Abstract
Emergency granulopoiesis occurs in response to severe microbial infection. However, whether and how other blood components, particularly monocytes/macrophages and their progenitors, including hematopoietic stem/progenitor cells (HSPCs), participate in the process and the underlying molecular mechanisms remain unknown. In this study, we challenged zebrafish larvae via direct injection of Escherichia coli into the bloodstream, which resulted in systemic inoculation with this microbe. The reaction of hematopoietic cells, including HSPCs, in the caudal hematopoietic tissue was carefully analysed. Both macrophages and neutrophils clearly expanded following the challenge. Thus, emergency myelopoiesis, including monopoiesis and granulopoiesis, occurred following systemic bacterial infection. The HSPC reaction was dependent on the bacterial burden, manifesting as a slight increase under low burden, but an obvious reduction following the administration of an excessive volume of bacteria. Pu.1 was important for the effective elimination of the microbes to prevent excessive HSPC apoptosis in response to stress. Moreover, Pu.1 played different roles in steady and emergency monopoiesis. Although Pu.1 was essential for normal macrophage development, it played suppressive roles in emergency monopoiesis. Overall, our study established a systemic bacterial infection model that led to emergency myelopoiesis, thereby improving our understanding of the function of Pu.1 in this scenario.
Similar content being viewed by others
Introduction
When vertebrates are infected by pathogens such as bacteria, the immune cells, particularly mononuclear (monocytes/macrophages) and polymorphonuclear (granulocytes) myeloid phagocytes, respond immediately1. A slight infection such as a localised bacterial challenge induces the recruitment of myeloid phagocytes, particularly granulocytes, from the bloodstream, but exerts a limited influence on their exhaustion and expansion2,3. However, severe infection, including systemic bacterial inoculation, usually results in a bacteraemia-like syndrome. Granulocytes are intensively involved in this process, resulting in their significant exhaustion and expansion, which is called emergency granulopoiesis2,3. Emergency granulopoiesis is achieved via the activation of granulocyte progenitors, including hematopoietic stem and progenitor cells (HSPCs)2,3,4.
As the ancestral cells of all blood components, HSPCs are a heterogeneous population, and only a limited portion comprises bona fide hematopoietic stem cells (HSCs), which reside quiescently in the bone marrow under steady conditions4,5,6,7,8,9. Although these physiologically dormant HSCs quickly enter the cell cycle upon challenge4,5,6,7,8,9, how they react to pathogens remains unclear, primarily because of the difficulty in isolating pure HSCs3,4. However, the response of HSPCs and their subsequent developmental potential under demand-derived emergency conditions, such as when encountering pathogens or when stimulated by cytokines, has been the focus of research recently1,4,6. Different pathogens induce different HSPC reactions6. Additionally, the route and severity of the infection lead to different HSPC outcomes1,4,6. Moreover, HSPCs are prone to granulocyte production by sacrificing lymphoid cells in infection-induced emergency granulopoiesis1,2. Although the reaction of bone marrow-derived HSPCs has been examined1,4,6, how HSPCs respond during the embryonic stages has rarely been addressed.
Zebrafish (Danio rerio) provides an ideal system for studying bacterial infection-stimulated immune response, particularly the interaction of phagocytes with microbes, owing to its optical transparency and the exclusive involvement of myeloid phagocytes in early embryos10,11,12,13. Using this model, the behaviours of phagocytes, including their response, mobility, movement, interaction, and engulfment, have been reported10,11,12,13. Recently, a study investigated emergency granulopoiesis through local infection of larvae in the hindbrain10. However, to our knowledge, myelopoiesis in systemically infected larvae and the role of HSPCs in this phenomenon have not been elucidated.
HSPCs emerge in zebrafish from approximately 36 hours post-fertilization (hpf) via endothelial-to-hematopoietic transition14,15. From 2 days post-fertilization (dpf), these HSPCs migrate to the caudal hematopoietic tissue (CHT), an organ transiently supporting hematopoiesis and the functional homolog of the fetal liver and placenta16,17,18,19. CHT refers to the lumen between the caudal artery (CA) and the definitive caudal vein (dCV). Structurally, it is a complex vascular network composed of a fibroblastic reticular-cell matrix, loose mesenchyme, and expanded blood progenitors16,17,18,19. After a transient stay, these HSPCs move to the kidney, which is the functional equipment of the bone marrow17,19,20, from 3–4 dpf on, and eventually, give rise to all blood components. Some of the HSPCs migrate to the thymus to generate T lymphoid cells21. Functional T lymphoid cells are detected after 6 dpf, and the adaptive immunity functions until the juvenile period22.
Taking advantage of the well-addressed process of hematopoiesis20, bacterial inoculation of early embryos provides an effective assay for investigating infection-induced emergency hematopoiesis, in addition to elucidating the behaviour of myeloid phagocytes. Consequently, an improved understanding of the cellular and molecular mechanisms responsible for emergency hematopoiesis can be achieved. In this study, we established a systemic infection model by injecting the non-pathogenic bacteria, Escherichia coli, into the bloodstream of zebrafish embryos. CHT16,17,18,19 was examined to elucidate the influence of the bacteria on various phagocytes and their progenitors, including HSPCs.
Results
Both macrophages and neutrophils in the CHT are involved in bacterial phagocytosis upon systemic infection
To investigate the hematopoietic reaction following systemic inoculation of larvae with microbes, 2 dpf larvae were challenged by intravenous injection of Dsred-labelled E. coli13,23. A site close to the ear, where vessels are enriched (Fig. 1A), was chosen as the injection site to facilitate rapid inoculation of the microbes into the blood circulation. When the microbes were administered, tremendous amounts of circulating bacteria were identified by intensive red fluorescent signals moving through the vessels (see supplementary Video S1 and S2). Gradually, the circulating microbes disappeared; instead, large clusters of Dsred+ bacterial foci were detected13,23 (Fig. 1B; see supplementary Video S1 and S2). In parallel with the disappearance of moving bacteria in the circulation, the number of large foci increased to approximately 16 in the CHT16,19,24 at 1 day post-injection (dpi). However, this number decreased thereafter, and the bacteria almost completely disappeared by 6 dpi (Fig. 1B). The alteration of foci reflected bacterial phagocytosis and digestion by the myeloid phagocytes, which are the only immunocytes that function at these stages13,23. To monitor the behaviour of both myeloid phagocytes, Tg(mpeg1:eGFP)25 and Tg(lyz:eGFP)nz117 26, which specifically label macrophages and neutrophils, respectively, were exploited. The mpeg1-GFP+ macrophages reacted immediately to interact with and engulf the microbes13,23 (Fig. 1D, white stars). At approximately 30 minutes post-injection, the macrophages had engulfed numerous microbes, resulting in the formation of large red foci in the CHT (Fig. 1D; see supplementary Video S1; white stars). Over time, the number of active macrophages increased markedly, and approximately 72% of them were observed to actively engulf microbes at 6 hours post-injection (hpi) (Fig. 1C), which was consistent with the numerous large E. coli foci observed in the CHT (Fig. 1B). Gradually, macrophages with a huge microbe burden underwent cell death, manifested by weakened and even lost GFP signals, and these scarified macrophages were quickly engulfed by their surrounding macrophages (see supplementary Video S1, white arrowheads). The lyz-GFP+ neutrophils also phagocytosed bacteria following treatment with Tg(lyz:eGFP)nz117 26. Interestingly, the phagocytic behaviour of neutrophils was distinct from that of macrophages. Neutrophils first aggregated the bacteria on their surface, resulting in their encircling by more extensive Dsred+ signals (Fig. 1E; see supplementary Video S2, white arrowheads), followed by engulfment of the bacteria13 (see supplementary Video S2, white arrowheads). However, the reaction sensitivity and phagocytic ability of neutrophils were less efficient than those of macrophages27. Only approximately 37% of the total lyz-GFP+ neutrophils in the CHT performed phagocytosis, which was half the rate determined for macrophages at similar time points (Fig. 1C). Therefore, both macrophages and neutrophils were involved in phagocytosis when encountering bacteria in the CHT.
