Abstract
Engineering the coenzyme specificity of redox enzymes plays an important role in metabolic engineering, synthetic biology, and biocatalysis, but it has rarely been applied to bioelectrochemistry. Here we develop a rational design strategy to change the coenzyme specificity of 6-phosphogluconate dehydrogenase (6PGDH) from a hyperthermophilic bacterium Thermotoga maritima from its natural coenzyme NADP+ to NAD+. Through amino acid-sequence alignment of NADP+- and NAD+-preferred 6PGDH enzymes and computer-aided substrate-coenzyme docking, the key amino acid residues responsible for binding the phosphate group of NADP+ were identified. Four mutants were obtained via site-directed mutagenesis. The best mutant N32E/R33I/T34I exhibited a ~6.4 × 104-fold reversal of the coenzyme selectivity from NADP+ to NAD+. The maximum power density and current density of the biobattery catalyzed by the mutant were 0.135 mW cm−2 and 0.255 mA cm−2, ~25% higher than those obtained from the wide-type 6PGDH-based biobattery at the room temperature. By using this 6PGDH mutant, the optimal temperature of running the biobattery was as high as 65 °C, leading to a high power density of 1.75 mW cm−2. This study demonstrates coenzyme engineering of a hyperthermophilic 6PGDH and its application to high-temperature biobatteries.
Similar content being viewed by others
Introduction
Biobatteries are a type of enzyme-catalyzed fuel cells that can directly convert chemical energy of a fuel into electricity in a closed system1,2,3. In biobatteries, electrons transfer from chemical compounds to electrodes via two means: mediated electron transfer (MET) via coenzymes (e.g., NAD+) and electron mediators (e.g., benzyl viologen, vitamin K3), which often produce high current and power densities; and direct electron transfer (DET) featuring high over-potential, mediator-free, and simple configuration4. Although sugar-powered biobatteries have advantages of running on renewable sugary fuels, high safety, high energy density potentials, quiet and mild operating conditions, three main technical challenges include low power density, short lifetime, and high cost, hindering its potential applications5,6.
The discovery and utilization of thermostable enzymes from thermophilic microorganisms are of importance to increase enzyme stability and potentially decrease enzyme purification cost. If used in biobatteries, these thermophilic enzymes can often lead to prolonged lifetime of enzymes7,8,9. Thermotoga maritima is an anaerobic, rod-shaped eubacterium, originally isolated from geothermally heated marine sediment at Valcano, Italy. Growing in the optimal temperature of ~80 °C, T. maritima is regarded as an invaluable source of intrinsically thermostable enzymes10. 6-phosphogluconate dehydrogenase (6PGDH, E.C.1.1.44), the third enzyme in the pentose phosphate pathway, converts 6-phophogluconate and NADP+ to ribulose 5-phosphate, NADPH and CO2. The thermostable 6PGDH from T. maritima is very stable with a half-life time of 140 h at 80 °C, and can be easily purified by heat precipitation10. However, this enzyme prefers NADP+ to NAD+ so that it has not been tested in biobatteries that should work on NAD+ as the coenzyme.
Rational design is one of the protein engineering approaches to engineer enzymes based on knowledge of their structures and catalytic mechanisms11,12. It may offer a rapid solution to tailor enzymes with desired properties. In bioelectrocatalysis, enzyme kinetics, stability, and electron transfer efficiency can be enhanced by rational design methods, including mutating several key amino acids, trimming off nonessential protein fragments, shuffling domains, and modifying protein surface13,14. Coenzyme engineering that changes coenzyme selectivity (i.e., NAD+ or NADP+) of oxidoreductases is one of the important tools in metabolic engineering, synthetic biology, and biocatalysis. Changing the coenzyme selectivity of dehydrogenases from NADP+ to NAD+ is highly desirable because (1) NAD+ is less costly than NADP+ 15,16 and (2) NADH is more stable than NADPH17,18,19. For biobatteries, changing coenzyme preference to NAD+ must be important because NADP+ cannot be efficiently used to generate electricity8,20. Intensive studies have been conducted to change or broaden coenzyme selectivity of redox enzymes from NADP+ to NAD+ 21,22,23,24. However, there are few studies pertaining to the coenzyme engineering of thermostable dehydrogenases with the application to biobatteries25.
