Abstract
Dengue virus (DENV) is a mosquito-transmitted virus imposing a significant burden on human health around the world. Since current control strategies are not sufficient, there is an urgent need to find alternative methods to control DENV transmission. It has been demonstrated that introduction of Wolbachia pipientis in Aedes aegypti mosquitoes can impede DENV transmission with the mechanism(s) not fully understood. Recently, a number of studies have found the involvement of chromodomain DNA binding helicases in case of Human Immunodeficiency virus (HIV) and Influenza A virus infection. In this study, we have identified three chromodomain helicase DNA binding protein (CHD) genes in Ae. aegypti and looked at their response in the case of Wolbachia and DENV infections. Foremost amongst them we have found that AeCHD7/Kismet is significantly downregulated in the presence of Wolbachia infection only in female mosquitoes. Furthermore, AeCHD7 levels showed significant increase during DENV infection, and AeCHD7 depletion led to severe reduction in the replication of DENV. Our data have identified AeCHD7 as a novel Ae. aegypti host factor that is important for DENV replication, and Wolbachia downregulates it, which may contribute towards the mechanism(s) of limiting DENV replication.
Similar content being viewed by others
Introduction
Among arboviruses, dengue virus (DENV) is one of the most important flaviviruses having the potential to affect two-thirds of the world’s population1,2. DENV is primarily transmitted to humans through the bit of mosquito vector Aedes aegypti, leading to dengue infection and potentially dengue haemorrhagic fever3,4,5. Lack of availability of an effective vaccine and proper medical care has narrowed DENV management strategies to vector control. One of the strategies used to overcome DENV vector Ae. aegypti is through the application of pesticides, but due to their severe consequences on the environment and the emergence of resistance to pesticides, their potential application seems bleak in the near future6. Therefore, new strategies for vector control are urgently needed. One of the novel options is the use of an endosymbiotic bacterium Wolbachia which has recently been demonstrated to limit DENV, West Nile virus (WNV), and Zika virus (ZIKV) replication in Ae. aegypti7,8,9,10.
Wolbachia pipientis is an alphaproteobacterium that naturally infects almost 40–60% of insect species11,12. This bacterium is maternally transmitted and is usually associated with manipulations of host reproduction, such as feminization13 and male killing14, to promote successful colonization of its host species. Wolbachia naturally infects several mosquito species, including Aedes albopictus and Culex pipiens15. However, there is no natural Wolbachia infection in the case of Ae. aegypti, which is the most notorious vector for several arboviruses. In order to exploit Wolbachia’s potential to limit arbovirus transmission, three strains of Wolbachia, wAlbB (from Ae. albopictus)16, wMel (from Drosophila melanogaster)17 and wMelPop-CLA (from D. melanogaster)18 have been successfully transinfected into Ae. aegypti. Among these three strains, wMel and wMelPop-CLA are the most promising ones for virus blocking7,8,9,10,19,20. However, the exact mechanism(s) by which Wolbachia blocks viral replication in Ae. aegypti mosquitoes is still elusive. Few studies that have looked into the transcriptional changes in Ae. aegypti mosquitoes upon Wolbachia infection have found increased redox and mitochondrial activity along with differential serine protease activity21,22,23. However, very little is known about the role of chromatin remodelers in the case of DENV-Aedes-Wolbachia molecular interactions.
Chromodomain helicase DNA binding proteins (CHD) represent a class of ATP-dependent chromatin remodelling enzymes that contribute towards invoking changes in the interaction between DNA and nucleosomes24, influencing a wide array of cellular processes such as replication, transcription, recombination, repair and development25. Members of the CHD family have been found to be involved in replication of Human Immunodeficiency virus (HIV) and Influenza A virus26,27. All the CHD protein family members have a pair of chromodomains at their N-terminus along with one sucrose non-fermenting (SNF2) domain in the centre25. In humans, the CHD family has nine members. These are further classified, on the basis of additional motif features, into three subfamilies: CHD1-2 (class I), CHD3-5 (Class II) and CHD 6-9 (class III)25,28. In D. melanogaster, there are three well characterized CHD members named CHD129, Mi230 and Kismet/CHD731. CHD1 is essential for the fecundity of both males and females and is indirectly involved in transcriptional elongation32, whilst Mi2 actively participates in transcriptional repression and is vital for expression of heat shock proteins33,34. Drosophila Kismet, that is a homolog of human CHD7, mediates transcriptional elongation35. Apart from characterization of the CHD family members’ role in development and chromatin modification, very little is known about their potential role in host-pathogen interactions.