Bacterial (E. coli) phagocytosis by both macrophages and neutrophils following intravenous injection.
(A), The intravenous injection site (red arrow) of E. coli (5–10 × 103 cfu) in 2 dpf embryo. Scale bars, 200 μm. (B), The numbers of Dsred+ E. coli foci in the CHT regions at different time points after injection (13.90 ± 1.31; 16.20 ± 2.35; 11.90 ± 1.33; 4.60 ± 0.65; 2.20 ± 0.33; 1.40 ± 0.40; 0.60 ± 0.22 at each time points; 10 embryos were counted in each group). (C), The percentage of macrophages (72.06 ± 4.44, N = 8) and neutrophils (36.93 ± 8.89, N = 8) involved in bacterial phagocytosis at 6 hpi in the CHT. (D,E), Time-lapse imaging of an infected Tg(mpeg1:eGFP) (D) or Tg(lyz:eGFP) (E) CHT from 0.5 hpi to 6 hpi. The white stars in (D) denote mpeg1-GFP+ macrophages that engulfed large amounts of Dsred+ E. coli. The white arrowheads in (E) indicate the initial aggregation of Dsred+ E. coli on the surface of lyz-GFP+ neutrophils, which were quickly phagocytosed. The red foci in (D,E) represent the phagocytosed bacteria. Scale bars, 20 μm. See also Video S1 and S2.
Inoculation of microbes into the bloodstream leads to emergency granulopoiesis
Emergency granulopoiesis has been detected in larvae infected through the hindbrain10. Whether a similar phenomenon was recapitulated in the CHT during systemic infection was investigated. We first examined several neutrophil markers, including cebp1, lyz, mpx, and Sudan Black (SB)28, at 2 dpi. All examined neutrophil markers displayed a drastic increase in the treated larvae compared with the controls (Fig. 2A; see supplemental Figure S1A–C). Moreover, the degree of neutrophil expansion depended on the bacterial burden. More significant expansion of SB+ cells was observed with 5–10 × 103 colony-forming units (cfu) than with 5–10 × 102 cfu E. coli (see supplemental Figure S1D). However, when the E. coli volume reached 5–10 × 104 cfu, the larvae showed remarkable mortality (see supplemental Figure S1E), and approximately half of the surviving larvae presented obvious morphological abnormalities, exemplified by pericardium oedema (see supplemental Figure S1F). A larger volume of microbes caused a stronger neutrophil reaction, but an excessive burden led to fish abnormality and lethality. Therefore, 5–10 × 103 cfu E. coli was chosen as the dosage in all further experiments because it caused a remarkable immune response with few morphological defects and low lethality, thus permitting continuous investigation of the challenge-induced emergency hematopoietic reaction.
Emergency granulopoiesis occurs following intravenous E. coli (5–10 × 103 cfu) infection.
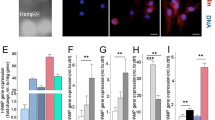
(A), The drastic expansion of SB+ neutrophils (red arrows) in the CHT of an infected embryo when compared with that treated with PBS at 2 dpi (4 dpf). (B), Calculation of the data obtained for SB+ neutrophils at different time points after infection (15.60 ± 2.80 vs 28.20 ± 5.27; 65.71 ± 10.60 vs 55.20 ± 4.76; 149.50 ± 8.70 vs 77.60 ± 9.45; 207.60 ± 39.15 vs 128.80 ± 15.00; 96.40 ± 23.80 vs 56.40 ± 6.33 in E. coli vs PBS group at each time point. N = 8 in each group). (C,D), Fluorescence images (C) and calculation (D) of lyz-GFP+ neutrophils in PBS (25.11 ± 2.58; N = 9) or E. coli (16.50 ± 1.84; N = 10) treated larval CHT at 6 hpi. The red signals indicate the bacteria phagocytosed by lyz-GFP+ neutrophils (white arrows). (E), The percentage of lyz-GFP+ neutrophils that are co-stained with TUNEL at 6 hpi (9.31 ± 1.87 vs 0.63 ± 0.37 in E. coli vs PBS group. N = 10 in each group). (F), Time-lapse imaging of an infected Tg(lyz:eGFP) CHT from 1 dpi to 1.5 dpi. Obvious generation, expansion and maturation of lyz-GFP+ neutrophils are observed. Scale bars, 20 μm. See also Video S4. 4a, 4b and 8a, 8b in (F) showing the dividing lyz-GFP+ cells. (G), SB+ signals (red arrows) in the runx1 mutant treated by either PBS or E. coli at 2 dpi (4 dpf). (H), WISH of pu.1 (red arrows) in the CHT of an embryo challenged with PBS or E. coli at 2 dpi (4 dpf). (I), The number of the pu.1+ myeloid progenitors at different time points after challenge (45.88 ± 5.87 vs 59.63 ± 4.29; 203.38 ± 5.02 vs 79.88 ± 8.70; 214.50 ± 13.98 vs 78.16 ± 6.89 in E. coli vs PBS group at each time point. N = 8 in each group). Scale bars, 20 μm.