In this study, we demonstrated to switch coenzyme preference of T. maritima 6PDGH (Tm6PGDH) from NADP+ to NAD+ by rational design. The best Tm6PGDH mutant was applied in the biobattery, exhibiting an improved performance and a high power density at the elevated temperature.
Results
Amino acid-sequence alignment and structure analysis of 6PGDH
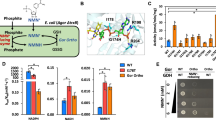
We analyzed the Rossmann fold domain of two kinds of 6PGDHs based on their coenzyme specificity (Fig. 1a). Most 6PGDHs in nature favor NADP+, such as those from Syanophora paradoxa (B2NIW2), Euglena gracilis (B2NIV9), Saccharomyces cerevisiae (P38720), Corynebacterium diphtheria (Q6NHC5), Lactococcus lactis (P96789), Bacillus subtilis (P80859), E. coli K12 (P00350), T. maritima (WP_004081528.1) and M. thermoacetica (WP_025774778.1). On the other hand, NAD+-preferred 6PGDHs are rare, including those from Haloferax volcanii (D4GST8), Gluconobacter oxydans (G5EBD7) and engineered M. thermoacetica 6PGDH. The alignment of the loop region shows that three amino acids (positions 32, 33, and 34) in NADP+-preferred 6PGDHs are highly conservative (Fig. 1b), which are asparagine, arginine and threonine (Asn32, Arg33 and Thr34) in Tm6PGDH. Via NADP+ docking, these residues are found in close contact with the 2′-phosphate of the adenosine ribose via hydrogen bonds (Fig. 1b). Additionally, Arg33 is able to form a cation-phosphate interaction with the NADP+ adenosine ring, further stabilizing the NADP-enzyme complex. However, NAD+-preferred 6PGDH enzymes have the acidic aspartate residue in the N-terminal end of loop region, which is also very conservative and completely different with that of NADP+ preferred-6PGDH. The other two residues exhibit three different combinations. According to the above information, we hypothesized that the changes in Asn32, Arg33, and Thr34 could enable Tm6PGDH to work on NAD+. Consequently, three different mutants (i.e., N32D, N32D/R33I/T34I and N32D/R33L/T34S) were firstly designed as mutation candidates.
(a) Amino acid sequence alignment of the coenzyme-binding motif of various 6PGDH enzymes. The residues composing the loop region and responsible for coenzyme recognition are boxed. Red stars represent M. thermoacetica wide-type NADP+-preferred 6PGDH and NAD+-preferred 6PGDH mutant. Blue star indicates T. maritima 6PGDH studied in this research. (b) Subalignments of key amino acid residues playing an important role in 2′–phosphate interaction. Colors in sequence logo refer to hydrophobic (black), positive charge (blue), negative charge (red) and polar (green) residues.
Structural basis of coenzyme preference
To identify key amino acids pertaining to coenzyme preference, a detailed analysis of the coenzyme-6PGDH complexes was performed based on the auto-docking program. The mutant N32D has the replacement of the alkaline asparagine with the acidic aspartate, deleting the former hydrogen bond between the 2′-phosphate of NADP+ and the asparagine residue of the wide type Tm6PGDH (Fig. 2a) and therefore resulting in a decrease in the binding affinity of the mutant N32D towards NADP+. To further enhance coenzyme preference of N32D, the amino acids with large and hydrophobic side chain (leucine and isoleucine) were introduced to the position 33 and 34 to construct the mutant N32D/R33L/T34S, N32D/R33I/T34I. The newly-introduced residues can further disrupt the other two original hydrogen bonds with 2′-phosphate of NADP+. Moreover, their large and hydrophobic side chains display much stronger steric exclusion effect and compress the space used to accommodate the 2′-phosphate group of adenosine monophosphate moiety of NADP+ (Fig. 2b,c). The combined effect of the deletion of original hydrogen bond and the enhanced steric exclusion should lead to a significant decrease in binding with NADP+ for the enzyme.
The representations of enzyme-coenzyme interactions.
