Abstract
Ubiquitination is a crucial post-translational modification; however, the functions of ubiquitin-coding genes remain unclear. UBA52 encodes a fusion protein comprising ubiquitin at the N-terminus and ribosomal protein L40 (RPL40) at the C-terminus. Here we showed that Uba52-deficient mice die during embryogenesis. UBA52-deficient cells exhibited normal levels of total ubiquitin. However, UBA52-deficient cells displayed decreased protein synthesis and cell-cycle arrest. The overexpression of UBA52 ameliorated the cell-cycle arrest caused by UBA52 deficiency. Surprisingly, RPL40 expression itself is insufficient to regulate cyclin D expression. The cleavage of RPL40 from UBA52 was required for maintaining protein synthesis. Furthermore, we found that RPL40 formed a ribosomal complex with ubiquitin cleaved from UBA52. UBA52 supplies RPL40 and ubiquitin simultaneously to the ribosome. Our study demonstrated that the ubiquitin-coding gene UBA52 is not just an ubiquitin supplier to the ubiquitin pool but is also a regulator of the ribosomal protein complex. These findings provide novel insights into the regulation of ubiquitin-dependent translation and embryonic development.
Similar content being viewed by others
Introduction
Ubiquitination is a common post-translational modification. The 8-kDa protein ubiquitin is covalently attached to one or more lysine (Lys) residues of a substrate protein1. Ubiquitin chains can attach at seven ubiquitin Lys residues (K6, K11, K27, K29, K33, K48, and K63) or at the ubiquitin amino-terminal methionine 1 residue (generating linear chains)2. Different ubiquitin linkage types have different functions in the cell cycle3, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and cell-death signaling4,5,6. Ubiquitin proteins are encoded by four genes (UBA52, UBA80, UBB, and UBC)7,8. UBB and UBC are polymers of ubiquitin linked in a “head-to-tail” manner. UBA52 and UBA80 comprise a single ubiquitin fused at the C-terminus to ribosomal protein (RP) L40 and S27a, respectively9,10 (Fig. 1A). UBA52 and UBA80 are mainly post-translationally processed11. A pool of free ubiquitin is maintained through synthesis by ubiquitin-coding genes, release from protein-conjugated ubiquitin chains, and release from unanchored ubiquitin chains12,13. It is assumed that the ubiquitin pool is under strict regulation in order to ensure appropriate responses to different cellular conditions. However, the function of ubiquitin-coding genes as a source of ubiquitin remains unknown.
UBA52 is required for embryonic development.
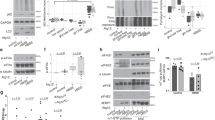
(A) Diagram of human ubiquitin genes and their chromosomal locations. (B) Schematic representation of the gene-targeting construct and screening strategy. Southern blots of EcoRI- and PstI-digested genomic DNA from embryonic stem cells showing the targeted allele. The probe was specific for exon 1. (C) Polymerase chain reaction (PCR) analysis of genomic DNA obtained from the tails of wild-type (Uba52+/+) and mutant (Uba52tm1a/+) mice using the PCR primers indicated. (D) Quantitative real-time reverse-transcription PCR of Uba52 mRNA expression in tissues (kidney, spleen, small intestine, and colon) of 3-week-old mice. mRNA levels were normalized to Actb levels. Uba52+/+ (n = 8), Uba52tm1a/+ (n = 8). Error bars indicate standard deviations. N.S. indicates not significant by two-tailed Student’s t-test. (E) Numbers of surviving offspring from Uba52tm1a/+ parents. Pictures show embryonic day (E) 10.5 and E13.5 littermates. Parentheses: expected numbers based on a Mendelian ratio.
Genetic approaches have been used to analyze the physiological function of ubiquitin-coding genes. Mice lacking one or both copies of UBB develop normally and are viable at birth14. Loss of UBB leads to a progressive degenerative disorder that affects the neurons15. In contrast, loss of UBC leads to embryonic death between embryonic days (E)12.5 and E14.5 in utero16. UBA52 and UBA80 are preferentially overexpressed during hepatoma cell apoptosis17. In addition, previous findings suggested that UBA52 functions in the pathogenesis of diabetic nephropathy18. Northern hybridization also suggested that UBA52 is expressed in human colorectal carcinoma19. However, the physiological functions of the ubiquitin hybrid gene UBA52 remain unclear.
Ribosome biogenesis and protein synthesis are tightly regulated process linked to other fundamental cellular processes20,21. Targeted disruption of the ribosomal protein genes (e.g., Rps19 and Rps6) is lethal22,23. Dysfunctional ribosome biogenesis is associated with developmental defects and an increased risk of cancer24,25. Recent studies have shown that regulatory 40 S ribosomal ubiquitination is an important phase of translational control26. In addition, translation reactions terminate with ub-dependent removal of defective nascent chain27,28. Nevertheless, the association between ubiquitin-dependent regulation of translation and the ubiquitin-ribosomal hybrid gene UBA52 remains unclear. To determine the physiological functions of UBA52, we generated mice lacking UBA52. The obtained findings highlight the central roles of ubiquitin-coding genes in the regulation of physiological ubiquitin and ribosomal function in vivo.
