Abstract
Auxin and cytokinin (CK) are both important hormones involved in many aspects of plant growth and development. However, the details of auxin biosynthesis and the interaction between auxin and CK are still unclear. Isolation and characterization of an auxin deficient mutant cytokinin induced root curling 2 (ckrc2) in this work reveal that CKRC2 encodes a previously identified member of YUCCA (YUC) flavin monooxygenase-like proteins (YUC8). Our results show that, like other YUCs, CKRC2/YUC8 is a rate-limiting enzyme for catalyzing the conversion of indole-3-pyruvic acid (IPyA) to indole-3-acetic acid (IAA), acting downstream of CKRC1/TAA1 in the IPyA pathway. Here we show that the transcription of both CKRC1/TAA and CKRC2/YUC8 can be induced by CK and that the phytochrome-interacting factor 4 (PIF4) is required for this upregulation. Transcription of PIF4 itself is induced by CK via the AHKs-ARR1/12 signalling pathway. These results indicate that PIF4 plays an essential role in mediating the regulatory effect of CK on the transcriptions of CKRC1 and CKRC2 genes in the IPyA pathway of auxin biosynthesis.
Similar content being viewed by others
Introduction
Auxin is an important phytohormone and influences many processes in plant growth and development, such as cell division and elongation, tropism, apical dominance, senescence and blooming1,2,3,4,5. In planta, auxin homeostasis is controlled by biosynthesis, transport and metabolism6. Based on biochemical and genetic evidence, it has been proposed that IAA, the predominant form of auxin in plants, is synthesized via two major pathways: Trp-dependent (TD) and Trp-independent (TI) pathways5,7,8. The TD pathway can further be divided into four branch pathways according to their first intermediate metabolites, i.e., the indole-3-acetamide (IAM) pathway, the indole-3-pyruvic acid (IPyA) pathway, the tryptamine (TAM) pathway and the indole-3-acetaldoxime (IAOx) pathway. So far only the IPyA pathway has been completely determined on genetic and biochemical levels. For many years, no gene or intermediate metabolite involved in the TI pathway has been identified. Most recently, however, careful studies on the mutant of the indole synthase (INS) gene provide evidence that the cytoplasmic protein INS is involved in auxin biosynthesis via the TI pathway9.
YUC genes were initially linked to auxin biosynthesis based on the finding that overexpression of YUC1 leads to an auxin overproduction phenotype10. There are 11 predicted members of YUC genes encoding YUCCA (YUC) flavin monooxygenase-like proteins in Arabidopsis. Overexpression of each of the YUCs results in high auxin phenotypes11. However, inactivation of a single YUC gene does not cause obvious developmental defects suggesting overlapping functions among YUC genes11,12. YUC1 was initially suggested to catalyze the conversion of TAM to N-hydroxylated tryptamine (HTAM) in the TAM pathway10 but recent studies have placed the YUC proteins downstream of CKRC1/TAA1, catalyzing the conversion of IPyA to IAA13,14,15,16. Further results showed that YUC can synthesize a quasi-stable 4-α-hydroperoxyl flavin intermediate from flavin adenine dinucleotide (FADH-) and acts on numerous substrates12,17,18It was reported that YUC6 utilizes NADPH and O2 to convert IPyA to IAA12. In this work we show that, like other YUCs, CKRC2/YUC8 is a rate-limiting enzyme in the IPyA pathway for catalyzing the conversion of IPyA to IAA. Together with CKRC1/TAA1, CKRC2/YUC8 plays an essential role in the CK-dependent regulation of auxin biosynthesis.
The interaction between auxin and CK plays a key role in plant growth and development19. Recent studies reveal that CK can regulate both the biosynthesis and the polar transport of auxin via its signaling pathway19,20,21. We previously reported that CK can stimulate auxin biosynthesis by up-regulating the transcription of CKRC1/TAA1 and other auxin biosynthesis genes including YUC821; however, the associated transcriptional factors/regulators have not been identified so far. Here we show that PIF4 is essential for CK-dependent regulation of CKRC1/TAA1 and CKRC2/YUC8 transcription.