(a) Interaction analysis of mutant N32D with NADP+; (b) Interaction analysis of mutant N32D/R33L/T34S with NADP+; (c) Interaction analysis of mutant N32D/R33I/T34I with NADP+; (d) Interaction analysis of mutant N32E/R33I/T34I with NADP+; (e) Interaction analysis of mutant N32D with NAD+; (f) Interaction analysis of mutant N32D/R33L/T34S with NAD+; (g) Interaction analysis of mutant N32D/R33I/T34I with NAD+; (h) Interaction analysis of mutant N32E/R33I/T34I with NAD+. The residues involved in defining coenzyme-specificity are shown as sticks. Hydrogen bonding between residues and cofactor were shown as red line. The distance between the two atoms were shown as yellow line. The figures were made using PyMol.
To better bind NAD+, it is also important to form productive hydrogen bonds. In our case, the distance between the hydrogen atom in 2′-hydroxyl group of NAD+ and the oxygen atom in carboxyl group of Asp32 in the mutant N32D and N32D/R33I/T34I is calculated as 3.15 Å (Fig. 2e,g), which equals to the threshold value of the hydrogen bond formation26. Therefore, their ability to bind NAD+ could be improved slightly. In the mutant N32D/R33L/T34S (Fig. 2f), however, such distance increases to 3.33 Å, which is too long to form a productive hydrogen bond. Consequently, this mutant may have no positive effect on NAD+ binding. Instead, when the residue aspartate at position 32 is mutated to a similar one, glutamate, this distance can be decreased to 2.10 Å (Fig. 2h). In this new mutant N32E/R33I/T34I, a new hydrogen bond is formed, increasing the binding with NAD+.
Enzyme kinetics analysis
According to the alignment result and structure analysis, the plasmids encoding mutants N32D, N32D/R33L/T34S, N32D/R33I/T34I and N32E/R33I/T34I were constructed by site-directed mutagenesis. The protein mutants were overexpressed in E. coli BL21 (DE3) and purified to homogeneity based on SDS-PAGE (data not shown). The kinetic constants on NADP+ and NAD+ of the wild type and mutant 6PGDHs are summarized in Table 1. The mutant N32D has a 460-time higher Km value of 2.3 mM on NADP+ and a slightly decreased Km value of 4.0 mM on NAD+ compared with those of the wide type. The triple-site mutant N32D/R33I/T34I exhibits a far higher Km value of 69.5 mM on NADP+ and a slightly decreased Km value of 3.9 mM on NAD+. Another mutant N32D/R33L/T34S has a Km value on NADP+ of more than 100 mM. Its Km value on NAD+ is also increased to 7.5 mM. However, these three mutants do not show significant differences of kcat on NADP+ with that of the wild-type.
The mutant N32E/R33I/T34I was found to be the best one. As shown in Table 1 and Fig. 3, an almost 2-fold declined Km (from 4.3 mM to 2.5 mM) and a 2-fold increased kcat on NAD+ (from 23.3 to 47.9 s−1) compared to that of wide type can be observed. The catalytic efficiency (kcat/Km) towards NADP+ decreases from 3,520 to 0.2 s−1, while the catalytic efficiency towards NAD+ increases from 5.4 to 19.2 s−1. Overall, the coenzyme catalytic coefficient ratio of NAD+ to NADP+ increases about 6.4 × 104 times for this mutant.
Biobattery tests
The effect of the best mutant (N32E/R33I/T34I) versus the wild type 6PGDH was evaluated electrochemically in an anodic reaction system containing two enzymes: 6PGDH and DI, a coenzyme (NADP+ or NAD+), an electron mediator AQDS, and a 6-phosphogluconate substrate (6PG). Cyclic voltammetry results clearly show that both 6PGDHs produce significant oxidation current peaks at −0.3 V versus Ag/AgCl. The mutant N32E/R33I/T34I exhibits a current density 25% higher than that generated by the wild type (Fig. 4a). The blank control samples including the one without the fuel, without the NAD+, or with denatured 6PGDH did not generate any oxidation current peak. The current densities of 6PG oxidation at various NAD+ concentrations were further evaluated, by plotting peak currents at −0.3 V versus Ag/AgCl against NAD+ concentrations varying from 0 to 2 mM (Fig. 4b). The current density increases along with the increase of the NAD+ concentration, while the mutant generates currents 20–30% higher than those by the wild-type. Both figures suggest that the mutant N32E/R33I/T34I is more effective than the wild-type on using NAD+ as the coenzyme in the electrochemical reaction system.
Electrochemical performances of the wild-type 6PGDH and mutant (N32D/R33I/T34I)-based biobatteries.