In this study, we have identified functional homologs of the CHD family members in Ae. aegypti and looked at the effect of Wolbachia infection on their expression. There was significant reduction in the expression of all CHD family members in the presence of Wolbachia. Furthermore, we found that AeCHD7 is highly induced during DENV infection in Ae. aegypti mosquitoes. A silencing assay demonstrated that AeCHD7 is required for the efficient replication and virion production of DENV. This study will help to understand the role of AeCHD7 in DENV-Aedes-Wolbachia interactions.
Results
Screening of the CHD family genes during Wolbachia infection
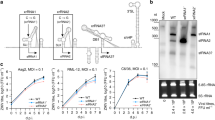
Three CHD genes were identified in the Ae. aegypti genome using Vectorbase36. Blastp was run to identify their homologs in D. melanogaster and Culex quinquefasciatus, and these were determined as AeCHD1 (AAEL004716) having 58% identity with D. melanogaster CHD1 protein (NP_477197.1), AeCHD3 (AAEL013136) that showed 70% identity with D. melanogaster CHD3 protein (AAD17276.1) and AeCHD7 (AAEL002230) showing 58% identity with D. melanogaster Kismet/CHD7 protein (NP_001245820.1). qPCR primers were designed for all the three AeCHD family members to experimentally validate their expression in Ae. aegypti mosquitoes by RT-qPCR, and the effect of Wolbachia (wMelPop) infection on their expression level. For this, we selected two age groups of Ae. aegypti mosquitoes, 4-day- and 12-day-old. While expression of all the three AeCHD genes was confirmed in the mosquitoes, they were all mostly downregulated in Wolbachia-infected mosquitoes (Fig. 1A–F), except for AeCHD3, which was found to be non-significantly upregulated in 4-day-old Wolbachia-infected mosquitoes (Fig. 1C). However, AeCHD7 showed the highest change of 2.9-fold downregulation in 4-day-old Ae. aegypti female mosquitoes (Fig. 1E), which led us to further characterise the gene.
Relative expression of AeCHD genes in uninfected and Wolbachia-infected Ae. aegypti mosquitoes.
RT-qPCR based quantification of (A,B) AeCHD1, (C,D) AeCHD3, and (E,F) AeCHD7 genes in both Wolbachia-infected (Pop) and uninfected (Tet) 4-day-old and 12-day-old female mosquitoes, respectively. Error bars represent standard error of mean (SEM) from three biological replicates (*p < 0.05; **p < 0.01).
AeCHD7 is ubiquitously expressed in all mosquito tissues
In order to determine the relative abundance of AeCHD7 across different tissues, the salivary gland, midgut, muscle, ovary and fat body were isolated from 3-day-old female Ae. aegypti mosquitoes. Following RT-qPCR detection of AeCHD7 mRNA transcripts, it was found that AeCHD7 is ubiquitously expressed in all tissues with the highest expression level in the salivary gland, which was 2.1-fold higher than its expression level in the fat body which showed the lowest relative abundance of AeCHD7 transcripts (Fig. 2). These results are consistent with the previous findings which showed that AeCHD7 is expressed in all human tissues37.
Specific Wolbachia-mediated downregulation of AeCHD7 in female Ae. aegypti
To find out whether Wolbachia-mediated downregulation of AeCHD7 is gender specific, we evaluated the transcript levels of AeCHD7 in 4-day-old female and male Ae. aegypti mosquitoes with and without Wolbachia infection. RT-qPCR results showed that Wolbachia downregulates AeCHD7 only in female mosquitoes and not in their male counterparts (Fig. 3A). This is interesting in the sense that Wolbachia has a gender specific effect on gene expression in the mosquitoes. To examine if the effect can consistently be seen in cell lines as well, we cross-validated the AeCHD7 mRNA expression levels in Ae. aegypti cell lines, Aag2 and Aag2 infected with wMelPop-CLA (Pop) and found a similar trend of AeCHD7 transcript downregulation in Wolbachia-infected cells (Fig. 3B).
Modulation of AeCHD7/Kismet by Wolbachia infection in male and female mosquitoes and flies, and mosquito cell lines.
(A) RT-qPCR analysis of AeCHD7 transcript levels in 4-day-old female and male mosquitoes, both uninfected (Tet) and infected with Wolbachia (Pop). (B) Relative expression of AeCHD7 in Aag2 and Aag2 cells infected with wMelPop-CLA (Pop). (C) Relative expression of the D. melanogaster Kismet gene in uninfected (Tet) and Wolbachia-infected (Pop) flies. Error bars represent SEM from three biological replicates (**p < 0.01; ***p < 0.001; ns, not significant).