Next, the number of SB+ neutrophils was calculated. A transient reduction of SB+ neutrophils was detected at 6 hpi (Fig. 2B), suggesting their early exhaustion. To confirm this phenomenon, the challenged Tg(lyz:eGFP)nz117 26 were examined carefully. The lyz-GFP+ population showed a similar reduction as that of SB+ neutrophils (Fig. 2D). Further investigation revealed obvious phagocytosis of the Dsred+ microbes by the lyz-GFP+ neutrophils (Fig. 2C, white arrows) and a corresponding increase in the portion of lyz-GFP+/terminal deoxynucleotidyl transferase dUTP nick-end labelling (TUNEL)+ cells in the microbe-treated larvae when compared with their controls (Fig. 2E). These results suggested that the early exhaustion of neutrophils was caused by their increased apoptosis while fighting the bacteria. Subsequently, the SB+ neutrophil population began to expand beginning at 1 dpi, reaching their maximal level at 2–4 dpi, and then returning to physiological baseline numbers from 6 dpi on (Fig. 2B). The expansion of neutrophils indicated that emergency granulopoiesis occurred in response to systemic infection. CHT is a transient organ for definitive hematopoiesis16,17,18,19, and therefore, the origin of expanded neutrophils in this region was explored. Time-lapse images were acquired from 1 dpi for the treated Tg(lyz:eGFP)nz117 26, and the data revealed the generation of nascent lyz-GFP+ neutrophils in the niche adjacent to the CA (Fig. 2F; see supplementary Video S4), where more immature progenitors resided24. The lyz-GFP+ neutrophils showed weak signals initially that gradually increased in strength, indicating differentiation of the neutrophils after challenge. Meanwhile, the lyz-GFP+ neutrophils divided more frequently in E. coli-treated larvae. An average of 8.00 ± 0.41 divisions were observed in four imaged larvae, which is approximately four-times higher than that in the control groups (only 2.33 ± 0.33 divisions were observed in three control larvae during the imaging time window) (Fig. 2F; see supplementary Video S3 and S4). As a result, the lyz-GFP+ neutrophil numbers increased. The in situ expansion of neutrophils in the CHT predicted a definitive hematopoietic origin10. To test our hypothesis, we utilised a runx1w84x mutant, in which definitive hematopoiesis is abolished24. The results revealed no detectable expansion of SB+ neutrophils in challenged runx1w84x larvae compared with the control (Fig. 2G). This finding suggested that the expanded neutrophils were largely generated from runx1-regulated definitive hematopoiesis24, in agreement with a previous study10. The enhanced output of neutrophils supported the activation and expansion of their progenitors. This hypothesis was verified by the significantly enhanced output of both pu.1+ and cebpα+ myeloid progenitors at 2 dpi (Fig. 2H; see supplemental Figure S2A). Further calculations demonstrated that the increase in myeloid progenitors was initiated as early as 1 dpi (Fig. 2I), a time point prior to the tremendous output of neutrophils (Fig. 2B). However, the myeloid progenitors did not display a reduction at 6 hpi (Fig. 2I), suggesting limited exhaustion of myeloid progenitors in the initial fight against the bacteria. The expansion of myeloid progenitors predicted their higher proliferation upon challenge. To support this hypothesis, Tg(coro1a:eGFP)29, which marks both myeloid phagocytes and their progenitors, was evaluated by anti-phospho-histone H3 (pH3) antibody staining30. A remarkable increase in pH3+/coro1a-GFP+ cells was detected in the microbe-treated larvae, when compared with the control, at 1 dpi (see supplemental Figure S2 B,C). Together, these results suggested that the myeloid progenitors were activated, and emergency granulopoiesis occurred in the larvae following systemic infection.
Distinct reactions of the HSPC compartment in response to different bacterial burdens
Because no specific HSC-labelling method is available in zebrafish, we mainly utilised Tg(runx1:en-GFP), in which runx1-GFP largely marks the HSPC compartment31, to dissect their reactions in our assay. To our surprise, runx1-GFP+ cells did not show an obvious increase in the CHT after challenge by 5–10 × 103 cfu E. coli (Fig. 3A–C), even when the myeloid progenitors were drastically expanded (Fig. 2H; see supplemental Figure S2A), suggesting distinct reactions of HSPCs and myeloid progenitors upon infection. To confirm this result, we employed another HSPC compartment marker, cmyb, using both whole-mount in situ hybridization (WISH) and its reporter line32. The results revealed a similar number of cmyb+ cells in both control and challenged larvae (Fig. 3D–F; see supplemental Figure S3A–C), suggesting that HSPCs maintained their homeostasis during this process. The reaction of HSPCs upon challenge was distinct from that observed previously10,33,34, and the underlying reasons were explored by first considering the bacterial burden6. Different volumes of E. coli were administered. When the volume was 5–10 × 102 cfu, the runx1-GFP+ HSPCs showed a notable expansion (Fig. 3A,C). Surprisingly, when the burden increased, the number of HSPCs decreased, and 5–10 × 104 cfu E. coli led to a significant reduction in runx1-GFP+ cells in surviving larvae with a normal appearance at 2 dpi (Fig. 3A,C). Similar alterations were also observed in cmyb+ cells (Fig. 3D,F). Together, these results indicated that the HSPC reaction was dependent on the bacterial burden. Next, the possible mechanisms underlying the HSPC reactions were examined. Several inflammatory cytokines have been suggested to be critical for the activation of HSPC proliferation5,35,36,37; however, their overproduction causes HSPC apoptosis5,6,38,39. Therefore, the expression levels of various cytokines were measured in response to different bacterial burdens by quantitative real-time polymerase chain reaction (qPCR). The results revealed an obvious increase in the expansion of these factors, including tnfα, ifng1–2, and il1b (see supplemental Figure S3D), following a more severe challenge at 2 dpi. This result was consistent with the reaction of HSPCs, further supporting that an optimal level of inflammatory factors is essential for their homoeostasis and that overdose might lead to their exhaustion5,6,37,38,39.
Reaction of HSPCs to different E. coli burdens after intravenous injection.
(A), Fluorescence images showing the runx1-GFP+ cells in Tg(runx1:en-GFP) larvae treated with different volumes of E. coli at 2 dpi (4 dpf). Scale bars, 20 μm. (B), The number of runx1-GFP+ cells in larvae treated with 5–10 × 103 cfu E. coli or PBS. (70.31 ± 4.18 vs 71.14 ± 4.42; 148.8 ± 7.14 vs 137.6 ± 8.65 in E. coli vs PBS group at each time points. N ≥ 7 in each group). (C), Calculation of the runx1-GFP+ cells in (A) (137.6 ± 8.65; 194.6 ± 15.63; 148.8 ± 7.14; 103.4 ± 8.38 in each group. N ≥ 8 in each group). (D), WISH of cmyb (red arrows) in the larval CHT treated with different volumes of E. coli at 2 dpi (4 dpf). (E), The data obtained for cmyb+ cells at different time points after treatment with 5–10 × 103 cfu E. coli or PBS (102.80 ± 11.67 vs 82.40 ± 6.59; 114.00 ± 4.97 vs 112.00 ± 6.60; 121.60 ± 9.70 vs 127.00 ± 6.69; 129.00 ± 13.71 vs 127.60 ± 8.20; 203.80 ± 17.06 vs 226.40 ± 20.07 in E. coli vs PBS group at each time point. N = 8 in each group). (F), Calculation of the cmyb+ cells in (D) (221.60 ± 18.56; 268.70 ± 17.77; 203.80 ± 17.06; 89.30 ± 12.54 in each group. N = 10 in each group).
Emergency monopoiesis occurs during systemic infection
Similar emergency granulopoiesis, but with a different HSPC reaction, was observed in our assay compared with the results of a previous study10. Whether this difference was caused by the distinct infection methods or variations in the microbes used was explored further. When the microbes, whether they were Salmonella typhimurium or Dsred-labelled E. coli13,23, were injected into the 2 dpf larval hindbrain or the blood circulation, emergency granulopoiesis occurred, as evidenced by clear expansion of SB+ neutrophils (see supplemental Figure S4A). Thus, emergency granulopoiesis occurred following either brain or systemic infection of the larvae. When the mpeg1+ macrophages were examined, no expansion of mpeg1+ macrophages was observed in the CHT when the larvae were challenged via the hindbrain by both microbes (see supplemental Figure S4B)10, indicating that brain infection did not lead to emergency monopoiesis in the CHT. However, intravenous injection of microbes caused a tremendous expansion of mpeg1+ macrophages in the CHT (see supplemental Figure S4B). Thus, emergency monopoiesis, in addition to granulopoiesis, likely occurred when the microbes were systemically inoculated. This conclusion was further confirmed by the drastic expansion of another macrophage marker, mfap4, at 2 dpi (Fig. 4A,B). The reasons for the different reactions of the macrophages in response to different infection methods were investigated further. Although an obvious restriction of the bacteria was detected in the injured brain, hindbrain administration led to limited circulating microbes in the CHT, in contrast to the drastic increase in circulating microbes after intravenous injection (see supplemental Figure S4C,D). Therefore, the tremendous number of bacteria in the circulation and the intensive involvement of macrophages in bacterial phagocytosis (Fig. 1D) probably provided cues that led to emergency monopoiesis. To carefully dissect the process of emergency monopoiesis, mfap4+ and mpeg1-GFP+ cells were quantified. Similar to the fluctuation of neutrophils, mfap4+ and mpeg1-GFP+ macrophages25 displayed an initial exhaustion (Fig. 4B,D), which was in agreement to their intensive involvement in phagocytosis and resultant increased apoptosis (Fig. 4C,E). Subsequently, a significant expansion of mfap4+ macrophages followed (Fig. 4B). However, their recovery to baseline was faster than that of SB+ neutrophils. At 4–6 dpi, the macrophage numbers had already declined to the levels of the control (Fig. 4B), approximately 2 days earlier than the neutrophils (Fig. 2B). Other lineage markers presented no obvious alterations (see supplemental Figure S5). Overall, systemic infection of microbes caused both emergency monopoiesis and granulopoiesis, which are collectively referred to as emergency myelopoiesis.