Results and Discussions
Comparison of root phenotypes among mutants in different YUC genes and their transcription
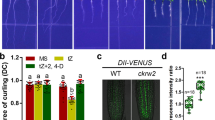
The ckrc2-1 mutant was isolated as one of the auxin-deficient mutants in a large-scale forward genetic screen for the so-called cytokinin induced root curling (ckrc) mutants (Supplementary Fig. S1)21,22. When grown on medium containing 0.1 μM trans-zeatin (tZ) these mutants display a root curling phenotype. Genetic and molecular analysis identified ckrc2-1 as a loss of function mutation in the YUC8 gene. The mutation is caused by a 3554 bp deletion in the promoter coding region (Supplementary Fig. S2). As YUC8 is one of 11 members of the YUC gene family functioning in auxin biosynthesis10,11,12,13,14,15,16, root phenotypes in the other 10 YUC genes were also analysed (Fig. 1a & Supplementary Fig. S3). We found that none of the single mutants in other YUC genes had the ckrc2/yuc8-like curling root phenotype on the medium containing 0.1 μM tZ (Fig. 1a). Furthermore only yuc8 displayed a significantly defective gravitropic response (GR) on MS medium. Yuc5, showed a weak root GR defect (Fig. 1a,b). Root measurements showed that most yuc mutants had decreased root length when grown on MS medium (Fig. 1c), and yuc5, ckrc2-1/yuc8 and yuc9 were less sensitive to 0.1 μM tZ in terms of relative root length compared to other yuc mutants (Fig. 1d).
Comparison of root phenotypes between yuc mutants.
(a) The phenotypes of all 11 yuc mutants on MS medium with (bottom) or without (top) 0.1 μM tZ 7d after germination (bar = 5 mm); (b) Gravitropic responses of yuc mutants. Seedlings were grown on MS medium for 6d, then transferred to fresh MS medium and reoriented 90 degrees for 24 hours (n = 75–100); (c,d) Root elongation was measured 7d after germination on MS with (c) or without (d) 0.1 μM tZ. All experiments were repeated 3 times. Shown are mean values ± SD with n = 40–45 in each repeat. ***P < 0.001.
To determine why only ckrc2-1/yuc8 showed a root curling phenotype, the relative transcription of YUC genes in roots and whole seedlings was analyzed by qRT-PCR (Fig. 2a & Supplementary Fig. S4). Consistent with data previously reported by Chen et al.23, YUC3, YUC5, YUC7, YUC8 and YUC9 were highly expressed in roots (Fig. 2a & Supplementary Figs S4 and S5). However, in our results also YUC2 and YUC6 were detected in high levels in roots (Fig. 2a & Supplementary Fig. S4). Analysing the relative transcription of the YUC gene family after tZ treatment, we found that out of the seven YUC genes with high transcription levels in roots, only CKRC2/YUC8 showed significant up-regulation by tZ (Fig. 2b). Up-regulation of the transcription of CKRC2/YUC8 after short time treatment with tZ was previously also shown in microarray data and qRT-PCR results ( http://www.weigelworld.org/resources/microarray/AtGenExpress/) (Supplementary Figs S6 and S7).
CKRC2/YUC8 is highly transcribed in roots and induced by tZ.
(a) Analysis of the relative transcription of YUC genes in roots by qRT-PCR; (b) Analysis of the relative transcription of YUC genes after 0.1 μM tZ treatment. Data are mean values of 3 replicates. Error bars indicate ± SD. 0.05 > *P > 0.01; ***P < 0.001.
The high abundance of YUC8 in roots and its up-regulation by CK could explain why ckrc2-1/yuc8 is the only yuc single mutant with a curled roots phenotype when grow on tZ containing medium and thus could be isolated in our CK forward genetic screen.