Results and Discussion
UBA52 is required for embryonic development
To investigate the potential physiological roles of UBA52, we generated Uba52-deficient mice. We confirmed insertion of the tm1a cassette into Uba52 genomic fragment in embryonic stem cells by Southern blotting (Fig. 1B) and in DNA obtained from the tail by polymerase chain reaction (PCR; Fig. 1C). We found that the deletion of one Uba52 allele in mice did not affect the expression of Uba52 mRNA (Fig. 1D). To further confirm the UBA52 tm1a allele, we consider that aberrant UBA52 proteins may act as dominant-negative molecules. We analyzed the UBA52 protein expression by immunoblotting; the truncated protein was not detected in Uba52tm1a/+ liver lysates (data not shown). However, no live-born Uba52-deficient (uba52tm1a/tm1a) mice were obtained from intercrossed Uba52tm1a/+ parents (Fig. 1E). Because Ubc-deficient mice died at around E13.5, we analyzed Uba52-deficient mice at this time. Uba52tm1a/+ normally developed, but we could not obtain uba52tm1a/tm1a at E13.5 (Fig. 1E). Rps6-heterozygous embryos die during gastrulation23. Therefore, we analyzed at E10.5, but we could not obtain uba52tm1a/tm1a at E10.5. Thus, these data indicate that one allele of the Uba52 gene is enough for development but UBA52 is required for embryonic development.
UBA52 regulates protein synthesis
To better understand how UBA52 sustains embryonic development, we noted that UBC is essential for fetal development16. Given that UBA52 is a ubiquitin hybrid gene, we hypothesized that UBA52 regulates the total ubiquitin mRNA expression. To investigate this possibility, we used a short interfering RNA (siRNA) approach for reducing UBA52 expression in a colon cancer cell line (DLD-1). Acute knockdown of UBA52 did not affect the total ubiquitin mRNA levels. Conversely, knockdown of UBC reduced the total amount of ubiquitin (Fig. 2A). Our finding that UBA52-deficient cells display similar levels of total ubiquitin mRNA as those displayed by wild-type cells indicates that changes in the level of ubiquitin mRNA can be compensated for by other ubiquitin-coding genes. Because previous reports have shown that UBA52 is rapidly cleaved11,17, we transfected a FLAG-UBA52-green fluorescent protein (GFP) vector into DLD-1 cells. We detected a band of approximately 34 kDa using anti-UBA52 and anti-GFP antibodies but did not detect this using anti-FLAG and anti-ubiquitin antibodies (Fig. 2B). The anti-FLAG antibody blot showed a high-molecular weight smear. These findings indicate that FLAG-UBA52–GFP is cleaved and that polyubiquitin is generated from FLAG-ubiquitin. In addition, we detected RPL40-GFP and endogenous RPL40 using an anti-UBA52 antibody. To confirm that ubiquitin was obtained from the cleavage of UBA52, we generated cleavage-resistant UBA52 (CR) expression vector, which was constructed by mutating the terminal region of ubiquitin (G75/76A)17. FLAG-UBA52-GFP (CR) expression vector showed decreased high-molecular weight smear as compared to FLAG-UBA52-GFP (WT) (Fig. 2C). FLAG-UBA52-GFP (CR) was detected at approximately 44 kDa by anti-FLAG antibody and anti-GFP antibody (Fig. 2C). Thus, we confirmed that ubiquitin was generated from UBA52. RPs are known to play many extra ribosomal roles29. For example, RPS3 regulates NF-κB signaling30 and RPL13a in macrophages resolves inflammation31. To examine the localization of UBA52, we transfected a FLAG-UBA52-GFP vector into DLD-1 cells. Microscopic analysis revealed that FLAG-GFP was expressed in both the cytosol and nucleolus. RPL40-GFP was not expressed in the nucleolus (Fig. 2D). To confirm the localization of endogenous RPL40, we lysed the cells and separated the soluble, ribosome-free cytosol (S100) from a particulate pellet containing ribosomes (P100) by ultracentrifugation30. RPL7a and RPS3 were expressed in the ribosomal fraction (P100) (Fig. 2E). In contrast, CDK6 and Actin were expressed in the cytosol (S100). We found that RPL40 was expressed not only in the ribosomal fraction (P100) but also in the cytosol (Fig. 2E). In addition, RPL40 expression in the ribosomal fraction (P100) gradually decreased over time after siRNA transfection. These findings showed that we could analyze ribosomal function by the siRNA approach. To better understand the roles of UBA52, we tested whether it regulates ribosomal function. A protein synthesis assay using O-propargyl-puromycin (OPP) revealed that cycloheximide (CHX) completely inhibited protein biosynthesis under these conditions. RPS3 and UBA52 knockdown decreased protein synthesis (Fig. 2F). To confirm the general role of UBA52, we tested Hela cells as well as DLD-1 cells (Fig. 2G). Along with DLD-1 cells, UBA52-deficient Hela cells showed decreased protein synthesis. Together, these data indicate that the loss of UBA52 does not regulate the total amount of ubiquitin mRNA but regulates protein synthesis functions in cells. Thus, Uba52-deficient lethality may be because of ribosomal dysfunction. Further investigation is required to clarify the mechanism by which UBA52 regulates embryonic development in vivo.
UBA52 regulates protein synthesis.