Emergency monopoiesis occurs upon intravenous E. coli (5–10 × 103 cfu) infection.
(A), WISH of mfap4 (red arrows) in the CHT of a larva treated with PBS or E. coli at 2 dpi (4 dpf). (B), The data obtained for mfap4+ macrophages at different time points after challenge (36.13 ± 4.52 vs 53.88 ± 3.90; 64.25 ± 6.58 vs 57.00 ± 3.29; 160.44 ± 9.92 vs 53.20 ± 1.90; 87.00 ± 15.67 vs 42.44 ± 4.21; 24.71 ± 2.14 vs 23.00 ± 2.30 in E. coli vs PBS group at each time point. N ≥ 8 in each group). (C,D), Fluorescence images (C) and calculation (D) of mpeg1-GFP+ macrophages in PBS (25.50 ± 2.94; N = 8) or E. coli (13.13 ± 1.09; N = 8) treated larval CHT at 6 hpi. The red signals indicate the bacteria phagocytosed by mpeg1-GFP+ macrophages (white arrows). (E), The percentage of mpeg1-GFP+ macrophages that co-stained with TUNEL at 6 hpi (21.13 ± 3.86 vs 2.82 ± 1.05 in E. coli vs PBS group. N ≥ 10 in each group). (F), Time-lapse imaging of an infected Tg(mpeg1:eGFP) CHT from 1 dpi to 1.5 dpi (see also Video S5). (G-H), WISH (G) and calculation data (H) for mfap4+ macrophages (red arrows) in the runx1 mutant treated with either PBS or E. coli at 2 dpi (4 dpf) (WT: 117.9 ± 9.63 vs 25.38 ± 0.96 E. coli vs PBS; runx1 mut: 110.3 ± 6.88 vs 44.40 ± 5.72 E. coli vs PBS; N ≥ 5 in each group). Scale bars, 20 μm.
Emergency monopoiesis is achieved through the expansion of primitive myeloid cells
The definitive hematopoietic origin of the emergency granulopoiesis suggested a similar origin for emergency monopoiesis. To verify this hypothesis, time-lapse imaging was performed in the infected Tg(mpeg1:eGFP) larval CHT. Expansion of mpeg1-GFPlow macrophages was detected, and their numbers increased upon infection (Fig. 4F; see supplementary Video S5). These mpeg1-GFPlow macrophages should have been nascent. However, they highly expressed GFP signals following engulfment of bacteria (Fig. 4F; see supplementary Video S5). The mpeg1-GFPlow macrophages suggested the definitive hematopoietic origin of emergency monopoiesis. However, when the mfap4+ macrophages were examined in the runx1w84x 24, they presented a surprisingly remarkable expansion following challenge, and the numbers of expanded macrophages were similar to those in the siblings (Fig. 4G,H). This macrophage phenomenon is the converse of that observed for neutrophils in similar mutant larvae, suggesting that the emergency monopoiesis was largely independent of runx1-mediated hematopoiesis and that these cells were probably generated from primitive myeloid cells. Thus, emergency granulopoiesis and monopoiesis at this stage had different origins.
The immune response and hematopoiesis-related factors are transcriptionally influenced after challenge
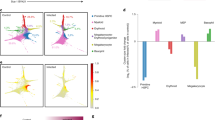
The molecular mechanisms underlying emergency myelopoiesis were further explored. To this end, deep-sequence analysis was performed using samples collected at successive time points after treatment. Three typical representative time points—6 hpi, 1 dpi, and 4 dpi—were chosen on the basis of both the alteration of myeloid phagocytes and the level of bacterial clearance. The results indicated that large amounts of factors changed during the different stages after challenge (Fig. 5A,B; Table S1 and S2). Altered factors functioning in bacterial defense and hematopoiesis were examined further. The heat-map results revealed that dozens of bacterial defense-related and hematopoiesis-related genes were transcriptionally modified throughout the process (Fig. 5C and Table S2). These genes could be classified mainly into two types. The first type showed a typical increase at 6 dpi but a quick reduction thereafter. Nos2b and Duox—two important factors that are closely involved in the formation of H2O2 and NO—were the representative examples (Fig. 5C), suggesting essential roles for small molecules as initial emergency signals10,34,40. Another type manifested an initial reduction followed by an obvious increase at later stages. This group of factors accounted for a large portion of the total members, and was typified by pro-inflammatory cytokines such as il1b and mmp9, as well as most hematopoiesis-related genes (Fig. 5C). qPCR was performed to validate the deep-sequence analysis data. The results revealed similar alterations in the expression of key factors, including tlr5a, mpx, il1b, mm9, irf8, csf3r, and pu.1, to that in the deep-sequence analysis results (Fig. 5D). However, the HSPC marker cmyb exhibited a slight upregulation (<2-fold) (Fig. 5D), which was consistent with its behaviour in the deep-sequence analysis, and this finding further supported the results suggesting limited alterations of HSPC numbers in our assay.
Comparison of bacterial defense-related and hematopoiesis-related response factors at different time points after intravenous injection of E. coli (5–10 × 103 cfu).
(A,B), Venn diagrams showing the overlap and differences between successive time points after E. coli infection in the numbers of downregulated (A) or upregulated (B) genes. (C), Gene expression profiles of E. coli-infected embryos at different time points are depicted in a heat map. Bacterial defense-related and hematopoiesis-related genes in the heat map are ordered in functional groups. All genes included in the heat map are represented by a minimum of two probes that showed significant up- or down-regulation (significance cut-offs for the ratios of infected versus control groups were set at 2-fold). Up- and down-regulation are indicated by increasingly bright shades of red and green, respectively. (D), qPCR analysis indicating the similar alterations of the expression levels of tlr5a, mpx, il1b, mmp9, irf8, csf3r, pu.1 and cmyb in (C).