Furthermore, to evaluate if Wolbachia has a similar effect on the CHD7 gene in its natural host D. melanogaster, four 7-day-old male and female flies infected with Wolbachia (wMelPop strain) were examined for the relative expression of CHD7. RT-qPCR results confirmed that there was no significant change in the level of CHD7 mRNA in both male and female D. melanogaster flies infected with Wolbachia (Fig. 3C).
AeCHD7 is upregulated upon DENV infection
Considering the virus blocking effect of Wolbachia in Ae. aegypti mosquitoes, we examined the transcript levels of AeCHD7 in the context of mosquito-DENV interaction. For this, the transcript levels of AeCHD7 in DENV-injected mosquitoes at three different time points of 2, 6 and 12 days post-infection (dpi) were analysed. To have a more consistent and high success rate of DENV infection, mosquitoes were injected rather than orally fed with the virus. The results revealed that there was an increase in the AeCHD7 transcript levels upon DENV infection at all the time points (Fig. 4A–C); however, the upregulation was only significant at 2 and 12 dpi, which was 2-fold (Fig. 4A) and 4-fold higher than that in uninfected mosquitoes (Fig. 4C), respectively. The aforementioned findings were further confirmed in the Ae. aegypti cell line, Aa20. Cells were infected with DENV2 at 0.1 multiplicity of infection (MOI) and were harvested at two different time points that were 1 and 5 dpi. RT-qPCR analysis showed significant increase in AeCHD7 transcript levels in the case of DENV infection at both 1 and 5 dpi (Fig. 4D). Infection was confirmed by relative quantification of DENV genomic RNA levels, which showed gradual increase in DENV genomic RNA over time (Fig. 4E).
Expression pattern of AeCHD7 in DENV infected female Ae. aegypti.
(A–C) RT-qPCR quantification of AeCHD7 transcript levels at 2, 6 and 12 days post DENV infection of female Ae. aegypti mosquitoes. (D) Relative transcript levels of AeCHD7 in Aa20 cells infected with 1 MOI of DENV2 analysed at 1 and 5 dpi. (E) RT-qPCR quantification of DENV2 genomic RNA in samples used in (D) confirming virus infection and replication. Error bars show SEM from three biological replicates (*p < 0.05; ***p < 0.001; ****p < 0.0001; ns, not significant).
AeCHD7 is required for efficient DENV replication
The upregulation of AeCHD7 in DENV-infected cells suggested that the gene could be beneficial for the virus. To investigate whether AeCHD7 is required for efficient DENV replication, we knocked down AeCHD7 transcripts in Aa20 cells and challenged these cells with DENV at 1 MOI for 72 h. The effect of CHD7 knockdown on DENV was evaluated both at the genomic and the virion levels using RT-qPCR and plaque assay. RT-qPCR results confirmed ~50% decrease in AeCHD7 mRNA level (Fig. 5A), which led to 2-fold reduction in DENV genomic RNA (Fig. 5B). RT-qPCR results were further validated with plaque assay, which confirmed reduction in DENV virion production in AeCHD7 knocked down cells as compared with dsGFP or mock-transfected Aa20 cells (Fig. 5C). These results indicate that AeCHD7 is a host factor that is used by DENV to facilitate its replication in Ae. aegypti female mosquitoes, and Wolbachia downregulates AeCHD7 as shown above, which may contribute to restricting DENV replication.
Depletion of AeCHD7 impairs DENV replication both at the genomic and the virion levels.
(A) RT-qPCR analysis of Aa20 cells transfected with either no RNA (Mock) or with dsRNA against GFP as a control or with dsRNA against AeCHD7. RPS17 was used to normalize the qPCR data. Error bars show SEM from three biological replicates (*p < 0.05; **p < 0.01). (B) RT-qPCR analysis of Aa20 cells treated as in (A) followed by DENV infection at 1 MOI using DENV-specific primers to quantify viral genomic RNA. Error bars show SEM from three biological replicates (**p < 0.01). (C) Viral plaque visualization by in vitro cell plaque assay conducted on the supernatant media from cells treated as in (A,B).
Discussion
There is accumulating experimental evidence showing the effectiveness of Wolbachia in suppressing the replication of several flaviviruses, including DENV, ZIKV, WNV, and the alphavirus chikungunya virus (CHIKV) in both mosquitoes and mosquitoes-derived cell lines8,10,38. Perhaps the most well studied is the case of DENV replication that is severely compromised in the presence of Wolbachia39,40,41. However, the exact mechanism(s) of how Wolbachia imparts this antiviral effect is not yet fully understood. In this study, we provide experimental evidence that chromodomain DNA binding helicase 7 (AeCHD7) is an Ae. aegypti host factor that is exploited by DENV to facilitate its replication, and its downregulation by Wolbachia may contribute to limit DENV replication.