(A) Ubiquitin expression in UBA52-deficient cells. Total RNA was isolated from DLD-1 cells 24 h after short interfering RNA (siRNA) transfection. UBC, UBB, UBA52, and UBA80 mRNA levels were measured by quantitative real-time reverse-transcription PCR and normalized to ACTB levels. Error bars indicate standard deviations. Data represent three independent experiments (n = 8). **P < 0.01 (one-way ANOVA followed by Tukey’s test). (B) UBA52 cleavage in DLD-1 cells. FLAG-UBA52-green fluorescent protein (GFP) expression in DLD-1 cells. DLD-1 cells were transfected with FLAG-GFP or FLAG-UBA52-GFP. After 45 h, cells were lysed and immunoblotted for the indicated proteins. Data are representative of more than three independent experiments. (C) Cleave-resistant mutants in HEK293T cells. HEK293T cells were transfected with three types of mutation vectors. After 24 h, cells were lysed and immunoblotted for the indicated proteins. G75/76A mutations lead to cleavage resistance (FLAG-UBA52-GFP (CR))17. Data are representative of more than three independent experiments. (D) UBA52 localization in DLD-1 cells. DLD-1 cells were transfected with FLAG-GFP or FLAG-UBA52-GFP. After 24 h, images were acquired using a confocal laser microscope. Data are representative of three independent experiments. GFP (green) and 4′,6-diamidino-2-phenylindole (blue). (E) Subcellular fractions of DLD-1 cells. DLD-1 cells were transfected with a UBA52 siRNA and lysed for ultracentrifugation at the indicated time. S100 (cytosol) and P100 (crude ribosome pellet) fractions were immunoblotted for the indicated proteins. Data are representative of more than three independent experiments. (F,G) Protein synthesis in UBA52-deficient cells. DLD-1 cells (F) or Hela cells (G) were transfected with UBA52 or ribosomal protein (RP) S3 siRNAs. The cells were treated with cycloheximide (100 μg/ml) for 3 h. Cells were incubated with O-propargyl-puromycin (20 μM) for 30 min and then harvested for the protein synthesis assay. Data are representative of more than two independent experiments. *P < 0.05, **P < 0.01 (one-way ANOVA followed by Tukey’s test). We confirmed knockdown efficiency by quantitative real-time (RT) PCR and normalized to ACTB levels. Data are representative of two independent experiments.
UBA52 regulates the cell cycle
Ribosomal stress can regulate the cell cycle by p53-dependent and -independent pathways32,33. To understand the role of UBA52, we analyzed cell proliferation. We found that UBA52-deficient DLD-1 cells displayed decreased cell numbers and cell-cycle arrest at G1/S (Fig. 3A,B). To determine whether UBA52 expression is sufficient to regulate the cell cycle, we mutated UBA52 expression vectors to induce siRNA resistance. We found that Myc-UBA52 ameliorated the cell-cycle arrest caused by UBA52 deficiency (Fig. 3C). Together, these findings indicate that UBA52 regulates the cell cycle. Next, to understand the mechanism underlying this, we consider that cyclin D promotes cell cycle as a main regulator34. We analyzed cyclin D1 and D3 gene expressions. There were no differences in cyclin D1 and D3 mRNA expressions between control and UBA52-deficient cells (Fig. 3D). In contrast, the levels of cyclin D1 and D3 protein expression decreased in UBA52-deficient cells (Fig. 3E). To examine the possibility that UBA52 regulates cyclin D expression, we analyzed cell division protein kinase 6 (CDK6), which is known to be a major partner of cyclin D and a key molecule in the regulation of G1/S phase. CDK6 expressed in the cytosol fraction (Fig. 2E). Using a proximity ligation assay, we found that endogenous RPL40 co-localized with CDK6 (Fig. 3F). To confirm physical interaction, we performed immunoprecipitation assay. Endogenous CDK6 associated with RPL40-GFP in DLD-1 cells and HEK293T cells (Fig. 3G). These data suggested that RPL40 may affect the CDK6 complex formation. Nevertheless, cyclin D1 and D3 quickly degraded after CHX treatment in DLD-1 cells (Fig. 3H). Moreover, similar observations that the normal expression of mRNAs and the decreased protein levels of cyclins D1 and D3 in UBA52-deficient cells were found in Rps6wt/del p53−/− embryos23. These findings indicated that decreased levels of cyclin D1 and D3 were provoked mainly by the suppression of protein synthesis in UBA52-deficient cells. Taken together, UBA52 modulates cyclin D1 and D3 protein expression and regulates the cell cycle.
UBA52 regulates the cell cycle.