Different roles of Pu.1 in steady and emergency monopoiesis
The function of Pu.1 in emergency myelopoiesis was explored because it presented significant increases in expression levels after infection (Fig. 5C,D). To this end, Pu.1 was functionally disrupted using either pu.1G242D/G242D hypomorphic alleles or morpholinos (MOs) knockdown41,42. In agreement with previous studies43,44, the large phagocytic foci that appeared in wild type (WT) embryos (see supplementary Video S6) were detected in small numbers, and the clearance of E. coli was much slower in challenged Pu.1-deficient embryos (Fig. 6A; see supplementary Video S7). Furthermore, these infected embryos showed the highest mortality (Fig. 6I). Together, these results suggested the presence of defective phagocytosis and immune responses in the absence of normal Pu.1 function43,44. Next, emergency myelopoiesis in challenged pu.1-deficient larvae was examined. Because deficiency in Pu.1 activity resulted in obvious defects in macrophage development but an expansion of the neutrophil population in larvae during the steady state41, we predicted that Pu.1 was probably dispensable for emergency granulopoiesis but played critical roles in emergency monopoiesis. To test this hypothesis, emergency granulopoiesis was first examined. The results indicated that, even without the normal function of Pu.1, emergency granulopoiesis took place, as evidenced by the fact that the SB+ and lyz+ neutrophils and pu.1+ and cebpα+ myeloid progenitors in pu.1-deficient larvae expanded noticeably to the level of their counterparts in infected WT larvae at 2 dpi (Fig. 6B–E; see supplemental Figure S6A,B). The baseline of neutrophils and their progenitor population in these larvae was higher than the baseline in WT41 (Fig. 6C,E), indicating that the expansion potential of neutrophils after infection was smaller than that in WT (Fig. 6H). However, emergency granulopoiesis still occurred, although the intensity was not as strong as that in WT. Thus, Pu.1 was largely dispensable for emergency granulopoiesis. However, when the macrophage marker mfap4 was checked, the mfap4+ cells that were markedly reduced in PBS-treated pu.1G242D/G242D larvae presented a surprisingly drastic expansion after infection (Fig. 6F,G). Their number was even higher than that in the infected WT larvae at 2 dpi (Fig. 6G). Consistently, the expansion potential of mfap4+ cells was much higher than that in WT (Fig. 6H). Other macrophage markers, including csf1ra and mpeg1, displayed similar expansion in the E. coli-challenged pu.1G242D/G242D larvae (see supplemental Figure S6C,D), further confirming that the macrophage lineage dramatically expanded in the emergency condition when Pu.1 was defective. Therefore, Pu.1 functioned differently during the demanding situation of monopoiesis. Although Pu.1 is essential for normal macrophage formation, this finding revealed its suppressive roles during emergency monopoiesis.
Pu.1 suppresses emergency monopoiesis after intravenous infection of E. coli (5–10 × 103 cfu).
(A), Time-lapse imaging of an infected 2 dpf pu.1G242D/G242D/Tg(lyz:eGFP) CHT from 6 hpi to 19 hpi. The white stars indicate that lyz-GFP+ neutrophils phagocytosed bacteria to form small foci. The white arrowheads represent free bacteria. The white cross star denotes the large E. coli foci. Scale bars, 20 μm (see also Video S7). (B,C), SB staining (B) and calculation of SB+ neutrophils (C) in the CHT of treated WT (172.90 ± 26.23 vs 77.10 ± 16.47 in E. coli vs PBS group. N = 8 in each group) or pu.1G242D/G242D larvae (198.00 ± 21.13 vs 140.80 ± 23.92 in E. coli vs PBS group. N = 8 in each group). (D,E), WISH (D) and calculation (E) of the pu.1+ myeloid progenitors in the CHT of stressed WT (214.50 ± 13.98 vs 78.10 ± 6.89 in E. coli vs PBS group. N = 8 in each group) or pu.1G242D/G242D larvae (230.30 ± 24.54 vs 187.00 ± 17.78 in E. coli vs PBS group. N = 8 in each group). (F,G), WISH (F) and calculation (G) of mfap4+ macrophages in the CHT of challenged WT (128.80 ± 7.32 vs 75.60 ± 5.09 in E. coli vs the group. N = 8 in each group) or pu.1G242D/G242D larvae (174.50 ± 14.03 vs 49.30 ± 3.12 in E. coli vs PBS group. N = 8 in each group). (H), The fold expansion of various cell types in both the infected WT (SB+: 2.24; pu.1+: 2.75; mfap4+: 1.70) or pu.1G242D/G242D (SB+: 1.41; pu.1+: 1.22; mfap4+: 3.54) at 2 dpi. (I), Kaplan–Meier survival analysis of PBS- and E. coli-treated WT and pu.1G242D/G242D embryos. The total numbers of animals used in each experiment are 100 (log-rank/Mantel–Cox statistic). Red arrows indicate WISH signals.
Protective roles of Pu.1 in HSPC survival following challenge
A deficiency in the efficient clearance of bacteria and the resulting greater severity of bacteraemia-like syndrome in Pu.1-deficient embryos probably increased the exposure of HSPCs to microbes in the CHT. We were interested in the reaction of the HSPCs in this scenario. To this end, Tg(runx1:en-GFP) larvae were treated with control and pu.1 MOs. The response of runx1-GFP+ cells following bacterial challenge was investigated. Approximately 4% of the runx1-GFP+ cells were observed to engulf bacteria in the control group. However, this population clearly expanded in the pu.1 morphants (Fig. 7A,B), suggesting that HSPCs could directly interact with microbes in the CHT and that during more severe infection, more HSPCs were involved. Consequently, more runx1-GFP+ cells underwent apoptosis, as evidenced by the increased percentage of TUNEL+/runx1-GFP+ apoptotic cells in the challenged pu.1 morphants, and the level was higher than that detected in either the infected control embryos or the PBS-treated pu.1 morphants (Fig. 7C,D). Therefore, Pu.1 was critical for HSPC survival following challenge. To confirm this conclusion, another HSPC marker, cmyb, was evaluated. In agreement with the findings in runx1-GFP+ HSPCs, cmyb-GFP+ cells underwent similar excessive apoptosis in infected pu.1 morphants (see supplemental Figure S6E,F). However, the cmyb-GFP+ cells underwent similar proliferation in both the control and pu.1 morphants treated with either PBS or E. coli (see supplemental Figure S6G,H) when examined using anti-pH3 antibody staining30, suggesting a dispensable role for Pu.1 in infection-induced HSPC proliferation. To accurately present the data, the cmyb+ cells were quantified. The cmyb+ cell number was markedly lower in the treated pu.1 morphants than in the control group at 2 dpi (Fig. 7E,F), which was consistent with their increased apoptosis (see supplemental Figure S6E,F). A similar phenomenon was recapitulated in pu.1G242D/G242D embryos, although the reduction of cmyb+ cells was less drastic in pu.1G242D/G242Dthan in pu.1 morphants(Fig. 7F,G). This result is likely a consequence of the partial disruption of Pu.1 activity in pu.1G242D/G242Dcompared with the more severe disruption resulting from a high dose of pu.1 MOs41. Thus, Pu.1 was essential for the efficient clearance of microbes, which in turn prevented over-exposure of HSPCs to microbes. This procedure is quite important for HSPC homeostasis after E. coli challenge.
Pu.1 deficiency causes excessive HSPC apoptosis after intravenous E. coli (5–10 × 103 cfu) infection.