CKRC2/YUC8 encodes an enzyme catalyzing a rate-limiting step in the IPyA pathway for IAA biosynthesis
Some members of YUC family, including YUC1, YUC2, YUC4 and YUC6, have been shown to function in the same biosynthetic pathway with CKRC1/TAA1 and are catalyzing the conversation of IPyA to IAA, a rate-limiting step in the TAA/YUC pathway12,13,14,16,22. To determine if this was also the case for YUC8, a double mutant of ckrc2/yuc8 ckrc1/taa1 was phenotypically compared with the two parent single mutants ckrc2/yuc8 and ckrc1/taa1. In terms of root curling when grown on tZ medium the double mutant showed a less-than-additive phenotype (Fig. 3a & Supplementary Fig. S1), a phenomenon often associated with two mutations in different steps of the same linear genetic pathway16,24. The curling roots phenotype of ckrc2-1/yuc8 on tZ medium can be rescued by expressing of CKRC2/YUC8 cDNA under the control of the CKRC1/TAA1 promoter (Fig. 3a). Overexpression of CKRC2/YUC8 cDNA under the control of a 35S or CKRC1/TAA1 promoter Col-0 plants results in high auxin phenotypes such as increased amounts of root hairs, epinastic leaves, and elongated hypocotyls (Fig. 3b)10 indicating that CKRC2/YUC8 is catalyzing a rate-limiting step in IAA biosynthesis. This is not the case when overexpressing CKRC1/TAA1 under a 35S promoter in a wildtype background (Fig. 3b)21,25. Notably, the high-auxin phenotypes in p35S::CKRC2 or pCKRC1::CKRC2 transgenic plants can be suppressed by addition of L-kynurenine (L-Kyn), a specific TAA1/TAR inhibitor. This is indicating that the CKRC2/YUC8 mediated IAA production depends on the function of CKRC1/TAA1 (Fig. 3b)26. Moreover, addition of endogenous IPyA in tZ medium can partially rescue the curling root phenotype of ckrc1/taa1 but not that of ckrc2/yuc8 (Fig. 3c). Those results support the hypothesis that CKRC2/YUC8 and CKRC1/TAA1 act in the same biosynthetic pathway and CKRC2/YUC8 catalyses a rate-limiting step downstream from CKRC1/TAA1.
CKRC2/YUC8 and CKRC1/TAA1 function in the same biosynthetic pathway and CKRC2/YUC8 catalyzes a rate-limiting step.
(a) The ckrc1-1 ckrc2-1 double mutant displays a less-than-additive phenotype and the curling root phenotype of ckrc2-1 on tZ medium can be rescued by overexpression of CKRC2/YUC8 cDNA under the control of the CKRC1/TAA1 promoter (pCKRC1::CKRC2 > ckrc2-1); (b) The high auxin phenotype of transgenic p35S::CKRC2 or pCKRC1::CKRC2 plants can be inhibited by the addition of 1 μM L-Kyn; (c) Phenotypic analysis of Col, ckrc1-1 and ckrc2-1 plants grown on 0.1 μM tZ medium with 0.01 μM IPyA is shown; (d) Both CKRC2/YUC8 and YUC1 catalyze the conversion of IPyA to IAA; (e) Schematic diagram of auxin biosynthetic pathways.
Recent work has established that in IAA biosynthesis CKRC1/TAA1 catalyzes the conversion of Trp to IPyA and YUCs catalyze the subsequent oxidation of IPyA to IAA12,13,14,15,16,24,25. Like flavin monooxygenases (FMO in plants and animals), YUC proteins contain conserved motifs for binding FAD (flavin-adenine dinucleotide) and NADPH (the reduced form of nicotinamide adenine dinucleotide phosphate)10. So far such enzyme activities have been confirmed for 3 YUC proteins (YUC2/4/6)13,14. Detailed catalytic mechanism have been shown for YUC612. YUC6 uses NADPH to catalyze the reduction of FAD to FADH−; FADH− then reacts with oxygen to form a flavin-C4α- (hydro) peroxy intermediate, which reacts with IPyA to produce IAA12.
To provide evidence that CKRC2/YUC8 is also capable of catalyzing the conversion of IPyA to IAA, the CKRC2/YUC8 cDNA was cloned into the expression vector pET28b+ and the protein was expressed in a cell-free protein expression system. The enzymatic activities of the expressed YUC proteins were analyzed (Fig. 3d & Supplementary Fig. S7)27. The results showed that YUC1 and YUC8 proteins could convert 54.28% and 50.34% of total IPyA into IAA, respectively (Fig. 3d) whereas in the control cells expressing Renilla luciferase only a small amount of IPyA was converted to IAA non-enzymatically (Fig. 3d). Hence as reported for other YUC proteins12,13,14 YUC8 is capable of catalyzing the conversion of IPyA to IAA.