(A) Cell viability assays in UBA52-deficient cells. DLD-1 cells were transfected with a UBA52 siRNA. Then, cells were harvested for the cell viability assay at the indicated time. The fluorescent score was normalized to the level at 0 h. Data are representative of three independent experiments. **P < 0.01 (two-tailed Student’s t-test). (B) Cell-cycle analysis in UBA52-deficient cells. DLD-1 cells were transfected with UBA52 siRNA. Twenty-four hours later, cells were harvested for cell-cycle analysis. Data are representative of more than three independent experiments. **P < 0.01 (one-way ANOVA followed by Tukey’s test). (C) Myc-UBA52 (WT) regulates the cell cycle. DLD-1 cells were transfected with Myc-UBA52 (WT) #7R and UBA52 siRNA simultaneously. Thirty-six hours later, cells were harvested for cell-cycle analysis. Data are representative of more than three independent experiments. **P < 0.01 (one-way ANOVA followed by Tukey’s test). (D) Cell-cycle-related mRNA expression in UBA52-deficient cells. DLD-1 cells were transfected with a UBA52 siRNA. Twenty-four hours later, cells were harvested for quantitative real-time reverse-transcription PCR and normalized to ACTB levels. Data are representative of three independent experiments. (E) Cell cycle-related protein expression in UBA52-deficient cells. DLD-1 cells were transfected with a UBA52 siRNA. Twenty-four hours later, cells were harvested for immunoblotting. Data are representative of more than three independent experiments. (F) RPL40 co-localises with CDK6. DLD-1 cells were harvested for the in situ proximity ligation assay. Anti-UBA52 and anti-CDK6 antibodies were used. Data are representative of four independent experiments. **P < 0.01 (two-tailed Student’s t-test). (G) UBA52 interacts with CDK6. DLD-1 cells and HEK293T cells were transfected with FLAG-UBA52-GFP. Twenty hours later, cells were lysed and protein extracts were immunoprecipitated with GFP antibody and immunoblotted for the indicated proteins. Data are representative of more than three independent experiments in DLD-1 cells and HEK293T cells. (H) CHX chase experiment for the cyclin D protein. DLD-1 cells were treated with 100 μg/ml CHX and cell lysates were harvested for immunoblotting at the indicated time points. Data are representative of more than three independent experiments.
Ubiquitin cleaved from UBA52 forms a molecular complex with RPL40
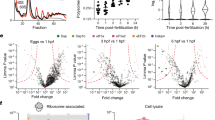
To understand how the ubiquitin-ribosomal hybrid gene regulates protein synthesis, we generated cleavage-resistant (CR) and siRNA resistant UBA52 expression vectors17. Ubiquitin, Myc-RPL40, and Myc-UBA52 (CR) overexpression in DLD-1 cells did not affect cyclin D expression, but Myc-UBA52 (WT) ameliorated the decreased cyclin D1 and D3 expressions (Fig. 4A, Supple Fig. 1). These findings indicated that RPL40 expression itself is not sufficient to regulate protein synthesis. The RPL40 cleavage mechanism and localization of ubiquitin cleaved from UBA52 may influence the function of UBA52. Thus, we tested whether UBA52 and UBA52 (CR) formed a molecular complex. GFP immunoprecipitation revealed that RPL40 bound molecules that had been ubiquitinated using ubiquitin generated from UBA52 cleavage (Fig. 4B). Furthermore, FLAG immunoprecipitation revealed that ubiquitin cleaved from UBA52 ubiquitinated the ribosomal complex (Fig. 4C). These findings indicated that UBA52 supplies ubiquitin to the ribosomal complex. To confirm whether UBA52 regulates the ribosomal protein complex, we tested the ribosomal fraction. FLAG-UBA52-GFP transfected cell showed that both FLAG-ubiquitin from UBA52 and RPL40-GFP expressed in the ribosomal fraction (Fig. 4D). In addition, UBA52-deficient cell showed reduced ubiquitin smear in the ribosomal fraction (Fig. 4E). Taken together, these data suggested that RPL40 formed a ribosomal protein complex with ubiquitin cleaved from UBA52 (Fig. 4F). Recent studies have shown that the regulation of ubiquitin-dependent translation is an important feature of cell-fate determination35. Regulatory 40 S ribosomal ubiquitination events play a critical role in protein biogenesis36. Our findings demonstrated that ubiquitin, RPL40, UBA52 (CR), and a mixture of ubiquitin and RPL40 could not ameliorate cyclin D1 and D3 expressions (Fig. 4A). However, full-length UBA52 abolished the cell-cycle-related UBA52-deficient phenotype. These findings indicate that the RPL40 cleavage is critical for the functions of UBA52. Thus, we hypothesize that UBA52 regulates ribosomal function by two steps. RPL40 influences the ribosomal biogenesis as a ribosomal protein, at the same time, ubiquitin cleaved from UBA52 generates ubiquitination of the ribosomal protein complex. These data reveal a novel mechanism that ubiquitin regulates translation by the ubiquitin–ribosomal hybrid gene. Future studies will be needed to identify enzymes that cleave ubiquitin from UBA52 under physiological conditions. Moreover, it is critical to analyze UBA52’s physiological functions in cell type-specific contexts.
Ubiquitin cleaved from UBA52 forms a molecular complex with RPL40.