Wolbachia is an endosymbiotic bacterium infecting 40–60% of insect species naturally11 by manipulating host reproduction12. Despite fitness costs, Wolbachia may benefit its host by blocking a variety of RNA viruses15,42. However, Wolbachia has not been found naturally infecting the most notorious vector Ae. aegypti, that is responsible for transmitting multiple viral diseases18. McMeniman et al. transinfected different strains of Wolbachia into Ae. aegypti mosquitoes43 and found that they successfully inhibited replication of DENV and CHIKV38. Further studies also demonstrated Wolbachia’s ability to block WNV and ZIKV in the mosquito8,9,10. Wolbachia’s potential to be used as an invaluable tool for disease control and prevention represents an increasingly promising approach to limit several mosquito-borne viral diseases, and it is fascinating to explore the exact mechanism(s) that induce the antiviral effect. Apart from one study shedding light on the effect of Wolbachia infection on the global DNA methylation pattern in mosquitoes44, there lies a huge grey area of the role of chromatin remodelers, in Wolbachia-host interactions and possibly the Wolbachia-mediated antiviral effect.
CHD proteins represent a class of proteins that belong to SNF2 superfamily of ATP-dependent chromatin modifiers. In mammals, there are 1–9 CHD proteins; however, in D. melanogaster, there are only three CHD proteins named CHD1, Mi2 and Kismet25. Members of the CHD family are involved in conducting a wide array of functions, including ATPase activity to maintain chromosome structure and regulation of heterochromatic elements29, nucleosome mobilization45, transcriptional regulation and elongation46,47,48,49, and development and differentiation31. Despite extensive characterization of CHD family proteins, their role in shaping host-pathogen interactions has not been much investigated. Yet, there are few reports supporting the involvement of CHD1 in the case of influenza A virus, and both CHD1 and CHD2 in the case of HIV as positive regulators26,27. Furthermore, RNAi screen carried out in D. melanogaster S2 cells identified the involvement of CHD7/Kismet in antimicrobial humoral response50. The aforementioned facts led us to investigate the possible role of the CHD family in Wolbachia-Aedes-DENV interactions. Data mining in VectorBase resulted in the identification of three potential AeCHD proteins encoded in the Ae. aegypti genome. Protein blast results identified them as AeCHD1, AeCHD3/Mi2 and AeCHD7/Kismet. In order to find out whether Wolbachia regulates the AeCHD genes during infection, RT-qPCR was performed to examine the transcript levels of all the three AeCHDs with and without Wolbachia infection in whole mosquitoes. Our results showed that there was a uniform trend of downregulation of the transcript levels of AeCHD genes in Wolbachia-infected mosquitoes (Fig. 1A–F), except those of AeCHD3 in 4-day-old Wolbachia-infected female mosquitoes, which showed a non-significant upregulation (Fig. 1C). The reduction in the CHD genes was more pronounced in 12-day-old mosquitoes, which could be due to increases in the Wolbachia load as the mosquitoes ages. In particular, the wMelPop strain is a virulent strain that may cause tissue damage and sickness. The reductions in the AeCHD genes at this late stage may not be of benefit in affecting DENV replication. However, AeCHD7 showed the highest fold change reduction (Fig. 1E,F) in both 4- and 12-day-old Wolbachia-infected mosquitoes, which prompted us to further investigate this gene in the context of Wolbachia-Aedes-DENV interactions.
AeCHD7/Kismet belongs to subfamily III of the CHD proteins that comprises CHD5-9 proteins. This subfamily is defined by the presence of two chromodomains at the N-terminus, one SNF2-like ATPase domain located in the central region of the protein structure and a Brahma and Kismet (BRK) domain at the C-terminus25. To find out whether the Ae. aegypti homolog fulfils this particular protein signature, NCBI conserved domain finder was used to detect the conserved domains51. Results confirmed the presence of all the domains characteristic of CHD7/Kismet proteins (Figure S1). Furthermore, in order to check the conservation of CHD7 across species, CHD7/Kismet amino acids were retrieved from Uniprot (Figure S2A) and subjected to maximum likelihood phylogenetic tree construction. Phylogenetic results showed that Ae. aegypti CHD7 is closely related to that of Cx. quinquefasciatus but not to that of D. melanogaster or Homo sapiens (Figure S2B). To find out the tissue-specific expression of AeCHD7, RT-qPCR was performed, which revealed that it is ubiquitously expressed across all main mosquito tissues (Fig. 2), which is consistent with the findings in humans37.