(A,B), Fluorescence images (A) and calculation (B) of runx1-GFP+ cells that phagocytose (white arrows) E. coli in the control (4.20 ± 0.79; N = 13) or pu.1 morphant (6.46 ± 1.0; N = 11) CHT at 6 hpi. (C), The fluorescence images of double staining by GFP and TUNEL in the CHT of PBS or E. coli-treated pu.1 morphants Tg(runx1:en-GFP) embryos at 2 dpi (4dpf). White arrows show the co-localization of TUNEL and GFP signals. (D), The percentage of TUNEL+/runx1-GFP+ in each group at 2 dpi (4 dpf) (Ctrl MO PBS: 0.48 ± 0.16; Ctrl MO E. coli: 0.36 ± 0.20; pu.1 MO PBS: 0.47 ± 0.14; pu.1MO E. coli: 1.07 ± 0.25. N ≥ 8 in each group). (E), WISH of cmyb (red arrows) in the CHT of the PBS or E. coli-treated pu.1 morphants at 2 dpi (4 dpf). (F), The numbers of cmyb+ cells in the CHT of treated control (230.80 ± 28.00 vs 248.80 ± 24.50 in E. coli vs PBS group. N = 10 in each group) or pu.1 morphants (154.40 ± 13.03 vs 306.60 ± 21.68 in E. coli vs PBS group. N = 10 in each group). (G) The numbers of cmyb+ cells in the CHT of the treated WT (203.80 ± 17.06 vs 226.40 ± 20.07 in E. coli vs PBS group. N = 10 in each group) or pu.1G242D/G242D embryos (164.80 ± 25.60 vs 291.70 ± 23.22 in E. coli vs PBS group. N = 10 in each group) at 2 dpi (4 dpf).
The reduction of HSPCs but expansion of myeloid progenitors in the challenged Pu.1-deficient larvae appeared to be contradictory, which led us to suspect that E. coli affects HSPCs and myeloid progenitors in distinct manners. To verify our hypothesis, co-staining of cmyb-GFP and pu.1 was performed in Tg(cmyb:eGFP). In the control group treated with PBS, the majority of the cmyb-GFP+ cells expressed pu.1 signals, resulting in a small ratio (approximately 23%) of pu.1+-only myeloid progenitors (see supplemental Figure S7). However, upon challenge, the pu.1+-only cell population increased dramatically to approximately 41% (see supplemental Figure S7), suggesting that the pu.1+-only myeloid progenitors themselves underwent notable expansion upon challenge. Concordantly, in E. coli-challenged pu.1 morphants, pu.1+-only myeloid progenitors showed drastic expansion, but cmyb-GFP+ HSPCs showed a clear reduction when compared with their control counterparts (see supplemental Figure S7). Consequently, the percentage of pu.1+-only myeloid progenitors increased to approximately 52%, which was much higher than that of the other groups (see supplemental Figure S7). Therefore, the HSPCs and myeloid progenitors separately responded to the microbes, and Pu.1 deficiency led to a reduction in HSPCs but showed a limited influence on the expansion of the myeloid progenitors.
Discussion
Taking advantage of the optical transparency of zebrafish larvae, an emergency myelopoiesis model was established through direct injection of Dsred+ E. coli13,23 into the circulatory system. Although intravenous injection of microbes has been employed by several groups10,11,12,13, infection-induced myelopoiesis has rarely been the experimental focus. Recently, Kathryn E. Crosier’s group dissected the role of the Cebpβ-Nos2a pathway in demand-adapted emergency granulopoiesis by injecting GFP+ Salmonella into the brains of larvae. In that study, emergency granulopoiesis was achieved by sacrificing lymphopoiesis, and HSPCs clearly increased under this condition; by contrast, macrophages showed no notable increase in the trunk region10. This work facilitated the initiation of research investigating infection-induced hematopoiesis using larval zebrafish45. However, the reaction of hematopoietic cells to systemic infection, particularly when HSPCs directly encountered microbes in CHT, has not been addressed. In our study, direct inoculation of microbes into the zebrafish bloodstream led to the expansion of both macrophages and neutrophils. Thus, emergency monopoiesis, in addition to emergency granulopoiesis, occurred with the use of this method, which could serve as a good supplementary assay to study emergency myelopoiesis10.
In contrast to locally infected larvae10, direct injection of bacteria into the circulation led to the development of a bacteraemia-like syndrome, which caused immediate and significant participation of macrophages and neutrophils in phagocytosis and digestion of microbes13,23,27. The intensive involvement of myeloid phagocytes led to their increased apoptosis and quick exhaustion, which was probably the cue for their subsequent expansion46. The expanded macrophages and neutrophils in the challenged larvae were probably of different origins, as suggested by the data obtained for runx1w84x 24. Almost no neutrophils were found in runx1-deficient larvae, regardless of whether they were challenged by microbes, thus supporting a runx1-dependent definitive hematopoietic origin of granulopoiesis under both physiological and stressed conditions. The macrophages slightly expanded in the steady state41, and emergency monopoiesis occurred normally in the runx1 mutant. This result suggested that the macrophages at this stage were largely generated from primitive hematopoiesis, which occurred independently of runx1. A recent study has demonstrated that the microglia, a subtype of macrophages in the brain, mainly originates from primitive myelopoiesis throughout the larval period47. In another study, a mutant fish line with compromised definitive hematopoiesis showed a limited influence on macrophages at later larval stages48. Thus, it is possible that larval macrophages have a largely primitive origin.
The HSPCs directly interacted with the microbes in the CHT in our assay. And their response was dependent on the bacterial burden. A relatively lower volume of E. coli led to a moderate expansion of HSPCs, which is consistent with previous reports10,33,34. However, excessive stress caused by a large microbial burden resulted in a drastic exhaustion of HSPCs. The distinct reactions of the HSPCs to different bacterial burdens were probably related to the severity of the direct exposure of HSPCs to the microbes and the overproduction of pro-inflammatory cytokines. Because a higher dose of E. coli would overcome the clearance by phagocytes and lead to the production of excessive levels of pro-inflammatory cytokines, it can be inferred that the longer and stronger influence of microbes on HSPCs in the CHT and the overproduction of pro-inflammatory cytokines probably facilitated their apoptosis. This hypothesis was further supported by the reduction of HSPCs in the infected Pu.1-deficient embryos. The functional defects of the macrophages in Pu.1-deficient embryos resulted in the slower clearance of E. coli43,44. Consequently, the interaction between HSPCs and pathogens was prolonged. Concordantly, increased apoptosis of HSPCs occurred, overcoming the cell proliferation and leading to a reduction of cell numbers. The increased apoptosis was correlated with excessive production of inflammatory factors, including IFNγ and TNFα (see supplemental Figure S6E). Appropriate levels of IFNγ and TNFα are essential for the activation of HSPC proliferation5,35,36,37. However, their overproduction causes rapid HSPC apoptosis5,6,38,39. Thus, the drastically altered levels of IFNγ and TNFα were probably responsible for the increased apoptosis of HSPCs.
Pu.1 is indispensable in the commitment of myeloid cells41,49,50 and in leukaemogenesis51,52. However, its function in infection-induced emergency myelopoiesis had not been addressed. Taking advantage of pu.1G242D/G242D and morpholino-mediated functional disruption, the roles of Pu.1 in infection-induced emergency myelopoiesis were carefully dissected. Surprisingly, compared with the insensitive expansion of neutrophils, macrophages with significant physiological shortcomings in the presence of defective Pu.1 presented drastic expansion after infection, and their numbers quickly exceeded the values determined in infected WT embryos. Pu.1 seemed to be an inhibitory regulator for the infection-induced expansion of macrophages. Thus, it played distinct roles in physiological and emergency monopoiesis. A previous study has demonstrated that Cebpβ plays different roles in physiological and emergency granulopoiesis because its deficiency leads to ineffective emergency granulopoiesis, although it is dispensable for the normal development of neutrophils2,10,53. The data obtained for cebpβ and pu.1 suggested that the regulatory networks underlying emergency myeloid cell development differed from that utilised in the steady state. Therefore, elucidation of the mechanisms responsible for emergency myelopoiesis is an interesting topic for further investigation.