(A) Myc-UBA52 (WT) regulates cyclin D expression. DLD-1 cells were transfected with a UBA52 siRNA. After 6 h, DLD-1 cells were transfected with the siRNA-resistant vectors indicated. Twenty-seven hours later, cells were harvested for immunoblotting. Data are representative of more than three independent experiments. (B) RPL40 binds ubiquitinated molecules. DLD-1 cells were transfected with FLAG-UBA52-GFP (WT) and other vectors. Twenty-four hours later, cells were lysed and protein extracts were immunoprecipitated with GFP antibody and immunoblotted for the indicated proteins. Data are representative of more than three independent experiments. (C) Ubiquitin, cleaved from UBA52, forms a molecular complex with ribosome. DLD-1 cells were transfected with FLAG-UBA52-GFP (WT) and other vectors. Twenty-four hours later, cells were lysed and protein extracts were immunoprecipitated with FLAG antibody and immunoblotted for the indicated proteins. Data are representative of two independent experiments. (D) Subcellular fractions of FLAG-UBA52-GFP expressed DLD-1 cells. DLD-1 cells were transfected with FLAG-UBA52-GFP vector or vehicle, and 24 h later, lysed for ultracentrifugation at 10,000 × g for 1 h. S100 (cytosol) and P100 (crude ribosome pellet) fractions were immunoblotted for the indicated proteins. Data are representative of two independent experiments. (E) Subcellular fractions of UBA52-deficient DLD-1 cells. DLD-1 cells were transfected with UBA52 siRNA or control siRNA, and 24 h later, ultracentrifugation was performed. Data are representative of three independent experiments. (F) Schematic showing that UBA52 is a dual regulator of ribosomal function. RPL40 influences the ribosomal biogenesis as a ribosomal protein, at the same time, ubiquitin cleaved from UBA52 generates ubiquitination of the ribosomal protein complex.
In conclusion, the generation of mice lacking Uba52 has allowed us to unveil the physiological function of the ubiquitin hybrid gene Uba52. In particular, we have discovered that UBA52 regulates cell fate and embryonic development. RPL40, the ribosomal domain of UBA52, generated a ribosomal protein complex, at the same time, the UBA52 ubiquitin domain simultaneously generated ubiquitination of the ribosomal complex. Thus, UBA52 is a dual regulator of ribosomal protein complex. These findings provide unique molecular insights into ubiquitin-related protein synthesis and embryonic development.
Methods
Mice
Uba52tm1a(EUCOMM)Wtsi embryonic stem cells were purchased from the European Conditional Mouse Mutagenesis Program (EUCOMM) and microinjected into the blastocysts of an albino C57BL6 strain. The chimeric mice were backcrossed with the same strain of albino C57BL6 mice to generate heterozygous Uba52 mutant mice. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Tokyo Medical and Dental University. Experiments were performed in compliance with Tokyo Medical and Dental University’s Animal Facility regulations. Genotypes were initially confirmed by Southern blotting using embryonic stem cells. In addition, genotypes were confirmed by PCR using DNA derived from the tail and the following primers: Primer4, F 5′-CTGCAGAGGGAGTTCAGGG-3′ and R 5′-GTTTGGTAAGTAGGGGCAGC-3′; Primer5, F 5′-FACAACCATGGAAGATCCCGT-3′ and R 5′-CCGTTGCACCACAGATGAAA-3′ and Primer6, F 5′-AGGAAGGAGTTGTGGCCAACCTGG-3′ and R 5′-TGAACTGATGGCGAGCTCAGACC-3′. Also, the following primers were used for long-range PCR: Primer1, F 5′-TCCAGACAGAACGACTATTCTCGC-3′ and R 5′-AACTGAAGGATCGGACAGCA-3′; Primer2, F 5′-ACAACCATGGAAGATCCCGT-3′ and R 5′-AACTGAAGGATCGGACAGCA-3′ and Primer3, F 5′-TCCAGACAGAACGACTATTCTCGC-3′ and R 5′-CCGTTGCACCACAGATGAAA-3′.
Southern blotting
A DNA template was extracted from embryonic stem cells purchased from EUCOMM. Probes were set to correspond with a sequence of 505 base pairs (bp) in the region containing exon 1 (forward primer, 5′-GCTCGGCCTAGGATTCATTT-3′; reverse primer, 5′-CGCCTCGTTGAAGAGAAAGA-3′). The DNA template was digested using EcoRI and PstI. The digested DNA was electrophoresed in a 1–1.5% agarose gel and transferred to a positively charged nylon membrane (Hybond-N+, RPN119B; GE Healthcare, Little Chalfont, Buckinghamshire, UK). DIG Easy Hyb (1603558; Roche Diagnostics, Basel, Switzerland) was used for hybridization, and ready-to-use disodium 3-{4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricycle(3.3.1.13,7)decan]-4-yl}phenyl phosphate (11755633001; Roche Diagnostics) was used for detection, as described in the manufacturer’s protocol.
Cell culture and reagents
The human colorectal cancer cell line DLD-1 was cultured in Roswell Park Memorial Institute-1640 medium (R8758; Sigma-Aldrich Corp., St. Louis, MO, USA) with 10% fetal bovine serum (FBS; S1820; Biowest SAS, Nuaillé, France) and 1% penicillin/streptomycin (26253-84; Nacalai Tesque, Inc., Kyoto, Japan) at 37 °C. HEK293T cells and Hela cells were cultured in Dulbecco’s Modified Eagle’s Medium (high glucose) (D5796; Sigma-Aldrich Corp., St. Louis, MO, USA) with 10% FBS and 1% penicillin/streptomycin at 37 °C. Cells were transiently transfected with siRNAs using Lipofectamine® RNAiMAX™ (13778150; Invitrogen, Carlsbad, CA, USA), as indicated by the manufacturer. DLD-1 cells were transiently transfected with plasmid vectors using Lipofectamine® LTX & Plus™ (15338-100; Invitrogen) or Lipofectamine® 3000 (L3000015; Invitrogen). DLD-1 cells were co-transfected with an siRNA and a plasmid vector for the cell-cycle assay using Lipofectamine® 3000 (L3000075; Invitrogen), as indicated by the manufacturer. DLD-1 cells were treated with CHX (06741-91; Nacalai Tesque, Inc.).