CKRC1/TAA1 and CKRC2/YUC8 are involved in the regulation of auxin biosynthesis by CK
The interaction between auxin and CK plays a key role in plant growth and development. Recently, it was suggested that the IPyA pathway catalyzed by TAA/TAR and YUC enzymes was the main pathway for auxin biosynthesis in Arabidopsis14,16,24. Our previous studies reveal that the ARABIDOPSIS HISTIDINE KINASE3 (AHK3)/ARABIDOPSIS RESPONSE REGULATOR1 (ARR1)/ARR12 involved in CK signaling can stimulate auxin biosynthesis via up-regulating the transcription of CKRC1/TAA1 and other auxin biosynthetic genes including YUC821. However, Dr5::GUS activity after CK treatment was reported to be decreased in the root tips of ckrc1 mutants but not changed significantly in WT root tips, a phenomenon suggested to be due to the dual effect of CK on auxin biosynthesis (positive) and polar transport (negative)21. Interestingly, a similar decrease in activity was also observed in ckrc2-1 mutant after tZ treatment (Fig. 4a,b) suggesting a similar role for CKRC2/YUC8 in auxin-CK crosstalk. Promoter-GUS staining and qRT-PCR analysis revealed that, like for CKRC1/TAA1 (Fig. 4c,d), the up-regulation of CKRC2/YUC8 was impaired in ahk3-1 and arr1-3/12-1 mutants (Fig. 4d,e). Therefore, CK can stimulate auxin biosynthesis in roots by up-regulating both CKRC1/TAA1 and CKRC2/YUC8 genes via the AHK3-ARR1/12 signaling pathway.
The IPyA pathway catalyzed by CKRC1/TAA1 and CKRC2/YUC8 is involved in CK induced auxin biosynthesis in roots.
Dr5::GUS expression (a) and transcription (qRT-PCR) (b) in root tips of wildtype Col plants as well as ckrc2-1 and ckrc1-1 mutants, shows the reduction of GUS activity in the two mutants after tZ treatment; (c) The GUS staining of pCKRC1::GUS and pCKRC2::GUS transgenic roots increases after tZ treatment; (d,e) The AHK3-ARR1/12 signalling pathway is involved in the process of induction of CKRC1 (d) and CKRC2 (e) by CK. The seedlings were grown on MS medium with or without 0.1 μM tZ for 7d and their roots were used for RNA extraction. Each staining shown in (a,c) represents data on at least 20–30 roots. Data show the mean of 3 biological replicates. Error bars indicate ± SD. Different letters indicate significant differences at P < 0.05 according to ANOVA followed by Tukey’s multiple comparison tests.
PIF4 is required for the regulation of auxin biosynthesis by CK
Auxin biosynthesis can be regulated by many factors, such as CK20,21,28,29, ethylene30,31, jasmonate32,33,34, temperature35,36,37 and light38,39. Both CK and high temperature can stimulate auxin biosynthesis by up-regulating the transcription of CKRC1 and CKRC2 in roots (Fig. 4c–e & Supplementary Fig. S9), and the effects of temperature are reported to be mediated by phytochrome-interacting factor 4 (PIF4). PIF4 acts as a transcription factor to promote the transcription of CKRC1 and CKRC235,36,37. To explore whether PIF4 also plays a role in the CK-dependent regulation of auxin biosynthesis, the CK-mediated inductions of CKRC1 and CKRC2 in the loss-of-function mutant of PIF4 (pif4) were compared with the wild-type Col-0. The results showed that CKRC1 and CKRC2 genes were no longer induced obviously in pif4 (Fig. 5b,c) indicating that PIF4 is essential for transcriptional regulation of CKRC1 and CKRC2 by CK. As for ckrc1 and ckrc2 the relative Dr5::GUS expression in pif4 mutants after tZ treatment was significantly reduced (Fig. 5a). Importantly, we found that tZ treatment also had a positive effect on the transcription of PIF4 suggesting that PIF4 itself is regulated by CK (Fig. 5d,e). qRT-PCR analysis showed that the induction of PIF4 by tZ is impaired in the CK-signaling mutants ahk3 and arr1/12 (Fig. 5e). Taken together, these results put the transcriptional factor PIF4 between AHK3-ARR1/ARR12 signaling and the transcriptional induction of CKRC1/2 for mediating CK-dependent regulation of auxin biosynthesis. However, the effect of the pif4 mutant was much weaker compared to the responses of ckrc1-1 and ckrc2-1 mutants to tZ and IAA treatment (Supplementary Figs S10 and S11). This might be due to the fact that in pif4 mutant roots the CKRC1-CKRC2 catalyzed IPyA pathway is functional and thus this mutant has more locally synthesized IAA than the ckrc1-1 or ckrc2-1 mutants. A comparison of the CK-induced CKRC1/2 transcription between WT and the mutants pif5, pif7 and erf109 -these proteins are reported to have binding activity to the YUC8 promoter to mediate responses to temperature35,36,37 or jasmonates32 - revealed no significant roles of these three proteins in CK regulation (Supplementary Fig. S12).
PIF4 is required for the regulation of auxin biosynthesis by CK.