(a) Cyclic voltammetry; (b) Peak current from cyclic voltammetry with the mutant or wild-type 6PGDH versus the concentration of NAD+; (c) Power curves of the biobattery at 23 °C; (D) Power of the biobattery equipped with the mutant 6PGDH at a temperature from 23 to 75 °C.
Moreover, both wide-type enzyme and mutant N32E/R33I/T34I were compared in biobatteries in terms of power output using linear sweep voltammetry. The whole-cell biobattery consisted of a carbon nanotube-coated carbon felt anode and a commercial membrane electrode assemble stacked in pile. An increased loading of 6PG, NAD+, AQDS, and 6PGDH was applied in order to get a high power output of the biobattery. The two polarization curves represent that the maximum power density and current density of the biobattery catalyzed by the mutant 6PGDH are 0.135 mW cm−2 and 0.255 mA cm−2 respectively at the room temperature, while for the biobattery catalyzed by the wild type 6PGDH, they are 0.122 mW cm−2 and 0.200 mA cm−2 (Fig. 4c). At elevated temperatures, the mutant 6PGDH-powered biobattery significantly increased power outputs of 0.61 mW cm−2 at 37 °C, 1.58 mW cm−2 at 50 °C, 1.75 mW cm−2 at 65 °C, and 1.58 mW cm−2 at 75 °C (Fig. 4d). It is noted that the biobattery has an optimal power density at 65 °C, probably due to the optimal enzyme kinetics and the increased mass transfer at this temperature. Too high temperature caused the deactivation of diaphorase, resulting in decreased power outputs in the biobattery.
Discussion
Most NAD(P)+-dependent dehydrogenases have the Rossmann fold, whose loop region contains three conservative amino acids. Alkaline amino acids, such as asparagine and arginine, in the first two positions play a critical role in NADP+-preferred 6PGDHs through the formation of stable hydrogen bonds with 2′-phosphate of NADP+ 27–29. In contrast, NAD+-preferred 6PGDH enzymes have acidic aspartate residues in the N-terminal end of loop region (Fig. 1b). The substrate docking result (Fig. 2) indicates that the formation of a new hydrogen bond with NAD+ through the replacement of alkaline amino acid by acidic one is favorable to increase the enzyme activity on NAD+, and even reverse the coenzyme preference from NADP+ to NAD+. Moreover, our previous research on the directed evolution of Moorella thermoacetica 6PGDH also demonstrated that the introduction of an acidic amino acid, aspartate, in the loop of the Rossmann fold to form a new hydrogen bond with 2′-hydroxyl group of NAD+ resulted in a 4,300-fold reversal of coenzyme selectivity from NADP+ to NAD+ 30. Reports on many other dehydrogenases also confirmed the positive effect of the formation of new hydrogen bond in reversing the coenzyme preference21,24,30,31,32,33,34.
Our results of kinetic constants of the wild-type enzyme and the mutant N32E/R33I/T43I are in good agreement with the structure analysis. For the mutant N32E/R33I/T43I, the coenzyme binding ability of 6PGDH on NADP+ is decreased greatly, because the mutant loses its original three hydrogen bonds with the 2′-phosphate of NADP+ and suffers from a strong steric exclusion effect from the hydrophobic side chain of two newly introduced isoleucines (Fig. 2d). Those significant structural changes result in a 15,800-fold increase in Km on NADP+ and a 17,600-fold decline in catalytic efficiency (kcat/Km) on NADP+ (Table 1). While on NAD+, the introduced acidic residue, glutamate, improves the binding of coenzyme due to the formation of the new productive hydrogen bond between Glu32 and 2′-hydroxyl group of NAD+. A 2-fold decline in Km and a 4-fold increase in catalytic efficiency on NAD+ compared to wide type Tm6PGDH are observed (Table 1). As a result, the overall coenzyme catalytic coefficient ratio of NAD+ to NADP+ of this mutant increases about 6.4 × 104 times.
As a thermostable enzyme with a half-life time of 140 h at 80 °C, 6PGDH from T. maritima has great potential in biobattaries. Firstly, it has a prolonged lifetime of biobattaries due to high stability of the enzymes35. Secondly, it allows to run biobattaries at elevated temperatures or tolerate harsh conditions, such as in deserts or extremely hot conditions. Third, high-temperature biobattaries would further generate high power density because of increased enzyme activity8,10 and mass transfer36. To our limited knowledge, this present biobattery is the one running on the second highest temperature among all the EFCs reported. The highest temperature of EFC was 80 °C by using a thermostable alcohol dehydrogenase but its current density was only 0.21 mA cm−2 (5% of this study)37.