Plasmids and short interfering RNA oligonucleotides
Human UBA52 cDNA was cloned from the HeLa cell line. Myc-ubiquitin was constructed by TAA (stop codon) insertion at 229–231 bp in UBA52 cDNA using the PrimeSTAR mutagenesis basal kit (R046A; Takara Bio Inc., Shiga, Japan). Myc-RPL40 was constructed by the deletion of ubiquitin at 4–228 bp. To make CR UBA52, alanine scanning was performed every two bases in the region connecting ubiquitin and RPL40. Finally, the UBA52 (CR) vectors were constructed by mutating the connecting region of ubiquitin and RPL40 (223–234 bp; ggtggcattatt) to gctgccattatt (G75/76A). ON-TARGETplus SMARTpool siRNA oligonucleotides specific for human UBA52, mouse UBA52, human RPS3, and a non-targeting pool siRNA were purchased from GE Dharmacon (Lafayette, CO, USA). The same human RPS3 siRNA sequence as that of the GE Dharmacon SMARTpool siRNAs was purchased from Hokkaido System Science Co., Ltd. Individual human UBA52 (J-011794-07, GCUGUCAACUGCCGCAAGA; UBA52 #7) (J-011794-05, CCUGCGAGGUGGCAUUAUU; UBA52 #5), siRNA-resistant Myc-UBA52 vectors [Myc-UBA52 (WT) #7R, Myc-UBA52 (CR) #7R, and Myc-RPL40 #7R] were constructed by mutation of the RPL40 region (319–337 bp) to GCTGTCAACTGTAGGAAGA, which had no impact on the encoded protein sequence. Myc-UBA52(WT) #5R vector was constructed by mutation of the connecting region of Ubiquitin and RPL40 (216–234 bp) to CTTAAGGGGTGGCATTATT, which had no impact on the encoded protein sequence too.
Flow cytometry
Cells were washed with phosphate-buffered saline (PBS) and dissociated using trypsin–ethylenediaminetetraacetic acid. Cells were then washed and resuspended in PBS. Iced 80% ethanol was added to a final concentration of 70%. The resuspended cells were incubated on ice for 30 min. Cells were then washed twice in cold PBS and stained with 4′,6-diamidino-2-phenylindole (DAPI)/TX-100 solution [0.1% Triton X-100, 1 μg/ml DAPI (D3571; Invitrogen) in PBS] for 30 min at room temperature. The constituent DNA amount was measured using a FACSCanto™ II flow cytometer (BD Biosciences) and cell-cycle status was analyzed using FlowJo Enterprise software (version 7.6.5; FlowJo, LLC, Ashland, OR, USA).
Quantitative real-time (RT) PCR
For in vitro assays, total RNA was isolated using the RNeasy® Mini Kit (Qiagen NV, Venlo, Limburg, The Netherlands). In vivo assays used 10–20 mg of tissue homogenized using PRECELLYS® 24 and CK14 ceramic beads in a tube (KT03961-1-003.2; PRECELLYS, Montigny-le-Bretonneux, France). Total RNA was isolated using the MAgNA Pure 96 System (Roche Diagnostics) or RNeasy® Mini Kit. cDNA was synthesized using the QuantiTect Reverse Transcription Kit (205313; Qiagen NV) or Transcriptor Universal cDNA Master Kit (5893151; Roche Diagnostics). Quantitative PCR was performed using the QuantiTect SYBR Green PCR Kit (204145; Qiagen NV) and the StepOnePlus™ RT-PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression was normalized to the expression of the housekeeping gene ACTB. The contributions of four ubiquitin-coding genes to total ubiquitin levels are shown after normalization by the number of ubiquitin moieties that each ubiquitin transcript generates as follows14,15,16:

Gene expression was determined using the following primers: human UBA52, 5′-GCGTCCCAAGAAGAAGGTCA-3′ and 5′-ACCAATTGCTGCTCCAGTCA-3′; human UBA80, 5′-ACTGTTTCAACAAACCAGAAGACA-3′ and 5′-AGGAAATGGTGTGACCATCAA-3′; human UBB, 5′-CCTGAGGGGTGGCTGTTA-3′ and 5′-TTAACATTTTGAACAGGTTCAGCTA-3′; human UBC 5′-CCACTCTGCACTTGGTCCTG-3′ and 5′-GGAATGCAACAACTTTATTGAAAGG-3′; human Cyclin D1 5′-AGCTGTGCATCTACACCGAC-3′ and 5′-GAAATCGTGCGGGGTCATTG-3′; human Cyclin D3 5′-TGCACATGATTTCCTGGCCT-3′ and 5′-CTGTAGCACAGAGGGCCAAA-3′; human RPS3 5′-GAGTCTCTGCGTTACAAACTCC-3′ and 5′-TTTCCCAGACACCACAACCT-3′; human ACTB 5′-GGATGCAGAAGGAGATCACTG-3′ and 5′-CGATCCACACGGAGTACTTG-3′; mouse Uba52 5′-GTCAGCTTGCCCAGAAGTAC-3′ and 5′-ACTTCTTCTTGCGGCAGTTG-3′; mouse Actb 5′-TTGGGTATGGAATCCTGTGG-3′ and 5′-GTACT TGCGCTCAGG AGGAG-3′.