(a) Dr5::GUS expressions in Col and pif4 after tZ treatment; Effects of CK on the transcriptions of CKRC1 (b) and CKRC2 (c) genes in WT and pif4; (d) GUS staining of pPIF4::GUS on MS medium with or without tZ; and (e) Relative transcription of PIF4 in Col and different CK signaling mutants. The roots of 7-day-old seedlings grown on MS medium with or without 0.1 μM tZ were used for qRT-PCR. Each staining shown in (a,d) are representative images of at least 20–30 roots. Data are mean values of 3 replicates. Error bars indicate ± SD. Different letters indicate significant differences at P < 0.05 according to ANOVA followed by Tukey’s multiple comparison tests.
In summary, CKRC2 encodes a previously identified member of YUCCA (YUC) flavin monooxygenase-like proteins (YUC8) and is expressed abundantly in roots. Like other YUCs, CKRC2/YUC8 is a rate-limiting enzyme for catalyzing the conversion of IPyA to IAA and is acting downstream of CKRC1/TAA1 in the IPyA pathway. The transcription of both CKRC1/TAA and CKRC2/YUC8 genes can be induced by CK, and the phytochrome-interacting factor 4 (PIF4) is required for this regulation via the classical AHKs-ARR1/12 signalling pathway. Thus, PIF4 plays an essential role in mediating the regulatory effect of CK on the transcription of CKRC1 and CKRC2 genes in the IPyA pathway of auxin biosynthesis. The appearance of an ARR1 recognition sequence (5′-AGATT-3′) in the 600 bp region upstream from the PIF4 translation start site implies that PIF4 may be directly targeted by ARR140. This possibility should be tested by yeast one-hybrid or Chip analysis in the future. These results together with previously reported data30,31,35,36,37,38,39, suggest that PIF4 is a central factor to mediate the regulation of auxin biosynthesis (Fig. 6).
Methods
Plant material and growth conditions
For mutant screening, the Arabidopsis thaliana activation-tagged T-DNA pools (CS31100, Col-2 background, composed of approximately 62,000 individual lines)41, were purchased from the Arabidopsis Biological Resource Center (ABRC) ( http://abrc.osu.edu/). Germination and plant growth was carried out at 25 °C with a 1 h light/8 h dark cycle. For growth analyses, seedlings were grown for 7 days on vertical Miller-Skoog plates (MS) (1.1% w/v agar and 10 g/L sucrose)21,42.
Arabidopsis accession Col-2 was used as wild-type control. The following mutants have been used in this study: pDR5::GUS marker line43; ckrc1-1 (ckrc1-1/pDR5::GUS; At1g70560)21,22, ckrc2-2 (N655757; At4g28720), yucca1 (N655809; At4g32540), yucca2 (N659779; At4g13260), yucca4 (N104041; At5g11320), yucca6 (N663363; At5g43890), yucca7 (N659416; At2g33230), yucca11 (N573485; At1g21430), arr1-3/12-1 (N6981; At3g16857/At2g25180) and ahk3-1 (N6562; At1g27320) were purchased from the The Nottingham Arabidopsis Stock Centre (NASC); Seeds for yucca3 (GABI_376G12; At1g04610), yucca5 (CSHL_GT6160; At5g43890), yucca9 (SAIL_762-D07; At1g04180) and yucca10 (FLAG_599G05; At1g48910) were kindly provided by Prof. Yunde Zhao (University of California, San Diego); pYUC8::GUS (line 132.1/132.2) and p35S::YUC8 (line 027.10.1W) by Prof. Stephan Pollmann (Campus de Montegancedo, Madrid) and the double homozygous pif4/pDR5::GUS (AGI codes: At2g43010) mutants by Prof. Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences)37.
To generate ckrc2-1/pDR5::GUS mutants, the ckrc2-1 mutant was crossed with Col/DR5::GUS and the double homozygous mutant in F2 was identified by the non-segregation of GUS staining in its seeds (F3).
Plant Vectors and Transformations
For constructing pCKRC2::CKRC2, the CKRC2 cDNA with stop codon and promoter were introduced into pCAMBIA1300 binary vector. The primers used for cloning pCKRC2::CKRC2 were YUC8-F/R (Supplementary Appendix S1). For pCKRC1::CKRC2, the full length cDNA was amplified using YUC8-cDNA-F/R (Supplementary Appendix S1) and introduced into pCKRC1::GUS vector21. After sequencing confirmation ( http://www.genomics.cn/), the constructed binary vectors were introduced into either wild-type Col plants (for pCKRC1::CKRC2) or ckrc2-1 mutant plants (for pCKRC1::CKRC2 and pCKRC2::CKRC2) via Agrobacterium tumefaciens-mediated (strain GV3101) floral-dip transformation method44. Selection for transgenic plants was performed as described in previous studies21.