In conclusion, this work reports a coenzyme specificity change of Tm6PGDH through rational design with its application to biobatteries. Via enzyme sequence alignment and structure analysis of the coenzyme-enzyme complexes, the key amino acid residues that are responsible for binding coenzymes were identified. The coenzyme preference was reversed from NADP+ to NAD+ through site-directed mutagenesis. The optimal temperature of the biobattary with the mutant was as high as 65 °C, resulting in a high power density of 1.75 mW cm−2.
Methods and Methods
Chemicals, Bacterial Strains and Medium
All chemicals were reagent grade or higher, purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA), unless otherwise noted. E. coli Top10 was used for general molecular cloning and E. coli BL21 (DE3) was used for recombinant protein expression. All enzymes for molecular biology experiments were purchased from New England Biolabs (NEB, Ipswich, MA). Strains, plasmids, and oligonucleotides primers used in this study are listed in Table 2.
Cetyltrimethylammonium bromide (CTAB) and glutaraldehyde (GA) were purchased from Sigma-Aldrich (Saint Louis, MO). 9,10-anthraquinone-2,7-disulphonic acid (AQDS) was purchased from Pfaltz & Bauer (Waterbury, CT). Carbon felt (C100 AvCarb) was purchased from Fuel Cell Earth (Woburn, MA). COOH-functionalized multi-wall carbon nanotubes (CNTs) with an outer diameter of <8 nm and a length of 10–30 μm were purchased from CheapTubes.com (Brattleboro, VA). Membrane electrode assemblies (MEAs) consisting of the Nafion 212 membrane and a carbon cloth cathode modified with 0.5 mg cm−2 Pt were purchased from Fuel Cell Store (San Diego, CA). The glassy carbon (GC) working electrode (3 mm diameter), Ag/AgCl reference electrode, Pt wire counter electrode were bought from BASi (West Lafayette, IN).
Site directed mutagenesis
The Tm6PGDH gene was amplified from plasmid10 by a primer pair of 6PGDH_IF and 6PGDH_IR. pET20b vector backbone was amplified with a primer pair of 6PGDH_VF and 6PGDH_VR. Plasmid pET20b-6pgdh based on the two DNA fragments was obtained using a Simple Cloning method38.
The QuickChangeTM site-directed mutagenesis method (Stratagene, La Jolla, CA) was used to introduce point mutations into the Tm6PGDH sequence according to the protocol of the NEB Phusion site-directed mutagenesis kit. The mutant N32D, N32D/R33I/T34I, N32D/R33L/T34S, and N32E/R33I/T34I were prepared by their respective primer pairs (Table 2). PCR reaction solution (50 μL) containing 1 ng of plasmid template (pET20b-6pgdh) was conducted as follows: 98 °C denaturation for 1 min; 20 cycles of 98 °C denaturation for 30 s, 60 °C annealing for 30 s and 72 °C extension for 2.5 min; and 72 °C extension for 5 min. The PCR product was digested by DpnI followed by the purification of gel electrophoresis using a Zymoclean™ Gel DNA Recovery Kit (Zymo Research, Irvine, CA) and then transformed into E. coli BL21 (DE3).
Overexpression and purification of wide-type Tm6PGDH and mutants
The transformants were grown overnight at 37 °C in Luria-Bertani medium plates containing 100 μg mL−1 ampicillin. For liquid culture, once A600 of the cultured cell reached ~0.8, 0.2 mM IPTG was added to induce protein expression at 37 °C for 6 h. Cell pellets were harvested by centrifugation and then were re-suspended in a 20 mM sodium phosphate and 0.3 M NaCl buffer (pH 7.5) containing 10 mM imidazole. After sonication and centrifugation, the His-tagged protein in the supernatant was loaded onto the column packed with HisPur Ni-NTA Resin (Fisher Scientific, Pittsburgh, PA) and eluted with 20 mM sodium phosphate buffer (pH 7.5) containing 300 mM NaCl and 250 mM imidazole. Mass concentration of protein was determined by the Bradford assay using bovine serum albumin (BSA) as the standard.
