci with miRDeep and ShortStack
When using ShortStack to annotate, we set the Dicer minimum and maximum size parameters to 18 and 36, minimum alignment coverage (mincov) to 5 and maximum distance to merge clusters (pad) to 50. To improve the predicted microRNA producing loci, we used MirDeep217. The mapper.pl and miRDeep2.pl modules were used to identify known and novel microRNAs. Reads between 18 and 36 nucleotides were used, with a maximum number of mismatches of 1, and 101 maximum number of locations for multi-mapping reads. The minimum alignment coverage (-a) was set to 5, the maximum number of precursors to analyze was set to 1000, and all the mature sequences from miRBase 21 were provided. Although both programs take small RNA sequencing reads mapped to a genome to predict microRNA loci, they produce slightly different results. They coincided in predicting 100 miRNAs, while miRDeep predicted an additional 162 and ShortStack 25 more.
Differential expression analysis of microRNAs
To focus the differential expression analysis on miRNAs, only reads in the 20–24 nucleotide length range were considered. After mapping these to the genome, on average 83% fell within miRNA hairpin and 63% within mature miRNA coordinates. For miRNA quantification, the featureCounts function of the Rsubread R package66 was used, asking for a minimum overlap of one nucleotide to any of the mature miRNA annotations. For differential expression analysis, the edgeR package was used67. miRNAs with less than 3 counts-per-million in at least 3 libraries were removed, leaving 302 out of the 574 annotated mature miRNAs. The trimmed mean of M-values was chosen as normalization method68. Genewise data dispersion was estimated with the function estimateGLMTagwiseDisp, which uses an empirical Bayes strategy67. Differentially expressed miRNAs were determined with a generalized linear model and gene-wise likelihood ratio tests. A False Discovery Rate threshold of 0.1 was selected to consider a miRNA to be significantly differentially expressed. According to this threshold, five miRNAs were overexpressed in response to hemolymph treatment and none were down regulated. Cluster_19164 was identified as miR-84 by a manual sequence search on the miRBase website58.
Gene prediction and synteny
The S. carpocapsae protein-coding gene set was inferred using de novo, homology- and evidence-based approaches (Supplementary Fig. S7). De novo gene prediction was performed on a repeat-masked genome using Augustus14. Training models were generated using hints from a compilation of S. carpocapsae gene structures (CEGMA13 [v2.4.010312] predictions, PASA assemblies from our RNA-seq data, and 2,269 publicly available ESTs from GeneBank). The homology-based prediction was conducted with Augustus algorithms for C. elegans and Brugia malayi. Synteny was assessed on scaffolds >1 Mb using pairwise alignments with E-value < 10−6 and homologous regions were visualized using CIRCOS69. Macro-synteny was analysed using the SynFind and SynMap tools from CoGe70,71.
Functional annotation of coding genes
Following the prediction of the protein-coding gene set, we conducted high-stringency BLASTp homology searches (E-value ≤ 10−5) against the NCBI non-redundant protein database. Functional annotation was performed using Blast2GO72. Gene ontology categories were summarized and standardized to level 2 and level 3 terms, defined using the GOslim hierarchy73. For the secretome prediction, the signal peptide was predicted by SignalP 4.074 and Phobious75 employing both Hidden Markov Models and Neural Networks. Proteins were then filtered for the presence of transmembrane regions using THMMN76 and Phobious75. Subcellular localizations were identified using TargetP (≥95% specificity)77 and WolfPSORT78 (score ≥30). Proteases and protease inhibitors were identified by homology searches to the MEROPS database79.
Orthologous proteins
Genes from different nematode species (Caenorhabditis angaria, C. briggsae, C. elegans, C. japonica, C. remanei, Pristionchus pacificus, Ascaris suum, Brugia malayi, Bursaphelenchus xylophilus, Heterorhabditis bacteriophora, Haemonchus contortus, Loa loa, Meloidogyne hapla, M. incognita, Onchocerca volvulus, Panagrellus redivivus, S. carpocasae, Strongyloides ratti, Trichinella spiralis, and Wuchereria bancrofti) were assigned to OrthoMCL80 orthologous groups, and the presence or absence of groups was compared among the different species using ad hoc scripts from scriptome (http://archive.sysbio.harvard.edu/csb/resources/computational/scriptome/UNIX/).