For CKRC2 protein expression vectors, the CKRC2 cDNA without stop codon were introduced into the vector pET28b+. The primers used for amplifying the YUC8 cDNA were YUC8-P-R/F (Supplementary Appendix S1). After confirming the amplified cDNA by sequencing, vectors were transformed into E. coli DH5α for analysis of the enzymatic activity.
Phenotype characterization
For root inhibition assays and biochemical complementation, seeds were germinated and grown vertically on MS medium with various hormones or compounds at 25 °C with a 16/8 h light/dark cycle for 7 days. Data shown are the mean values of three separate experiments using at least 40 seedlings.
For analyzing root gravitropism germinated seedlings were transferred to fresh MS media and grown vertically on MS plates 5 days at 25 °C with a 16/8 h light/dark cycle. Three hours later, the plates were rotated 90 degrees, further incubated for 24 h and the degree of gravitropic response was measured for each root. Approximately 100 seedlings were measured for each genotype and treatment.
TAIL-PCR, genetic mapping and identification of mutated gene
TAIL-PCR was performed according to Liu et al.45. In brief, tail-PCR was used to clone the T-DNA flanking sequences in isolated mutants45,46. All PCR products were electrophoretically separated on a 1% agarose gel and the expected TAIL-3 products were purified and sequenced. DNA sequences were aligned with Blastn ( http://www.ncbi.nlm.nih.gov/BLAST/) and Tair10 ( http://www.arabidopsis.org/Blast/) software. Map-based cloning was performed using F2 populations generated by backcrossing the ckrc2-1 mutant (Col background) with Landsberg (Ler) ecotype wild-type plants, as previously described47. High-throughput sequencing (ShangHai Biotechnology Corporation, China, http://www.shbiotech.org/ and Hangzhou Guhe Information and Technology Co., Ltd, China, http://www.guheinfo.com/) was used to reveal the exact DNA mutation in the mapped region of the mutation.
RNA preparation and real-time qRT-PCR analysis
RNA was extracted using Trizol agent (Sangon, http://www.sangon.com/). Reverse transcription was performed by a reverse transcription kit (DRR047A) (Takara, http://www.takara-bio.com/). For synthesizing first-strand cDNA 1 μg of total RNA was used after the pretreatment by RNase free-DNase. The cDNA was diluted 5 times for Real-Time PCR.
For quantitative RT-PCR (qRT-PCR) 20 μL amplification reactions (10 μL SYBR Premix Ex Taq (Takara, http://www.takara-bio.com/), 0.8 μL of each primer (Supplementary Appendix S1), 1.6 μL cDNA and 6.4 μL ddH2O) were used. The results of each primer pair were normalized relative to ACTIN 8 (AGI codes: At1g49240). All realtime qRT-PCR amplifications were performed in a Bio-Rad CFX96TM Real-time System (Bio-Rad, http://www.bio-rad.com). The following PCR program was used: An initial denaturation at 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s. During the melting curve analysis, PCR reactions were denatured at 95 °C for 15 min. Each experiment was repeated three times and each reaction was performed in triplicates48.
For analyzing the CK- and auxin-induced ARR5/15 (AGI codes: At3g48100/At1g74890) and IAA1/2 (AGI codes: At4g14560/At3g23030) transcription, 7-day-old seedlings grown on MS medium were treated in liquid MS medium with 10 μM trans-zeatin (tZ) for 30 minutes or 20 μM IAA for 1.5 hours49,50.
For analyzing the CK induced gene transcription, 7-day-old seedlings grown on MS and/or on 0.1 μM tZ medium were used.
Histochemical GUS assay
7-day-old seedlings containing a GUS marker were grown on medium with or without 0.1 μM tZ and then incubated in 1 mM X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide) and 50 mmol/L potassium phosphate buffer, pH7.5, with 0.1% v/v Triton X-100 for GUS staining at 37 °C for 45 minutes (Col/Dr5::GUS, ckrc1-1/Dr5::GUS, ckrc2-1/Dr5::GUS and pif4/Dr5::GUS), 1 h (pCKRC1::GUS) and 3 h (pCKRC2::GUS and pPIF4::GUS).