In this study we found that AeCHD7 was significantly downregulated in Wolbachia-infected mosquitoes. To further investigate whether this Wolbachia-mediated downregulation of AeCHD7 in female Ae. aegypti is gender specific and can also be seen in its natural host D. melanogaster, RT-qPCR was employed. Interestingly, we found that in the presence of Wolbachia AeCHD7 was specifically downregulated in female Ae. aegypti only (Fig. 3A), and there was no change in CHD7/Kismet transcript levels in both female and male D. melanogaster with and without Wolbachia (Fig. 3C). This difference in Wolbachia-mediated regulation of CHD7/Kismet in Ae. aegypti and D. melanogaster may be due to the fact that Wolbachia is a natural symbiont in D. melanogaster with a long association, while it has been recently transinfected into Ae. aegypti52. In regards to the mechanism by which Wolbachia infection may affect expression of AeCHD7, one can only speculate at this stage as there is very little information in regards to how Wolbachia manipulates its host at the molecular level. Only very recently, some molecular data have become available showing that Wolbachia infection leads to changes in the transcriptome or small RNA profiles of infected mosquitoes23,53. Regulation of host gene expression could be due to components secreted from the endosymbiont, including small non-coding RNAs54, or host response to accommodating the endosymbiont, in particular in new associations. However, how Wolbachia infection leads to these changes in the host remains to be investigated.
Viruses are the master manipulators of their host environment for their own benefit. Recently, it has been reported that CHD1 and CHD2 proteins play a pivotal role in the replication of influenza and HIV viruses26,27. Both viruses replicate inside the nucleus. Interestingly, it has further been demonstrated that CHD1 interacts with RNA polymerase II to facilitate the replication of influenza A virus27. Little is known about the role of CHD7/Kismet in the context of virus infection. However, the presence of conserved chromodomains and a SNF2 domain makes it highly likely that all the CHDs share similar functions25. We were intrigued to find what happens to AeCHD7 during DENV infection. RT-qPCR performed in DENV-infected mosquitoes at different time points suggested a continuous trend of upregulation during DENV infection (Fig. 4A–C), which points to the fact that it might play an important role in DENV replication. To investigate the role of AeCHD7 in DENV replication further, AeCHD7 knockdown study was carried out, which revealed that AeCHD7 is vital for DENV replication and virion production (Fig. 5A–C). Very few studies that have been carried out on the involvement of CHDs in virus replication have predominantly focused on viruses that replicate inside the nucleus. Therefore, the role of CHDs in the replication of viruses that multiply in the cytoplasm (such as DENV) is not known. However, it has been demonstrated that DENV capsid55,56 and NS5 proteins go inside the nucleus57,58 with NS5 known to be involved in disrupting nucleosome formation59. Therefore, these viral proteins may play a role in modulating AeCHD7 expression during virus infection. While we have found the involvement of AeCHD7 in mosquito-DENV interaction, the exact mechanism(s) that govern the interaction need further investigation.
In summary, we have demonstrated that AeCHD7 facilitates DENV replication, and Wolbachia-mediated downregulation of AeCHD7 in female Ae. aegypti may contribute to restriction of DENV replication. However, this mechanism is highly specific to female Ae. aegypti mosquitoes and does not appear to be a universal mechanism which Wolbachia employs across different hosts to block viral replication.
Materials and Methods
Mosquitoes and flies
For Wolbachia studies, mosquitoes had been previously generated by McMeniman et al.43 by transinfecting wMelPop-CLA strain of Wolbachia (Pop) into Ae. aegypti embryos, and uninfected mosquitoes were obtained through tetracycline (Tet) treatment of the infected mosquitoes43. w1118 fly line stably infected with wMelPop-CLA and the tetracycline cured line were generated by Min et al.60 and kindly provided by Dr Karyn Johnson from the University of Queensland.
For DENV infection studies, Ae. aegypti eggs were collected in Townsville in August 2015 and reared in insectary at Public Health Virology FSS. Five-day-old Ae. aegypti (F3) were used for the experiments. DENV2 NGC strain obtained from Prof. Roy Hall’s Lab (University of Queensland, School of Chemistry & Molecular Biosciences, Brisbane, Australia), was diluted in Opti-MEM (GIBCO Life Technologies, Grans Island, NY) supplemented with 3% foetal bovine serum (FBS, In Vitro Technologies, Australian origin) and intrathoracically injected (200 μl) in 5-day-old mosquitoes at 105.8/mL (102.1 per dose). Mosquitoes were placed into netted 900 mL containers at 28 °C with light:dark (L:D) 12:12 hours cycle and at high humidity. Mosquitoes were offered 15% honey water ad libitum. Mosquitoes were collected at 2, 6 and 12 dpi for downstream applications.