Tm6PGDH activity assay
The activities of wide type Tm6PGDH and mutants were measured in 100 mM HEPES buffer (pH 7.5) with the final concentration of 2 mM 6-phosphogluconate (6PG), 2 mM NAD(P)+, 5 mM MgCl2 and 0.5 mM MnCl2 at 70 °C for 5 min. The formation of NAD(P)H was measured at 340 nm by a UV/visible spectrophotometer (Beckman Coulter, Fullerton, CA). The enzyme unit was defined as one μmole of NAD(P)H produced per min. To determine enzyme kinetic parameters on coenzymes, the enzyme activity was measured in the same buffer above except changing the concentration of NAD(P)+ (2 μM to 10 mM). The result was regressed by GraphPad Prism 5 (Graphpad Software Inc., La Jolla, CA) and the apparent Km and kcat for NAD(P)+ of wide type and mutant Tm6PGDH were given based on Michaelis-Menten nonlinear regression. All the reactions contained three independent triplicates and the data were fitted within the linear range.
Electrochemical measurement
All electrochemical measurements were conducted using a 600D Potentiostat from CH Instruments Inc. (Austin, TX) interfaced to a PC. The reaction solution was flushed by argon gas for 20 minutes before each measurement. Each measurement was conducted in triplicate.
Cyclic voltammetry was performed in the anodic compartment at a scan rate of 20 mV s−1 using a 3-electrode system with a GC working electrode, an Ag/AgCl reference electrode and a Pt wire counter electrode. The anolyte contained up to 2 mM NAD+, 5 mM 6PG, 2.5 mM AQDS, 100 mM HEPES (pH 7.3), 10 mM MgCl2, 0.5 mM MnCl2, 100 mM NaCl, 0.0133 mg mL−1 of mutated or wild type Tm6PGDH, and 0.015 mg mL−1 of Geobacillus stearothermophilus diaphorase (DI).
Linear sweep voltammetry was performed at a scan rate of 5 mV s−1 in a stacked fuel cell configuration. A 1-cm2 carbon felt (CF) was coated with 0.25 mL of CNTs solution (CNTs: CTAB: H2O = 1 mg: 1 mL: 1 mL) and 30 μL of GA, dried at the room temperature, and used as the anode. An air-breathing carbon cathode was coated with 0.5 mg cm−2 Pt. A Nafion 212 membrane was used to separate two electrodes. The anolyte contained increased loadings of 8 mM NAD+, 40 mM 6PG, 16.7 mM AQDS, 0.05 mg mL−1 of mutated or wild type Tm6PGDH, and 0.06 mg mL−1 of DI. The performance of the biobattery with mutated Tm6PGDH was further evaluated at increased temperatures up to 75 °C.
Sequence alignment
Manually annotated and reviewed sequence data for 6PGDH were retrieved from the National Center for Biotechnology Information and Uniprot Database39. Clustal Omega40,41 was used to perform a multiple sequence alignment. The sequence logo plot42 was created with the WebLogo 3.3 interface43.
Structure analysis
The three-dimensional (3D) structure modeling of the wild-type Tm6PGDH and mutants were built by SWISS-MODEL based on the Lactococcus lactis 6PGDH (PDB: 2IYP) as a template (sequence identity: 46.5%). The final model was validated using the PROCHECK program44. The structures of NADP+ and NAD+ were built by using Chemdraw (PerkinElmer, Waltham, MA). The conformation space of the corresponding coenzyme binding area was analyzed using the Autodock program (Scripps Research Institute, La Jolla, CA).
Additional Information
How to cite this article: Chen, H. et al. Coenzyme Engineering of a Hyperthermophilic 6-phosphogluconate Dehydrogenase from NADP+ to NAD+ with Its Application to Biobatteries. Sci. Rep. 6, 36311; doi: 10.1038/srep36311 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Sakai, H. et al. A high-power glucose/oxygen biofuel cell operating under quiescent conditions. Energy Environ. Sci. 2, 133–138 (2009).
Zebda, A. et al. Mediatorless high-power glucose biofuel cells based on compressed carbon nanotube-enzyme electrodes. Nat. Commun. 2, 370 (2011).
Zhu, Z.-G., Kin Tam, T., Sun, F., You, C. & Zhang, Y.-H. P. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway. Nat. Commun. 5, 3026 (2014).