Microscopic analysis
Cells were washed with PBS and fixed in 4% paraformaldehyde for 15 min at 4 °C. Fixed cells were washed with PBS twice and dehydrated in 100% methanol for 10 min at 4 °C. The nucleolus was stained using VECTASHIELD® Mounting Medium with DAPI (H-1200; Vector Laboratories, Inc., Burlingame, CA, USA). Images were acquired using a confocal laser microscope (FV10i; Olympus Corp., Tokyo, Japan) with a 60× oil immersion objective lens.
Immunoblotting
Cells were incubated in lysis buffer {either 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and 10% glycerol or 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.2% NP-40, and 10% glycerol} with added Halt protease and phosphatase inhibitor cocktail (1861280; Pierce Biotechnology, Rockford, IL, USA), 2 mM N-ethylmaleimide (15512-24; Nacalai Tesque Inc.), and 10 μM MG-132 on ice for 20 min and centrifuged at 14,000 × g for 20 min. For FLAG or GFP immunoprecipitation, cell lysates were incubated with Anti-DDDDK-tag mAb-Magnetic Beads (M185-9; MBL International Corporation, Nagoya, Japan) or Anti-GFPmAb-Magnetic Beads (D153-9; MBL International Corporation). Cell lysates were incubated with the pre-coupled beads for 3 h in GFP-IP or 1 h in FLAG-IP at 4 °C and then washed twice with lysis buffer with added NaCl (final concentration, 450 mM). Samples were resolved on NuPage precast 4–12% Bis-Tris gels (NP0323; Invitrogen) and transferred to polyvinylidene difluoride membranes. The following antibodies and reagents were used for immunoprecipitation and immunoblotting studies: anti-ACTB (A5441; Sigma-Aldrich Corp.); anti-FLAG (F7425; Sigma-Aldrich Corp.); anti-Myc (A-14; sc-789; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); anti-GFP (A11122; Life Technologies, Carlsbad, CA, USA); anti-UBA52 (EPR4546, ab109227, Abcam, Cambridge, UK; and EPR4547, ab109230, Abcam); anti-ubiquitin (P4D1; sc-8017; Santa Cruz Biotechnology, Inc.); anti-RPS3 (D50G7; 9538; Cell Signaling Technology, Inc.); anti-RPL7a (E109; 2415; Cell Signaling Technology, Inc.); anti-cyclin D1 (M-20; sc-718; Santa Cruz Biotechnology, Inc.); anti-cyclin D3 (DCS22; 2936; Cell Signaling Technology, Inc.); anti-CDK2 (78B2; 2546; Cell Signaling Technology, Inc.); anti-CDK4 (C-22; sc-260; Santa Cruz Biotechnology, Inc.); and anti-CDK6 (C-21; sc-177; Santa Cruz Biotechnology, Inc.).
Ultracentrifugation
Cells were incubated in lysis buffer without glycerol [20 mM HEPES (pH 7.5), 150 mM NaCl, 0.5% TritonX-100, and 0.5% CHAPS] with added Halt protease and phosphatase inhibitor cocktail on ice for 20 min and centrifuged at 14,000 × g for 20 min to remove the nuclear fraction. Then, the supernatant was ultracentrifuged at 4 °C and 100,000 × g for 1 h. The supernatant was preserved as the S100 fraction. The pellet was washed with PBS and ultracentrifuged again at 4 °C and 100,000 × g for 20 min. The supernatant was discarded and the pellet, which was the P100 fraction, was refused in lysis buffer using an ultrasonic homogenizer.
Protein synthesis assay
To analyze protein synthesis in cells, a Click-iT® Plus OPP Protein Synthesis Assay Kit (Alexa Fluor® 488; C104567570; Molecular Probes, Eugene, OR, USA) was used in accordance with the manufacturer’s protocol. The GloMax® Discover System (Promega Corp.) was used to scan the plates.
Cell viability assay
CellTiter-Glo Luminescent Cell Viability Assay (G7570; Promega Corp.) was used to count viable cells in wells, as indicated by the manufacturer.
In situ proximity ligation assay
To detect protein interactions in cells, a Duolink® PLA in-situ kit (92101; Sigma–Aldrich Corp.) was used according to the manufacturer’s instructions. The primary antibodies were as follows: a rabbit anti-UBA52 antibody (EPR4547; ab109230; Abcam) and a mouse anti-CDK6 antibody (ab54576; Abcam). The primary antibody rabbit anti-Immunoglobulin G was used as a control (ab125938; Abcam). Images were acquired with a confocal laser microscope using a ×60 oil immersion objective lens. Dots within cells were counted using Duolink ImageTool (OLINK Bioscience, Uppsala, Sweden). Cells were only counted when the whole cell was in the field of vision.
Statistical Analysis
Data were analyzed using the two-tailed unpaired Student t-test or one-way ANOVA followed by Tukey’s test using Prism software (GraphPad Software, Inc.).
Additional Information
How to cite this article: Kobayashi, M. et al. The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci. Rep. 6, 36780; doi: 10.1038/srep36780 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zinngrebe, J., Montinaro, A., Peltzer, N. & Walczak, H. Ubiquitin in the immune system. EMBO Rep 15, 28–45 (2014).