Cell cultures
Ae. aegypti Aag2 cell line and Aag2 cells infected with wMelPop-CLA, previously described by61, were maintained in 1:1 Mitsuhashi-Maramorosch and Schneider’s insect medium (Invitrogen) supplemented with 5–10% FBS (Bovogen Biologicals, French origin), while Aa20 cells were maintained in L15 medium (Invitrogen) supplemented with 10% tryptose phosphate broth (TPB) and 5% FBS. All mosquito cell lines were kept at 28 °C and passaged every 3–4 days.
Vero cells were maintained in OptiMEM medium supplemented with 2% FBS and were kept at 37 °C in the presence of 5% CO2.
RT-PCR and qPCR analyses
Total RNA was extracted from mosquitoes (1–5 mosquitoes per biological replicate) or flies (1–5 flies per biological replicate) using Qiazol (Qiagen) and then treated with Turbo DNase (Ambion) according to the manufacturers’ instructions. 750–1000 ng of total RNA was then used to make the 1st strand cDNA using Superscript III (Invitrogen) with either oligo-dT primer for cellular transcripts or with DENV-qR primer in order to amplify the DENV genomic RNA.
For qPCR, cDNA produced as above was diluted in 1:5 ratios with nuclease free water. 2 μl of the diluted cDNA was used for downstream qPCR reaction. Both forward and reverse gene-specific primers were used to amplify the target genes (primer sequences in Table S1), using QuantiFast SYBR Green (Qiagen) in a Rotorgene qPCR machine (Qiagen). For Ae. aegypti samples, RPS17 transcript levels were used for the normalization of RNA templates, while RPL32 was used for normalization of D. melanogaster samples. Each qPCR reaction was performed in duplicates with at least three biological replicates. All qPCR data were normalized with Qiagen analysis templates and were further analysed by Prism 7.0. Unpaired t-test was used to determine statistical significance between two individual groups while one-way ANOVA with Tukey’s post-hoc test was performed to find statistical significance between more than two groups of data.
RNAi-mediated gene silencing
In order to knockdown the AeCHD7 gene for functional analysis in DENV life cycle, primers were designed to amplify a 586 bp product from the AeCHD7 gene with the addition of the T7 promoter sequences at both ends (Table S1). MEGAscript T7 Transcription kit was then used according to the manufacturer’s instructions in order to synthesize dsRNA targeting the AeCHD7 transcripts. A similar approach was followed to synthesize dsRNA against GFP RNA. For knockdown experiments, Aa20 cells were double transfected with 2–5 μg of dsRNA per well against the target gene. dsGFP RNA was used as non-specific control.
Virus infection and plaque assay
For virus inoculation experiments, Ae. aegypti Aa20 were seeded at the density of 3 × 105 cells per well in 12-well plates. Cells were first double transfected with dsRNA against the target gene or GFP control and after 6 h cells were infected with DENV2 (New Guinea strain) at a multiplicity of infection (MOI) of 1. Media were collected 72 h post-infection for plaque assay.
To perform plaque assay, Vero cells were seeded in a 96-well plate and were allowed to form monolayers. Virus containing media from the experiments were serially diluted into 100, 101, 102, 103 dilutions and added to Vero cells in duplicates. Cells were incubated with virus at room temperature with continuous shaking on shaker for 1 h and then incubated at 37 °C for one additional hour. After 2 h of incubation, media were aspirated and an overlay was added to the cells which comprised of 1.5% carboxymethyl cellulose (CMC) and 2.5% FBS in Opti-MEM medium (Sigma). Cells were then incubated for 72 h at 37 °C and 5% CO2 and fixed with 80% ice-cold acetone in 1X PBS for 20 min at −20 °C. Plates were then air dried overnight and blocked with 5% skimmed milk in 1 × PBST at 37 °C for 30 min. Cells were then incubated with the primary antibody against DENV2-Envelope (human) in 1:1000 dilution in 0.1% skimmed milk in 1 × PBST for 2 h at 37 °C as described previously62. Plates were washed 3 times with 1 × PBST and incubated with the secondary antibody (IRDye 800 CW goat anti-human LICOR) for 1 h at 37 °C. Plates were washed and dried as above and were dried and scanned on the Odyssey imager (LI-COR Biosciences) at 41 μM resolution.