Cooney, M. J., Svoboda, V., Lau, C., Martin, G. & Minteer, S. D. Enzyme catalysed biofuel cells. Energy Environ. Sci. 1, 320–337 (2008).
Calabrese Barton, S., Gallaway, J. & Atanassov, P. Enzymatic biofuel bells for implantable and microscale devices. Chem. Rev. 104, 4867–4886 (2004).
Minteer, S. D., Liaw, B. Y. & Cooney, M. J. Enzyme-based biofuel cells. Curr. Opin. Biotechnol. 18, 228–234 (2007).
Okuda-Shimazaki, J., Kakehi, N., Yamazaki, T., Tomiyama, M. & Sode, K. Biofuel cell system employing thermostable glucose dehydrogenase. Biotechnol. Lett. 30, 1753–1758 (2008).
Zhu, Z., Sun, F., Zhang, X. & Zhang, Y.-H. P. Deep oxidation of glucose in enzymatic fuel cells through a synthetic enzymatic pathway containing a cascade of two thermostable dehydrogenases. Biosens. Bioelectron. 36, 110–115 (2012).
Suraniti, E., Tsujimura, S., Durand, F. & Mano, N. Thermophilic biocathode with bilirubin oxidase from Bacillus pumilus. Electrochem. Commun. 26, 41–44 (2013).
Wang, Y. & Zhang, Y. P. Overexpression and simple purification of the Thermotoga maritima 6-phosphogluconate dehydrogenase in Escherichia coli and its application for NADPH regeneration. Microb. Cell Fact. 8, 1 (2009).
Bornscheuer, U. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Davids, T., Schmidt, M., Böttcher, D. & Bornscheuer, U. T. Strategies for the discovery and engineering of enzymes for biocatalysis. Curr. Opin. Chem. Biol. 17, 215–220 (2013).
Güven, G., Prodanovic, R. & Schwaneberg, U. Protein Engineering – An Option for Enzymatic Biofuel Cell Design. Electroanal 22, 765–775 (2010).
Wong, T. S. & Schwaneberg, U. Protein engineering in bioelectrocatalysis. Curr. Opin. Biotechnol. 14, 590–596 (2003).
Rollin, J. A., Tam, T. K. & Zhang, Y.-H. P. New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green. Chem. 15, 1708–1719 (2013).
Woodyer, R., van der Donk, W. A. & Zhao, H. Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry 42, 11604–11614 (2003).
Wu, J. T., Wu, L. H. & Knight, J. A. Stability of NADPH: effect of various factors on the kinetics of degradation. Clin. Chem. 32, 314–319 (1986).
Banta, S., Swanson, B. A., Wu, S., Jarnagin, A. & Anderson, S. Alteration of the specificity of the cofactor-binding pocket of Corynebacterium 2, 5-diketo-D-gluconic acid reductase A. Protein. Eng. 15, 131–140 (2002).
Wong, C.-H. & Whitesides, G. M. Enzyme-catalyzed organic synthesis: NAD (P) H cofactor regeneration by using glucose-6-phosphate and the glucose-5-phosphate dehydrogenase from Leuconostoc mesenteroides. J. Am. Chem. Soc. 103, 4890–4899 (1981).
Zhang, Y.-H. P., Myung, S., You, C., Zhu, Z. & Rollin, J. A. Toward low-cost biomanufacturing through in vitro synthetic biology: bottom-up design. J. Mater. Chem. 21, 18877–18886 (2011).
Brinkmann-Chen, S. et al. General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH. Proc. Natl. Acad. Sci. USA. 110, 10946–10951 (2013).
Lerchner, A., Jarasch, A., Meining, W., Schiefner, A. & Skerra, A. Crystallographic analysis and structure‐guided engineering of NADPH‐dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity. Biotechnol. Bioeng. 110, 2803–2814 (2013).
Katzberg, M., Skorupa-Parachin, N., Gorwa-Grauslund, M.-F. & Bertau, M. Engineering cofactor preference of ketone reducing biocatalysts: a mutagenesis study on a γ-diketone reductase from the yeast Saccharomyces cerevisiae serving as an example. Int. J. Mol. Sci 11, 1735–1758 (2010).
Meng, H. et al. Engineering a d-lactate dehydrogenase that can super-efficiently utilize NADPH and NADH as cofactors. Sci. Rep. 6 (2016).