Methods
Ethics statement
All experimental protocols were approved by the School of Life Sciences, Southwest University (Chongqing, China), and the methods were carried out in accordance with the approved guidelines. The zebrafish facility and study were approved by the Institutional Review Board of Southwest University (Chongqing, China). Zebrafish were maintained in accordance with the Guidelines of Experimental Animal Welfare from Ministry of Science and Technology of People’s Republic of China (2006) and the Institutional Animal Care and Use Committee protocols from Southwest University (2007).
Fish lines
AB, pu.1G242D41, runx1w84x 24, Tg(runx1:en-GFP)31, Tg(cmyb:eGFP)32, Tg(coro1a:eGFP)29, Tg(mpeg1:eGFP) and Tg(lyz:eGFP)nz11726 strains were used and maintained under standard conditions.
Generation of Tg(mpeg1:eGFP) lines
4.1-kb DNA sequence upstream of the mpeg1 translation start site amplified with the primers 5′- ACATGCATATCTTGCAGTATA-3′/5′- GATCGCCAGATGGGTGTTTT-3′ was used as a promoter to drive eGFP expression in the pTol2 vector. The pTol2-mpeg1-eGFP construct was injected into the wild-type fish embryos at one-cell stage. The embryos with an appropriate GFP expression were selected and raised to adults. The founder lines were identified based on their eGFP expression pattern.
Phagocytosis assays and time-lapse live imaging
The Dsred-labeled E. coli23 were cultured as previously described43. The cultured E. coli were collected in filter-sterilized PBS prior to the injection. To quantify the burdens, the volume of E. coli for injection was added to 1 ml LB and then plated at 1:10 and 1:100 dilutions on LB agar supplemented with 50 mg/ml kanamycin. Colonies were counted in plates incubated at 37 °C overnight to quantify the actual infection doses. The E. coli volume of each concentration was then microinjected into the circulation of each anesthetized embryo. The injected embryos were anesthetized, mounted in 1% agarose, and subsequently imaged under an LSM700 confocal microscope (Carl Zeiss) (X20 objective). Images were captured every 5 min, extracted, and converted into a movie using ZEN2012 software. Movie Maker was used to create the movies.
WISH and Sudan Black (SB) staining
Antisense RNA probes were prepared according to the standard protocol. The following digoxigenin-labeled antisense probes were used: cmyb, pu.1, cebpα, csf1ra, mpx, cebp1, lyz, mpeg1, mfap4, rag1, ccr9a, αe1-globin and gata1. Single-colour whole-mount in situ hybridization (WISH) was performed24. Sudan black (Sigma, 380B) solution was used to treat the fixed embryos28. The signals were observed under SteREO Discovery.V20 microscope (Carl Zeiss).
Double staining for RNA (pu.1) and protein (GFP)
The double staining process was mainly performed according to a previous study43. Briefly, WISH staining of pu.1 was first developed with Cy3 tyramide (PerkinElmer Life and Analytical Sciences). Afterward, the embryos were washed and incubated with goat anti-GFP antibody (1:400, 4 °C, overnight) (Abcam, ab6658) and visualized with Alexa Fluor 488 donkey anti–goat secondary antibodies (1:400, 4 °C, overnight) (Invitrogen).
Double fluorescence immunohistochemistry staining and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)
Double fluorescence immunohistochemistry staining of larvae was performed24. For eGFP and pH3 double staining, Tg(cmyb:eGFP) embryos treated with control or pu.1 morpholinos41 were fixed in 4% paraformaldehyde at the desired stages. The fixed embryos were incubated with primary rabbit anti–phospho-histone H3 (1:250, 4 °C, overnight) (pH3; Santa Cruz Biotechnology, sc-8656-R) and goat anti-GFP (1:400, 4 °C, overnight) (Abcam, ab6658) antibodies according to the manufacturer’s protocol and subsequently stained with Alexa Fluor 647 anti–rabbit and Alexa Fluor 488 anti–goat secondary antibodies (Invitrogen). For the TUNEL assays, the in situ cell death detection kit, TMR Red (Roche 12156792910), was applied. The staining process was performed as indicated in the protocol. All fluorescence images were obtained using an LSM700 confocal microscope (Carl Zeiss).
Differentially expressed gene (DEG) analysis
To explore the molecules involved after infection, E. coli-treated embryos were selected at 6 hpi, 24 hpi and 4 dpi. Their total RNA was extracted for deep sequencing by the Biomarker Company, Beijing. The differentially expressed genes (DEGs) between any 2 samples were identified based on the following two criteria. 1) The expression value (FPKM) of the DEG must be larger than 1 in both samples, which indicates that the gene is active in these samples and that the detected expression values are not caused by background noise (for example, read mismatching or multi-hit alignment). 2) The variation in gene expression between the two conditions should be larger than 2-fold. Based on these two criteria, we identified 1441 DEGs in at least one comparative case (see Supplementary Table S1 and S2). Gene set enrichment analyses were performed for the functional annotation of the DEGs. Functional annotation tools in DAVID Bioinformatics Resources54 were used to conducted these analyses.
Real-time quantitative polymerase chain reaction (qPCR)
The total RNA of the infected embryos at different time points was isolated for the qPCR55. Each sample was tested in triplicate. Elongation factor 1α (ef1α) expression was measured and used to normalize signals for each queried transcript using the ΔΔCt method. The primers used (5′-3′): TLR5a-GAGGTGCCAAAGATTTCCACTTAC/TGGTGCATCAGGATGAGGACT;mpx-CCTCAACGACAGCACTCTGA/TACTCCAGGTAGGGTTGAGCA; il1b-CCCCAATCCACAGAGTTT/TTCACTTCACGCTCTTGG; mmp9-CATTAAAGATGCCCTGATGTATCCC/AGTGGTGGTCCGTGGTTGAG; cmyb-TTTCTACCGAATCGAACAGATG/CAATCACCCGTTGGTCTTCT; irf8-CCATTTTCAAAGCGTGGGCA/CTGGCACAATCCGGTACACT; csf3r-TGAAGGATCTTCAACCACAC/GGGAATTATAGGCCACAAAC;pu.1-AGAGAGGGTAACCTGGACTG/AAGTCCACTGGATGAATGTG.;ifng1-2-CTATGGGCGATCAAGGAAAA/CTTTAGCCTGCCGTCTCTTG. Other inflammatory cytokines were designed as according to a previous study37.
Quantification, calculation and statistical methods
To quantify the WISH, SB, fluoresce and immunohistochemistry signals, the positive signals on the images of the larval CHT regions (WISH and SB: 10 somites; fluoresce and immunohistochemistry: 6 somites) were manually counted. All the quantified data were double confirmed and analysed by GraphPad Prism 6. Student’s t test (one tailed) was mainly used (Mean ± SEM). Survival was calculated with Kaplan-Meier calculations.
Additional Information
How to cite this article: Hou, Y. et al. Systemic inoculation of Escherichia coli causes emergency myelopoiesis in zebrafish larval caudal hematopoietic tissue. Sci. Rep. 6, 36853; doi: 10.1038/srep36853 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Glatman, Z. A., Engiles, J. B. & Hunter, C. A. Infection-induced changes in hematopoiesis. J. Immunol. 192, 27–33 (2014).
Manz, M. G. & Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014).
Takizawa, H., Boettcher, S. & Manz, M. G. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood 119, 2991–3002 (2012).