Additional Information
How to cite this article: Asad, S. et al. Downregulation of Aedes aegypti chromodomain helicase DNA binding protein 7/Kismet by Wolbachia and its effect on dengue virus replication. Sci. Rep. 6, 36850; doi: 10.1038/srep36850 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Bhatt, S. et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013).
Gubler, D. J. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp 277, 3–16; discussion 16–22, 71–13, 251–253 (2006).
Diosa-Toro, M., Urcuqui-Inchima, S. & Smit, J. M. Arthropod-borne flaviviruses and RNA interference: seeking new approaches for antiviral therapy. Adv Virus Res 85, 91–111 (2013).
Gubler, D. J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11, 480–496 (1998).
Lambrechts, L., Scott, T. W. & Gubler, D. J. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl Trop Dis 4, e646 (2010).
Marcombe, S. et al. Insecticide resistance in the dengue vector Aedes aegypti from Martinique: distribution, mechanisms and relations with environmental factors. PLoS One 7, e30989 (2012).
Iturbe-Ormaetxe, I., Walker, T. & O’Neill, S. L. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep 12, 508–518 (2011).
Hussain, M. et al. Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. J Virol 87, 851–858 (2013).
Aliota, M. T., Peinado, S. A., Velez, I. D. & Osorio, J. E. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci Rep 6, 28792 (2016).
Dutra, H. L. et al. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe 19, 771–774 (2016).
Zug, R. & Hammerstein, P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7, e38544 (2012).
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A. & Werren, J. H. How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiol Lett 281, 215–220 (2008).
Kageyama, D., Nishimura, G., Hoshizaki, S. & Ishikawa, Y. Feminizing Wolbachia in an insect, Ostrinia furnacalis (Lepidoptera: Crambidae). Heredity 88, 444–449 (2002).
Riparbelli, M. G., Giordano, R., Ueyama, M. & Callaini, G. Wolbachia-mediated male killing is associated with defective chromatin remodeling. PLoS One 7, e30045 (2012).
Caragata, E. P., Dutra, H. L. & Moreira, L. A. Exploiting intimate relationships: Controlling mosquito-transmitted disease with Wolbachia. Trends Parasitol 32, 207–218 (2016).
Xi, Z., Khoo, C. C. & Dobson, S. L. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310, 326–328 (2005).
Walker, T. et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453 (2011).
McMeniman, C. J. et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323, 141–144 (2009).
Frentiu, F. D. et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis 8, e2688 (2014).
Hoffmann, A. A. et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457 (2011).
Kambris, Z., Cook, P. E., Phuc, H. K. & Sinkins, S. P. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326, 134–136 (2009).
Pan, X. et al. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci USA 109, E23–E31 (2012).
Rances, E., Ye, Y. H., Woolfit, M., McGraw, E. A. & O’Neill, S. L. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathogens 8, e1002548 (2012).
Hopfner, K. P., Gerhold, C. B., Lakomek, K. & Wollmann, P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr Opin Struct Biol 22, 225–233 (2012).
Marfella, C. G. & Imbalzano, A. N. The Chd family of chromatin remodelers. Mutat Res 618, 30–40 (2007).
Rodgers, M. J., Banks, D. J., Bradley, K. A. & Young, J. A. CHD1 and CHD2 are positive regulators of HIV-1 gene expression. Virol J 11, 180 (2014).
Marcos-Villar, L., Pazo, A. & Nieto, A. Influenza virus and chromatin: Role of the CHD1 chromatin remodeler in the virus life cycle. J Virol 90, 3694–3707 (2016).
Yap, K. L. & Zhou, M. M. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50, 1966–1980 (2011).
Bugga, L., McDaniel, I. E., Engie, L. & Armstrong, J. A. The Drosophila melanogaster CHD1 chromatin remodeling factor modulates global chromosome structure and counteracts HP1a and H3K9me2. PLoS One 8, e59496 (2013).
Khattak, S., Lee, B. R., Cho, S. H., Ahnn, J. & Spoerel, N. A. Genetic characterization of Drosophila Mi-2 ATPase. Gene 293, 107–114 (2002).
Daubresse, G. et al. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126, 1175–1187 (1999).
McDaniel, I. E., Lee, J. M., Berger, M. S., Hanagami, C. K. & Armstrong, J. A. Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics 178, 583–587 (2008).
Kehle, J. et al. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900 (1998).