Additional bioinformatics analyses and use of software
Data analysis was conducted in a UNIX environment or Microsoft Excel 2007 using standard commands. Bioinformatic scripts required to facilitate data analysis were designed using bash, GNU coreutils, Perl, and Python.
Proteomic analysis
Sample preparation
Nematodes were induced as described64 with slight modifications. A pool of approximately 25,000 nematodes were induced with either hemolymph or intestine of Galleria mellonella. To obtain hemolymph we grinded G. mellonella larvae in liquid nitrogen, added a volume of cold Tyrode’s solution (NaCl 0.8%, KCl 0.02%, CaCl2 0.02%, MgCl2, 0.02%, NaH2PO4, 0.005%, NaHCO3, 0.1%, and glucose, 0.1%) and sonicated at a frequency of 20 kHz. Then we centrifuged at 1700 rcf for 15 min at 4 °C, obtaining three phases, of which the middle one corresponded to hemolymph. Intestines were dissected from insect larvae, collected on a watch glass and rinsed several times with sterile saline solution (NaCl 0.8%) to eliminate any trace of hemolymph or other remains. Nematode infecting juveniles (IJs) were superficially disinfected with 2% sodium hypochlorite during 10 min, and rinsed three times with sterilized water. According to our experience, this treatment is enough to kill all surface bacteria and fungi from the nematodes while keeping them viable. Washed nematodes were then transferred to a 90 × 15 mm Petri dish containing 7 ml of Tyrode’s solution with 10% of G. mellonella hemolymph (v/v) or 10% of intestines (w/v), and 1% Nalidixic acid, to avoid contamination. Previous experience in the Simões lab has shown that 10% of hemolymph is needed to induce recovery of the IJs and to allow them to complete their life cycle. The same concentration of intestines was used as a first approximation to understand the effects of insect intestines on protein expression. Different pools of nematodes were incubated under agitation (40 rpm) at 25 °C for 1, 2, 4 and 8 hours, and analysed separately. To determine these time points we followed the infection process in vivo, by conducting dissections at regular intervals, to understand the kinetics of the infection. We observed that after entering the intestine, it took the nematodes one hour to start traversing the intestine wall and at two hours, most of them were in the hemocoel. We included two additional time points to capture proteins expressed lately in the infection process; stopping at 8 hours because at this point the nematodes start to release the symbiotic bacteria81. Nematodes without induction were used as a negative control. Nematodes were grinded in liquid nitrogen and suspended in lysis buffer (7 M urea, 2 M thiourea, 3% CHAPS) with protease inhibitor mix GE and sonicated at a frequency of 20 kHz. After centrifugation (1 min at 16,000 rcf) and filtering (0.45 μm Millex-HV PVDF, Millipore), the supernatant was precipitated with the 2D Clean-up kit (GE Healthcare), resuspended in DeStreak solution (GE Healthcare Cat. No. 17600318), and quantified with the Bradford Method82 (BioRad Protein Assay Dye Cat. No. 500-0006).
SDS-PAGE and 2D electrophoresis
Protein samples (80 μg) were loaded onto the SDS-PAGE and ran at 100 V in a vertical Mini-PROTEAN Tetra cell BioRad. Gels were stained with Coomassie blue. Isoelectric focusing was performed using GE Healthcare Immobiline strips pH 3-10 and DryStrip pH 4–7, in both cases of 7 cm of length. Isoelectric focusing was performed on a Multiphor II (GE Healthcare) using the conditions recommended by the manufacturer. The second dimension was run on 13% polyacrylamide gels.
Shotgun proteomics
Two hundred μl of the total protein extract was fractionated by isoelectric focusing in an IEF ZOOM® Fractionator system (Life Technologies) using the protocol described by the manufacturer. The protein pellet was passed through a reduction/alkylation process with urea (6 M), dithiothreitol (DTT) at a final concentration of 5 mM, and iodoacetamide (IAA) at a final concentration of 15 mM. Peptides were digested with trypsin (Promega) overnight at 37 °C and desalted with a Macro Spin Column (Nest Group). Thirty μg of protein from each fraction were then analyzed by LC-MS/MS in the Proteomics Facility of the UC Davis Genome Center. A Thermo Scientific Q Exactive Orbitrap MS spectrometer was used in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Scientific) and a source Proxeon nanospray using a column 100 micron × 25 mm Magic C18 5U 100 Å reverse phase. The MS/MS spectra were acquired using the TOP15 method following the equipment manufacturer’s instructions. All analyses were run in duplicates, including treatment samples and controls. ProteinPilot (v4.5), Mascot (v2.4), MaxQuant (v1.3.0.5), and Sequest (v1.3) software were used to identify peptides and proteins in each sample (see software references in Supplementary Table S2). In all cases, a tolerance in the mass measurement of 50 ppm in MS mode and 0.5 Da for MS/MS ions was used, with a significant threshold set to p < 0.05 and a confidence value ≥95%, with the exception of MaxQuant, in which the peptide mass tolerance was of 20 ppm, the fragment mass tolerance of 0.5 Da, and the confidence value was ≥99%. Modifications allowed were carbamidomethylation C (fixed), deamination NQ (variable), and oxidation M (variable).