Campbell, E., Meredith, M., Minteer, S. D. & Banta, S. Enzymatic biofuel cells utilizing a biomimetic cofactor. Chem. Commun. 48, 1898–1900 (2012).
Tuomi, W. V. & Kazlauskas, R. J. Molecular basis for enantioselectivity of lipase from Pseudomonas cepacia toward primary alcohols. Modeling, kinetics, and chemical modification of Tyr29 to increase or decrease enantioselectivity. J.Org.Chem. 64, 2638–2647 (1999).
Tetaud, E. et al. 6-Phosphogluconate dehydrogenase from Lactococcus lactis: a role for arginine residues in binding substrate and coenzyme. Biochem. J. 338, 55–60 (1999).
Chen, Y.-Y. et al. Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism. J. Struct. Biol. 169, 25–35 (2010).
Li, L. & Cook, P. F. The 2′-phosphate of NADP is responsible for proper orientation of the nicotinamide ring in the oxidative decarboxylation reaction catalyzed by sheep liver 6-phosphogluconate dehydrogenase. J. Biol. Chem. 281, 36803–36810 (2006).
Huang, R., Chen, H., Zhong, C., Kim, J.-E. & Zhang, Y. P. High-throughput screening of coenzyme preference change of thermophilic 6-phosphogluconate dehydrogenase from NADP+ to NAD+. Sci. Rep. 6, 32644 (2016).
Sharkey, M. A., Oliveira, T. F., Engel, P. C. & Khan, A. R. Structure of NADP+‐dependent glutamate dehydrogenase from Escherichia coli–reflections on the basis of coenzyme specificity in the family of glutamate dehydrogenases. FEBS J. 280, 4681–4692 (2013).
Watanabe, S., Kodaki, T. & Makino, K. Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J. Biol. Chem 280, 10340–10349 (2005).
Brinkmann-Chen, S., Cahn, J. & Arnold, F. H. Uncovering rare NADH-preferring ketol-acid reductoisomerases. Metab. Eng. 26, 17–22 (2014).
Bastian, S. et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13, 345–352 (2011).
Wang, Y., Huang, W., Sathitsuksanoh, N., Zhu, Z. & Zhang, Y.-H. P. Biohydrogenation from biomass sugar mediated by in vitro synthetic enzymatic pathways. Chem. Biol. 18, 372–380 (2011).
Fogler, H. S. Elements of chemical reaction engineering (1999).
Kontani, A. et al. A bioanode using thermostable alcohol dehydrogenase for an ethanol biofuel cell operating at high temperatures. Electroanalysis 26, 682–686 (2014).
You, C., Zhang, X.-Z. & Zhang, Y.-H. P. Simple cloning via direct transformation of PCR product (DNA Multimer) to Escherichia coli and Bacillus subtilis. Appl. Environ. Microbiol. 78, 1593–1595 (2012).
Consortium, U. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, 1–5 (2012).
Goujon, M. et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38, W695–W699 (2010).
Sievers, F. et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr 26, 283–291 (1993).
Acknowledgements
This project cannot be carried out without support of the Biological System Engineering Department, Virginia Polytechnic Institute and State University, Virginia. It was mainly funded by DOE EERE award (DE-EE0006968) and partially supported by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture.
Author information
Authors and Affiliations
Contributions
P.Z., H.C. and Z.Z. wrote the main manuscript text, tables and figures. H.C. conducted major experiments which including sequence alignment, structure analysis, mutants construction, purification and activity assay of wide type and mutant Tm6PGDH. Z.Z. conducted experiments of electrochemical measurement in Figure 4. R.H. was involved project discussion and his previous research provided important reference to this project. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, H., Zhu, Z., Huang, R. et al. Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP+ to NAD+ with Its Application to Biobatteries. Sci Rep 6, 36311 (2016). https://doi.org/10.1038/srep36311
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36311
This article is cited by
-
Bio-inspired Green Power: A Thermocurrent Generator
Transactions on Electrical and Electronic Materials (2021)
-
Biochemical and structural insights into 6-phosphogluconate dehydrogenase from Leishmania donovani
Applied Microbiology and Biotechnology (2021)
-
Engineering a diaphorase via directed evolution for enzymatic biofuel cell application
Bioresources and Bioprocessing (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.