Murawska, M., Hassler, M., Renkawitz-Pohl, R., Ladurner, A. & Brehm, A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet 7, e1002206 (2011).
Srinivasan, S., Dorighi, K. M. & Tamkun, J. W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet 4, e1000217 (2008).
Giraldo-Calderon, G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43, D707–D713 (2015).
Vissers, L. E. et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36, 955–957 (2004).
Moreira, L. A. et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139, 1268–1278 (2009).
Bian, G., Zhou, G., Lu, P. & Xi, Z. Replacing a native Wolbachia with a novel strain results in an increase in endosymbiont load and resistance to dengue virus in a mosquito vector. PLoS Negl Trop Dis 7, e2250 (2013).
Hughes, H. & Britton, N. F. Modelling the use of Wolbachia to control dengue fever transmission. Bull Math Biol 75, 796–818 (2013).
Zhang, G., Hussain, M., O’Neill, S. L. & Asgari, S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc Natl Acad Sci USA 110, 10276–10281 (2013).
Johnson, K. N. The impact of Wolbachia on virus infection in mosquitoes. Viruses 7, 5705–5717 (2015).
McMeniman, C. J. et al. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl Environ Microbiol 74, 6963–6969 (2008).
Ye, Y. H. et al. Infection with a virulent strain of Wolbachia disrupts genome wide-patterns of cytosine methylation in the mosquito Aedes aegypti. PLoS One 8, e66482 (2013).
Brehm, A., Tufteland, K. R., Aasland, R. & Becker, P. B. The many colours of chromodomains. Bioessays 26, 133–140 (2004).
Stokes, D. G., Tartof, K. D. & Perry, R. P. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci USA 93, 7137–7142 (1996).
Stokes, D. G. & Perry, R. P. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 15, 2745–2753 (1995).
Srinivasan, S. et al. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132, 1623–1635 (2005).
Tai, H. H. et al. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun 308, 170–176 (2003).
Kleino, A. et al. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J 24, 3423–3434 (2005).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43, D222–D226 (2015).
Hughes, G. L. & Rasgon, J. L. Transinfection: a method to investigate Wolbachia-host interactions and control arthropod-borne disease. Insect Mol Biol 23, 141–151 (2014).
Hussain, M., Frentiu, F. D., Moreira, L. A., O’Neill, S. L. & Asgari, S. Wolbachia utilizes host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci USA 108, 9250–9255 (2011).
Mayoral, J. M. et al. Wolbachia small non-coding RNAs and their role in cross-kingdom communications. Proc Natl Acad Sci USA 111, 18721–18726 (2014).
Bulich, R. & Aaskov, J. G. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J Gen Virol 73, 2999–3003 (1992).
Wang, S. H. et al. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J Gen Virol 83, 3093–3102 (2002).
Brooks, A. J. et al. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem 277, 36399–36407 (2002).
Pryor, M. J. et al. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic 8, 795–807 (2007).
Colpitts, T. M., Barthel, S., Wang, P. & Fikrig, E. Dengue virus capsid protein binds core histones and inhibits nucleosome formation in human liver cells. PLoS One 6, e24365 (2011).
Min, K.-T. & Benzer, S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci USA 94, 10792–10796 (1997).
Frentiu, F. D., Robinson, J., Young, P. R., McGraw, E. A. & O’Neill, S. L. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS One 5, e13398 (2010).
Watterson, D. et al. A generic screening platform for inhibitors of virus induced cell fusion using cellular electrical impedance. Sci Rep 6, 22791 (2016).
Acknowledgements
We are thankful to Dr Karyn Johnson and Verna Hearne from UQ for providing D. melanogaster wMelPop-infected and tetracycline cured flies, Prof Paul Young from UQ for providing the anti-DENV antibody, and Dr Andrew van den Hurk from Queensland Health for providing Ae. aegypti eggs. Special thanks to Solomon-Osei-Amo (UQ) for critical reading of the manuscript. This project was supported by a National Health Medical Research Council grant (APP1062983) to S Asgari and a UQ International scholarship to S Asad.
Author information
Authors and Affiliations
Contributions
S.Asa. conceived and designed research, carried out experiments, analysed the data and drafted the manuscript. S.H.M. carried out experiments and edited the manuscript. S.Asg. designed research and edited the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Asad, S., Hall-Mendelin, S. & Asgari, S. Downregulation of Aedes aegypti chromodomain helicase DNA binding protein 7/Kismet by Wolbachia and its effect on dengue virus replication. Sci Rep 6, 36850 (2016). https://doi.org/10.1038/srep36850
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36850
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.