Proteins detected in at least one sample replica and undetected in the two control replicates were considered as differentially expressed proteins. Proteins detected in at least one of the control replicates, but undetected in the two sample replicates were considered differentially suppressed proteins. Annotation and functional enrichment of differentially expressed proteins were performed with Blast2GO72. The Fisher Exact Test was used to compare the GO terms identified under the different induction conditions (hemolymph or gut) with the nematodes without induction.
Phylogenetic analysis
Protein sequences of all organisms were downloaded from WormBase (ftp.wormbase.org release WS241 29-Nov-2013). Orthologous genes for nine species were identified with OrthoMCL (v5)80. A total of 245 orthologous proteins were aligned with MUSCLE (v3.8.31)83. Phylogenetically informative blocks were recovered with Gblocks84 and the best-fit evolutionary model for each aligned protein was predicted by ProtTest85. MrBayes86 was used for phylogenetic reconstruction using concatenated alignments. Partitions were created by grouping proteins according to their best-fit model, i.e. each partition contained all the proteins evolving under the same model. A mixed model was applied to each partition, with different G, I, and F parameters and unlinking the model between partitions. To check for convergence, two runs with four chains each were performed. The analysis was run for 1,000,000 generations, and a burn-in of 25% was used. Brugia malayi and Ascaris suum were used to root the tree because these species were the most phylogenetically basal of the nine nematode species in the phylogenies obtained by both Blaxter et al.38 and Nadler et al.41.
Analysis of positive selection
We used protein-coding genes from Panagrellus redivivus, Strongyloides ratti and Steinernema carpocapsae, all nematodes from phylogenetic clade IV, according to Blaxter et al.38. All nucleic and amino acid sequences, except for S. carpocapsae, were downloaded from WormBase (ftp.wormbase.org release WS241 29-Nov-2013). Orthologues proteins obtained with OrthoMCL (v5)80 were aligned with ClustalW2 (v2.1)87. After selecting phylogenetically informative sites with Gblocks84, and estimating the best-fit model with ProtTest85, we reconstructed a consensus phylogenetic tree with PhyML (v.3.0)88. A nucleotide alignment based on the complete amino acid alignment was obtained with RevTrans (v1.4)89 to preserve codon homology. The tree and the nucleotide alignments of each orthologous gene were used to assess signatures of natural selection with Codeml from the PAML package (v.4.6)90, using the Branch-site model to identify genes with sites under positive selection. Annotation and functional enrichment in genes with positively selected sites were performed with Blast2GO72.
Additional Information
How to cite this article: Rougon-Cardoso, A. et al. The genome, transcriptome, and proteome of the nematode Steinernema carpocapsae: evolutionary signatures of a pathogenic lifestyle. Sci. Rep. 6, 37536; doi: 10.1038/srep37536 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Oerke, E.-C. Crop losses to pests. The Journal of Agricultural Science 144, 31–43, doi: 10.1017/S0021859605005708 (2006).
Aktar, W., Sengupta, D. & Chowdhury, A. Impact of pesticides use in agriculture: their benefits and hazards. Interdisciplinary Toxicology 2, 1–12 (2009).
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M. & Kenerley, C. M. Trichoderma research in the genome era. Annual Review of Phytopathology 51, 105–129 (2013).
Lu, D., Baiocchi, T. & Dillman, A. R. Genomics of entomopathogenic nematodes and implications for pest control. Trends in Parasitology 32, 588–598, doi: 10.1016/j.pt.2016.04.008 (2016).