Kulathu, Y. & Komander, D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13, 508–523 (2012).
Sivakumar, S. & Gorbsky, G. J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol 16, 82–94 (2015).
Oshima, S. et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457, 906–909 (2009).
Wertz, I. E. TNFR1-activated NF-κB signal transduction: regulation by the ubiquitin/proteasome system. Curr Opin Chem Biol 23, 71–77 (2014).
Onizawa, M. et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol 16, 618–627 (2015).
Lund, P. K. et al. Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J Biol Chem 260, 7609–7613 (1985).
Wiborg, O. et al. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J 4, 755–759 (1985).
Redman, K. L. & Rechsteiner, M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature 338, 438–440 (1989).
Baker, R. T. & Board, P. G. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res 19, 1035–1040 (1991).
Grou, C. P., Pinto, M. P., Mendes, A. V., Domingues, P. & Azevedo, J. E. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep 5, 12836 (2015).
Komander, D., Clague, M. J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10, 550–563 (2009).
Kimura, Y. et al. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137, 549–559 (2009).
Ryu, K. Y. et al. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol Cell Biol 28, 1136–1146 (2008).
Ryu, K. Y., Garza, J. C., Lu, X. Y., Barsh, G. S. & Kopito, R. R. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc Natl Acad Sci USA 105, 4016–4021 (2008).
Ryu, K. Y. et al. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J 26, 2693–2706 (2007).
Han, X. J. et al. Altered dynamics of ubiquitin hybrid proteins during tumor cell apoptosis. Cell Death Dis 3, e255 (2012).
Sun, L. et al. Isolation and functional analysis of mouse UbA52 gene and its relevance to diabetic nephropathy. J Biol Chem 277, 29953–29962 (2002).
Barnard, G. F. et al. Ubiquitin fusion proteins are overexpressed in colon cancer but not in gastric cancer. Biochim Biophys Acta 1272, 147–153 (1995).
Lafontaine, D. L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22, 11–19 (2015).
Sanchez, C. G. et al. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell. 18, 276–290 (2016).
Matsson, H. et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol 24, 4032–4037 (2004).
Panić, L. et al. Ribosomal Protein S6 Gene Haploinsufficiency Is Associated with Activation of a p53-Dependent Checkpoint during Gastrulation. Mol Cell Biol. 26, 8880–8891 (2006).
Narla, A. & Ebert, B. L. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205 (2010).
Armistead, J. & Triggs-Raine, B. Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS Lett 588, 1491–1500 (2014).
Higgins, R. et al. The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40 S Ribosomal Proteins. Mol Cell 59, 35–49 (2015).
Duttler, S., Pechmann, S. & Frydman, J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 50, 379–393 (2013).
Wang, F., Durfee, L. A. & Huibregtse, J. M. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell 50, 368–378 (2013).
Warner J. R. & McIntosh K. B. How common are extraribosomal functions of ribosomal proteins? Mol Cell 34, 3–11 (2009).
Wan, F. et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131, 927–939 (2007).
Poddar D., Basu A., Baldwin W. M., Kondratov R. V., Barik S. & Mazumder B. An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J Immunol 190, 3600–3612 (2013).
Pestov, D. G., Strezoska, Z. & Lau, L. F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 21, 4246–4255 (2001).
Iadevaia, V. et al. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene 29, 5490–5499 (2010).
Musgrove, E. A., Caldon, C. E., Barraclough, J., Stone, A. & Sutherland, R. L. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 11, 558–572 (2011).
Werner, A. et al. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527 (2015).
Higgins, R. et al. The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40 S Ribosomal Proteins. Mol Cell 59, 35–49 (2015).
Acknowledgements
The authors thank Yasushi Saeki and Averil Ma for critical discussions and Ryoji Yao for technical assistance. They also thank the EUCOMM Consortium for providing Uba52tm1a(EUCOMM)Wtsi embryonic stem cells. Generation of Uba52-deficient mice was supported by the Scientific Support Program for Cancer Research, Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology. This work was also supported by the Takeda Science Foundation, The Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Daiichi Sankyo Foundation of Life Science. In addition, this study was supported by MEXT/JSPS KAKENHI (Grant Numbers 25460946, 26221307, 15H04808, 16K15423), the Research Center Network Program for Realization of Regenerative Medicine from the Japan Science and Technology Agency (JST) and Japan Agency for Medical Research and Development (AMED), the Practical Research Project for Rare/Intractable Diseases (15AeK0109047h0002) from AMED, and the Practical Research Project for Innovative Cancer Control (15Ack0106017h0002) from AMED.
Author information
Authors and Affiliations
Contributions
M.K. designed and performed experiments and assisted with the manuscript. S.O. designed experiments, supervised the overall study, and wrote the manuscript. C.M., Y.N., K.O., Y.M., Y.N., T.N., R.O., K.T. and T.N. provided technical assistance. M.W. supervised the overall study.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kobayashi, M., Oshima, S., Maeyashiki, C. et al. The ubiquitin hybrid gene UBA52 regulates ubiquitination of ribosome and sustains embryonic development. Sci Rep 6, 36780 (2016). https://doi.org/10.1038/srep36780
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36780
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.