Cell-free protein expression and enzyme activity tests
For protein expression, a S30 T7 High-Yield Protein Expression System (Promega) was used according to the manufacturer’s instructions. This kit contains all components used for expressing proteins in prokaryotic expression system and plasmid DNA encoding Renilla Luciferase as control protein.
To test the enzyme activities about 2 μg protein (YUCCA1, CKRC2 and Renilla Luciferase) (the amount was estimated from SDS-PAGE by comparison with the protein marker), NADPH (50 mM) 20 μL, FAD (2 mM) 2 μL, IPyA (50 mM) 0.4 μL and add nuclease-free water to a final volume of 100 μL. The mixture was incubated at 30 °C for 2 hours with vigorous shaking. For quantitative analysis, the reaction mixture was diluted 100 times before the measurement. The fluorescence intensities were measured at λex/λem = 363 nm/277 nm in a 1 cm quartz cell and with a slit at 2 nm for the excitation and 5 nm for the emission. Scan speed were 350 nm/min. The fluorescence values were quantified using a standard curve (y = 380.8x + 36.31).
Additional Information
How to cite this article: Di, D.-W. et al. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci. Rep. 6, 36866; doi: 10.1038/srep36866 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Di, D. W., Zhang, C., Luo, P., An, C. W. & Guo, G. Q. The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regulation 78, 275–285 (2016).
Mano, Y. & Nemoto, K. The pathway of auxin biosynthesis in plants. J Exp Bot 63, 2853–2872 (2012).
Shi, H. et al. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant physiology and biochemistry: PPB/Societe francaise de physiologie vegetale 82, 209–217 (2014).
Teale, W. D., Paponov, I. A. & Palme, K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7, 847–859 (2006).
Woodward, A. W. & Bartel, B. Auxin: regulation, action, and interaction. Annals of botany 95, 707–735 (2005).
Korasick, D. A., Enders, T. A. & Strader, L. C. Auxin biosynthesis and storage forms. J Exp Bot 64, 2541–2555 (2013).
Chandler, J. W. Auxin as compere in plant hormone crosstalk. Planta 231, 1–12 (2009).
Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harbor perspectives in biology 2, a001594 (2010).
Wang, B. et al. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112, 4821–4826 (2015).
Zhao, Y. et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 (2001).
Cheng, Y., Dai, X. & Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & development 20, 1790–1799 (2006).
Dai, X. et al. The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288, 1448–1457 (2013).
Kriechbaumer, V., Wang, P., Hawes, C. & Abell, B. M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70, 292–302 (2012).
Mashiguchi, K. et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108, 18512–18517 (2011).
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008).
Won, C. et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108, 18518–18523 (2011).
Tivendale, N. D. et al. Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol 154, 1957–1965 (2010).
Ziegler, D. M. Flavin-containing monooxygenases: enzymes adapted for multisubstrate specificity. Trends in pharmacological sciences 11, 321–324 (1990).
Schaller, G. E., Bishopp, A. & Kieber, J. J. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63 (2015).
Jones, B. et al. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22, 2956–2969 (2010).
Zhou, Z. Y. et al. Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J 66, 516–527 (2011).
Wu, L. et al. Forward genetic screen for auxin-deficient mutants by cytokinin. Sci Rep 5, 11923 (2015).
Chen, Q. et al. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant & cell physiology 55, 1072–1079 (2014).
Stepanova, A. N. et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 (2011).
Tao, Y. et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008).
He, W. et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960 (2011).
Sharma, H., Jain, V. K. & Khan, Z. H. Use of constant wavelength synchronous spectrofluorimetry for identification of polycyclic aromatic hydrocarbons in air particulate samples. Spectrochim Acta A 108, 268–273 (2013).
Choi, Y. I., Noh, E. W., Kim, H. J. & Park, W. J. Differential regulation of cytokinin oxidase genes and cytokinin-induced auxin biosynthesis by cellular cytokinin level in transgenic poplars. Plant Cell Rep 33, 1737–1744 (2014).
Zhu, J. et al. Low Temperature Inhibits Root Growth by Reducing Auxin Accumulation via ARR1/12. Plant & cell physiology 56, 727–736 (2015).
Alarcon, M. V., Lloret, P. G. & Salguero, J. Synergistic action of auxin and ethylene on root elongation inhibition is caused by a reduction of epidermal cell length. Plant Signal Behav 9, e28361 (2014).