Murfin, K. E. et al. Nematode-bacterium symbioses—cooperation and conflict revealed in the “Omics” age. The Biological Bulletin 223, 85–102 (2012).
Thaler, J.-O., Duvic, B., Givaudan, A. & Boemare, N. Isolation and entomotoxic properties of the Xenorhabdus nematophilus F1 lecithinase. Applied and Environmental Microbiology 64, 2367–2373 (1998).
Caldas, C., Cherqui, A., Pereira, A. & Simões, N. Purification and characterization of an extracellular protease from Xenorhabdus nematophila involved in insect immunosuppression. Applied and Environmental Microbiology 68, 1297–1304, doi: 10.1128/aem.68.3.1297-1304.2002 (2002).
Herbert, E. E. & Goodrich-Blair, H. Friend and foe: the two faces of Xenorhabdus nematophila. Nature Reviews Microbiology 5, 634–646 (2007).
Binda-Rossetti, S., Mastore, M., Protasoni, M. & Brivio, M. F. Effects of an entomopathogen nematode on the immune response of the insect pest red palm weevil: Focus on the host antimicrobial response. J Invertebr Pathol 133, 110–119 (2016).
Toubarro, D. et al. An apoptosis-inducing serine protease secreted by the entomopathogenic nematode Steinernema carpocapsae. International Journal for Parasitology 39, 1319–1330, doi: http://dx.doi.org/10.1016/j.ijpara.2009.04.013 (2009).
Dillman, A. R. et al. Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biology 16, 1–21 (2015).
Blaxter, M. & Koutsovoulos, G. The evolution of parasitism in Nematoda. Parasitology 142, S26–S39 (2015).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067, doi: 10.1093/bioinformatics/btm071 (2007).
Stanke, M. & Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Research 33, W465–W467 (2005).
Douzery, E. J., Snell, E. A., Bapteste, E., Delsuc, F. & Philippe, H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? PNAS 101, 15386–15391 (2004).
Quentin, M., Abad, P. & Favery, B. Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Frontiers in Plant Science 4, 53, doi: 10.3389/fpls.2013.00053 (2013).
Friedländer, M. R. et al. Discovering microRNAs from deep sequencing data using miRDeep. Nature biotechnology 26, 407–415 (2008).
Axtell, M. J. ShortStack: comprehensive annotation and quantification of small RNA genes. Rna 19, 740–751 (2013).
Abbott, A. L. et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Developmental cell 9, 403–414 (2005).
Toubarro, D. et al. Serine protease-mediated host invasion by the parasitic nematode Steinernema carpocapsae. Journal of Biological Chemistry 285, 30666–30675 (2010).
Hennebry, S. C., Law, R. H., Richardson, S. J., Buckle, A. M. & Whisstock, J. C. The crystal structure of the transthyretin-like protein from Salmonella dublin, a prokaryote 5-hydroxyisourate hydrolase. Journal of molecular biology 359, 1389–1399 (2006).
Lee, Y. et al. Transthyretin-related proteins function to facilitate the hydrolysis of 5-hydroxyisourate, the end product of the uricase reaction. FEBS Lett 579, 4769–4774 (2005).
Furlanetto, C., Cardle, L., Brown, D. & Jones, J. Analysis of expressed sequence tags from the ectoparasitic nematode Xiphinema index. Nematology 7, 95–104, doi: 10.1163/1568541054192180 (2005).
Britton, C. 18 Proteases of Nematodes: From Free-living to Parasite. Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology 351 (2013).
Blaxter, M., Page, A., Rudin, W. & Maizels, R. Nematode surface coats: actively evading immunity. Parasitology Today 8, 243–247 (1992).
Yoshida, A., Nagayasu, E., Horii, Y. & Maruyama, H. A novel C-type lectin identified by EST analysis in tissue migratory larvae of Ascaris suum. Parasitol Res 110, 1583–1586 (2012).
Hewitson, J. P., Grainger, J. R. & Maizels, R. M. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Molecular and biochemical parasitology 167, 1–11, doi: 10.1016/j.molbiopara.2009.04.008 (2009).
Schwarz, E. M. et al. The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection-specific gene families. Nat Genet 47, 416–422, doi: 10.1038/ng.3237 (2015).
Delannoy-Normand, A., Cortet, J., Cabaret, J. & Neveu, C. A suite of genes expressed during transition to parasitic lifestyle in the trichostrongylid nematode Haemonchus contortus encode potentially secreted proteins conserved in Teladorsagia circumcincta. Vet Parasitol 174, 106–114 (2010).