King, K. Y. & Goodell, M. A. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 (2011).
Baldridge, M. T., King, K. Y., Boles, N. C., Weksberg, D. C. & Goodell, M. A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 465, 793–797 (2010).
Baldridge, M. T., King, K. Y. & Goodell, M. A. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32, 57–65 (2011).
Busch, K. et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015).
Takizawa, H., Regoes, R. R., Boddupalli, C. S., Bonhoeffer, S. & Manz, M. G. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208, 273–284 (2011).
Wilson, A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008).
Hall, C. J. et al. Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell 10, 198–209 (2012).
Herbomel, P., Thisse, B. & Thisse, C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745 (1999).
Meijer, A. H., van, d. V. & Spaink, H. P. Real-time imaging and genetic dissection of host-microbe interactions in zebrafish. Cell Microbiol. 16, 39–49 (2014).
Colucci-Guyon, E., Tinevez, J. Y., Renshaw, S. A. & Herbomel, P. Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J. Cell Sci. 124, 3053–3059 (2011).
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010).
Kissa, K. & Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010).
Burns, C. E. & Zon, L. I. Homing sweet homing: odyssey of hematopoietic stem cells. Immunity. 25, 859–862 (2006).
Jin, H., Xu, J. & Wen, Z. Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 109, 5208–5214 (2007).
Tamplin, O. J. et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015).
Murayama, E. et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 25, 963–975 (2006).
Jagannathan-Bogdan, M. & Zon, L. I. Hematopoiesis. Development 140, 2463–2467 (2013).
Kissa, K. et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147–1156 (2008).
Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J. & Sin, Y. M. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp Immunol. 28, 9–28 (2004).
van der Sar, A. M. et al. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 5, 601–611 (2003).
Jin, H. et al. Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 136, 647–654 (2009).
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A. & Lieschke, G. J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56 (2011).
Hall, C., Flores, M. V., Storm, T., Crosier, K. & Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC. Dev. Biol. 7, 42 (2007).
Li, Y. J. & Hu, B. Establishment of multi-site infection model in zebrafish larvae for studying Staphylococcus aureus infectious disease. J. Genet. Genomics 39, 521–534 (2012).
Le, G. D. et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 111, 132–141 (2008).
Li, L., Yan, B., Shi, Y. Q., Zhang, W. Q. & Wen, Z. L. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 287, 25353–25360 (2012).
Hendzel, M. J. et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 (1997).
He, Q. et al. Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 125, 1098–1106 (2015).
North, T. E. et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007).
Zhang, P. et al. The lineage-c-Kit+Sca-1+cell response to Escherichia coli bacteremia in Balb/c mice. Stem Cells 26, 1778–1786 (2008).
Kwak, H. J. et al. Myeloid cell-derived reactive oxygen species externally regulate the proliferation of myeloid progenitors in emergency granulopoiesis. Immunity. 42, 159–171 (2015).
Kim, J. M., Oh, Y. K., Kim, Y. J., Youn, J. & Ahn, M. J. Escherichia coli up-regulates proinflammatory cytokine expression in granulocyte/macrophage lineages of CD34 stem cells via p50 homodimeric NF-kappaB. Clin. Exp. Immunol. 137, 341–350 (2004).
Rezzoug, F. et al. TNF-alpha is critical to facilitate hemopoietic stem cell engraftment and function. J. Immunol. 180, 49–57 (2008).
Espin-Palazon, R. et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 159, 1070–1085 (2014).
Selleri, C., Sato, T., Anderson, S., Young, N. S. & Maciejewski, J. P. Interferon-gamma and tumor necrosis factor-alpha suppress both early and late stages of hematopoiesis and induce programmed cell death. J. Cell Physiol 165, 538–546 (1995).
Snoeck, H. W. et al. Interferon gamma selectively inhibits very primitive CD342+. J. Exp. Med. 180, 1177–1182 (1994).
Niethammer, P., Grabher, C., Look, A. T. & Mitchison, T. J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009).
Jin, H. et al. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 119, 5239–5249 (2012).
Sun, J. et al. Suppression of Pu.1 function results in expanded myelopoiesis in zebrafish. Leukemia 27, 1913–1917 (2013).
Li, L., Jin, H., Xu, J., Shi, Y. & Wen, Z. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood 117, 1359–1369 (2011).
Prajsnar, T. K., Cunliffe, V. T., Foster, S. J. & Renshaw, S. A. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 10, 2312–2325 (2008).
Herbomel, P. Infection-induced hematopoiesis: a zebrafish perspective. Cell Stem Cell 10, 105–106 (2012).
Cain, D. W., Snowden, P. B., Sempowski, G. D. & Kelsoe, G. Inflammation triggers emergency granulopoiesis through a density-dependent feedback mechanism. PLoS. One. 6, e19957 (2011).
Xu, J. et al. Temporal-Spatial Resolution Fate Mapping Reveals Distinct Origins for Embryonic and Adult Microglia in Zebrafish. Dev. Cell 34, 632–641 (2015).
Murayama, E. et al. NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat. Commun. 6, 8375 (2015).
DeKoter, R. P., Kamath, M. B. & Houston, I. B. Analysis of concentration-dependent functions of PU.1 in hematopoiesis using mouse models. Blood Cells Mol. Dis. 39, 316–320 (2007).
Xu, J., Du, L. & Wen, Z. Myelopoiesis during zebrafish early development. J. Genet. Genomics 39, 435–442 (2012).
Metcalf, D. et al. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc. Natl. Acad. Sci. USA 103, 1486–1491 (2006).
Rosenbauer, F. et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36, 624–630 (2004).
Hirai, H. et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7, 732–739 (2006).
Dennis, G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3 (2003).
Bertrand, J. Y., Kim, A. D., Teng, S. & Traver, D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135, 1853–1862 (2008).
Acknowledgements
We thank Z. Wen for antibody and discussions; F. Liu for the Tg(runx1:en-GFP); HT Li for bacteria. This work was supported by the National Natural Science Foundation of China (31301198, 31271568 and 31571500); The National Key Basic Research Program of China (2015CB942800); The Fundamental Research Funds for the Central Universities (XDJK2014A007 and XDJK2016E102); and the Special Support Plan of Excellent Young Talents, Guangdong, China (YueRenCaiBan [2015] 8).
Author information
Authors and Affiliations
Contributions
L.L. designed the experiments and wrote the manuscript. Y.H. performed most experiments. Z.S. did the transcriptional data analysis. X.M. performed the bacteria injection by different methods. C.L. characterized the myeloid phenotypes upon hindbrain infection. J.C. performed the bacteria infection in transgenic lines. J.Z. gave suggestions in the experiments. H.H. discussed the results. H.R. commented on the manuscript. L.F.L. designed the experiments and supported the transgenic lines.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hou, Y., Sheng, Z., Mao, X. et al. Systemic inoculation of Escherichia coli causes emergency myelopoiesis in zebrafish larval caudal hematopoietic tissue. Sci Rep 6, 36853 (2016). https://doi.org/10.1038/srep36853
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36853
This article is cited by
-
Zebrafish: a convenient tool for myelopoiesis research
Cell Regeneration (2023)
-
Spatiotemporal imaging and pharmacokinetics of fluorescent compounds in zebrafish eleuthero-embryos after different routes of administration
Scientific Reports (2021)
-
Zebrafish embryo sensitivity test as in vivo platform to anti-Shiga toxin compound screening
Brazilian Journal of Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.