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A. & Alonso, J. M. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242 (2005).
Cai, X. T. et al. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5, 5833 (2014).
He, Y. & Zhao, Y. A key link between jasmonic acid signaling and auxin biosynthesis. Science China. Life sciences 58, 311–312 (2015).
Hentrich, M. et al. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74, 626–637 (2013).
Franklin, K. A. et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108, 20231–20235 (2011).
Stavang, J. A. et al. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60, 589–601 (2009).
Sun, J., Qi, L., Li, Y., Chu, J. & Li, C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8, e1002594 (2012).
Ma, D. et al. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113, 224–229 (2016).
Pedmale, U. V. et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 164, 233–245 (2016).
Sakai, H., Aoyama, T. & Oka, A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24, 703–711 (2000).
Weigel, D. et al. Activation tagging in Arabidopsis. Plant Physiol 122, 1003–1013 (2000).
Zhang, C. et al. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 22, 2353–2369 (2010).
Ulmasov, T., Hagen, G. & Guilfoyle, T. J. AuxREs and AuxRE transcription factors: a model for auxin-responsive gene expression. Plant Physiology 114, 1255–1255 (1997).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743 (1998).
Liu, Y. G., Mitsukawa, N., Oosumi, T. & Whittier, R. F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8, 457–463 (1995).
Wu, L. et al. Frequent problems and their resolutions by using thermal asymmetric interlaced pcr (tail-pcr) to clone genes in arabidopsis t-dna tagged mutants. Biotechnology & Biotechnological Equipment 29, 260–267 (2015).
Weigel, D. G., J. Arabidopsis: a laboratory manual (Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press, c2002).
Di, D. W. et al. Analysis the role of arabidopsis CKRC6/ASA1 in auxin and cytokinin biosynthesis. J Plant Biol 59, 162–171 (2016).
Laxmi, A., Paul, L. K., Raychaudhuri, A., Peters, J. L. & Khurana, J. P. Arabidopsis cytokinin-resistant mutant, cnr1, displays altered auxin responses and sugar sensitivity. Plant molecular biology 62, 409–425 (2006).
Tian, Q., Uhlir, N. J. & Reed, J. W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319 (2002).
Acknowledgements
We thank Prof. Jiayang Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for critical reading of the manuscript. We thank Dr Jane Murfett (Department of Biochemistry, University of Missouri, Columbia, MO) for providing DR5::GUS, Prof. Stephan Pollmann (Centro de Biotecnología y Genómica de Plantas, Campus de Montegancedo, Madrid, Spain) for p35S::YUC8 and pYUC8::GUS, Prof. Yunde Zhao (Cell and Developmental Biology, University of California San Diego, La Jolla, CA) for yuc3, yuc5, yuc9 and yuc10; Prof. Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for pif4/pDR5::GUS; Prof. Peter H. Quail (Plant & Microbial Biology, University of California, Albany, CA) for pPIF4::GUS, the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre for mutant pools and individual lines. This work was supported by grants from the National Natural Science Foundation of China (31030045 and 31671458).
Author information
Authors and Affiliations
Contributions
G.Q.G. designed the research. D.W.D. isolated the mutant and performed major parts of the experiments. L.W., L.Z., C.W.A., T.Z.Z., P.L. and H.H.G. performed parts of the research. D.W.D., G.Q.G. and L.W. analyzed data, tested statistics, and coordinated the figures. G.Q.G. and D.W.D. wrote the article. D.W.D., V.K. and G.Q.G. revised the article.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Di, DW., Wu, L., Zhang, L. et al. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci Rep 6, 36866 (2016). https://doi.org/10.1038/srep36866
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36866
This article is cited by
-
PIN5 is involved in regulating NH4+ efflux and primary root growth under high-ammonium stress via mediating intracellular auxin transport
Plant and Soil (2023)
-
Significance of NatB-mediated N-terminal acetylation of auxin biosynthetic enzymes in maintaining auxin homeostasis in Arabidopsis thaliana
Communications Biology (2022)
-
Function of histone H2B monoubiquitination in transcriptional regulation of auxin biosynthesis in Arabidopsis
Communications Biology (2021)
-
Adipose tissue hyaluronan production improves systemic glucose homeostasis and primes adipocytes for CL 316,243-stimulated lipolysis
Nature Communications (2021)
-
Emerging functions of chromatin modifications in auxin biosynthesis in response to environmental alterations
Plant Growth Regulation (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.