Jing, Y., Toubarro, D., Hao, Y. & Simões, N. Cloning, characterisation and heterologous expression of an astacin metalloprotease, Sc-AST, from the entomoparasitic nematode Steinernema carpocapsae. Molecular and Biochemical Parasitology 174, 101–108 (2010).
Delcroix, M. et al. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. Journal of Biological Chemistry 281, 39316–39329 (2006).
Geldhof, P., Claerebout, E., Knox, D., Agneessens, J. & Vercruysse, J. Proteinases released in vitro by the parasitic stages of the bovine abomasal nematode Ostertagia ostertagi. Parasitology 121, 639–647 (2000).
Williamson, A. L. et al. Hookworm aspartic protease, Na-APR-2, cleaves human hemoglobin and serum proteins in a host-specific fashion. Journal of Infectious Diseases 187, 484–494 (2003).
Aguileta, G., Refregier, G., Yockteng, R., Fournier, E. & Giraud, T. Rapidly evolving genes in pathogens: methods for detecting positive selection and examples among fungi, bacteria, viruses and protists. Infection, Genetics and Evolution 9, 656–670 (2009).
Rausell, A. & Telenti, A. Genomics of host-pathogen interactions. Curr Opin Immunol 30, 32–38 (2014).
Zhang, J., Nielsen, R. & Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular biology and evolution 22, 2472–2479 (2005).
Yang, Z., Wong, W. S. & Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Molecular biology and evolution 22, 1107–1118 (2005).
Blaxter, M. L. et al. A molecular evolutionary framework for the phylum Nematoda. Nature 392, 71–75 (1998).
Holterman, M. et al. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol Biol Evol 23, 1792–1800 (2006).
Montiel, R., Lucena, M. A., Medeiros, J. & Simoes, N. The complete mitochondrial genome of the entomopathogenic nematode Steinernema carpocapsae: insights into nematode mitochondrial DNA evolution and phylogeny. J Mol Evol 62, 211–225 (2006).
Nadler, S. A. et al. Phylogeny of Cephalobina (Nematoda): Molecular evidence for recurrent evolution of probolae and incongruence with traditional classifications. Molecular Phylogenetics and Evolution 40, 696–711, doi: 10.1016/j.ympev.2006.04.005 (2006).
Hoffmann, M. et al. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS ONE 4, e7644 (2009).
Addo, M. G. et al. Caenorhabditis elegans, a pluricellular model organism to screen new genes involved in mitochondrial genome maintenance. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1802, 765–773 (2010).
Calderone, R., Li, D. & Traven, A. System-level impact of mitochondria on fungal virulence: to metabolism and beyond. FEMS yeast research 15, fov027 (2015).
Bedding, R. Low cost in vitro mass production of Neoaplectana and Heterorhabditis species (Nematoda) for field control of insect pests. Nematologica 27, 109–114 (1981).
Neves, J., Simoes, N. & Mota, M. Evidence for a sex pheromone in Steinemema carpocapsae. Nematologica 44, 95–98 (1998).
Wright, S. Systems of mating. V. General considerations. Genetics 6, 167 (1921).
Collins, G. G. & Symons, R. H. Extraction of nuclear DNA from grape vine leaves by a modified procedure. Plant molecular biology reporter-ISPMB (USA) (1992).
Sambrook, J., Fritsch, E. & Maniatis, T. Molecular cloning: a laboratory manual. (Cold Spring Harbor, 1989).
Bennett, M. D., Leitch, I. J., Price, H. J. & Johnston, J. S. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25% larger than the Arabidopsis genome initiative estimate of∼125 Mb. Annals of Botany 91, 547–557 (2003).
Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0http:/www.repeatmasker.org (2013).
Smit, A. F. A. & Hubley, R. RepeatModeler Open-1.0.http:/www.repeatmasker.org (2008).
Nawrocki, E. P. et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43, 11 (2015).
Nawrocki, E. P. & Eddy, S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935, doi: 10.1093/bioinformatics/btt509 (2013).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25, 0955–0964 (1997).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research 35, 3100–3108 (2007).
Guerra-Assunção, J. A. & Enright, A. J. MapMi: automated mapping of microRNA loci. BMC bioinformatics 11, 133 (2010).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42(D41), D68–D73 (2013).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118 (2013).
Lawrence, M., Gentleman, R. & Carey, V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–1842 (2009).
Nguyen, K. & Hunt, D. Entomopathogenic nematodes: systematics, phylogeny and bacterial symbionts. (Brill, 2007).
Grabherr, M. G. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature biotechnology 29, 644 (2011).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research 31, 5654–5666, doi: 10.1093/nar/gkg770 (2003).
Hao, Y.-J., Montiel, R., Abubucker, S., Mitreva, M. & Simões, N. Transcripts analysis of the entomopathogenic nematode Steinernema carpocapsae induced in vitro with insect haemolymph. Molecular and Biochemical Parasitology 169, 79–86 (2010).
Davis, M. P., van Dongen, S., Abreu-Goodger, C., Bartonicek, N. & Enright, A. J. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63, 41–49 (2013).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Research 41, e108–e108 (2013).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research, gks042 (2012).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, 1 (2010).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645 (2009).
Tang, H. et al. SynFind: compiling syntenic regions across any set of genomes on demand. Genome biology and evolution 7, 3286–3298 (2015).
Eric, L., Matthew, D. B., Shannon, L. O. & rew, J. L. In Handbook of Plant and Crop Physiology, Third Edition Books in Soils, Plants, and the Environment 797–816 (CRC Press, 2014).
Götz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research 36, 3420–3435, doi: 10.1093/nar/gkn176 (2008).
Camon, E. et al. The Gene Ontology Annotation (GOA) Project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Research 13, 662–672, doi: 10.1101/gr.461403 (2003).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. Journal of molecular biology 340, 783–795 (2004).
Kall, L., Krogh, A. & Sonnhammer, E. Advantages of combined 613 transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res 35, W429–W432 (2007).
Krogh, A., Larsson, B., Von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology 305, 567–580 (2001).
Emanuelsson, O., Nielsen, H., Brunak, S. & Von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of molecular biology 300, 1005–1016 (2000).
Horton, P. et al. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587 (2007).
Rawlings, N. D., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Research 40, D343–D350 (2012).
Li, L., Stoeckert, C. J. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research 13, 2178–2189 (2003).
Snyder, H., Stock, S. P., Kim, S.-K., Flores-Lara, Y. & Forst, S. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Applied and Environmental Microbiology 73, 5338–5346 (2007).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry 72, 248–254 (1976).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular biology and evolution 17, 540–552 (2000).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 52, 696–704 (2003).
Wernersson, R. & Pedersen, A. G. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Research 31, 3537–3539 (2003).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution 24, 1586–1591 (2007).
Acknowledgements
This work was supported by a research grant from FOMIX-Hidalgo to RM (Fomix-Hgo-2008-C01-97032), and from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007/2013/ under REA grant agreement No. 612583. ARC received a postdoctoral fellowship from Conacyt (CVU 39220). MFP received a Conacyt fellowship for Master and Ph.D. studies (Reg. No. 219899). We are indebted to Sánchez-delPino M.M. and Valero L. from the proteomic service of the University of Valencia (Proteored ISCIII) for their support in proteomic data analysis. Clarisse, a script for massive scans of positive selection using PAML, was developed and kindly provided by Victor Villa-Moreno, from the “Laboratorio de la Diversidad Biomolecular” (Langebio) under the direction of Dr. Mauricio Carrillo-Tripp.
Author information
Authors and Affiliations
Contributions
A.R.C., M.F.P., R.M., designed the study; M.F.P., H.E.R.A., L.C., conducted experiments; A.R.C., M.F.P., H.E.R.A., C.E.M.G., Y.-J.H., J.A.R.M., C.O.V., J.R.B.B., C.A.G., R.M., analysed data; N.Ch.H., N.S., R.M., contributed materials and reagents; A.R.C., M.F.P., R.M., wrote the paper, with contributions from H.E.R.A., Y.-J.H., C.O.V., J.R.B.B., C.A.G. All authors reviewed and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Rougon-Cardoso, A., Flores-Ponce, M., Ramos-Aboites, H. et al. The genome, transcriptome, and proteome of the nematode Steinernema carpocapsae: evolutionary signatures of a pathogenic lifestyle. Sci Rep 6, 37536 (2016). https://doi.org/10.1038/srep37536
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37536
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.