Abstract
Litchi chinensis is a subtropical fruit crop, popular for its nutritional value and taste. Fruits with small seed size and thick aril are desirable in litchi. To gain molecular insight into gene expression that leads to the reduction in the size of seed in Litchi chinensis, transcriptomes of two genetically closely related genotypes, with contrasting seed size were compared in developing ovules. The cDNA library constructed from early developmental stages of ovules (0, 6, and 14 days after anthesis) of bold- and small-seeded litchi genotypes yielded 303,778,968 high quality paired-end reads. These were de-novo assembled into 1,19,939 transcripts with an average length of 865 bp. A total of 10,186 transcripts with contrast in expression were identified in developing ovules between the small- and large- seeded genotypes. A majority of these differences were present in ovules before anthesis, thus suggesting the role of maternal factors in seed development. A number of transcripts indicative of metabolic stress, expressed at higher level in the small seeded genotype. Several differentially expressed transcripts identified in such ovules showed homology with Arabidopsis genes associated with different stages of ovule development and embryogenesis.
Similar content being viewed by others
Introduction
Litchi chinensis Sonn. is a subtropical evergreen fruit tree of family Sapindaceae and sub family Nepheleae. Sapindaceae family comprises of about 150 genera and 2,000 species. The genus Litchi has three subspecies: Litchi chinensis ssp. chinensis, philippinensis and javensis. The subspecies chinensis bears arillate fruit, unlike long thorny protuberances with inedible flesh in ssp philippinensisis and thinner aril in ssp javensis. The genus originated in China, where it has been cultivated for more than 2,300 years.
Three types of flowers are found in litchi, type I staminate flowers, type II hermaphrodite female flowers and type III hermaphrodite flowers. Litchi fruits develop from bicarpellary ovary of type II hermaphrodite female flowers. Generally, only one carpel of type 2 flower develops into fruit. Each ovary contains an anatropous ovule, which develops into a seed. The edible part of the fruit, called aril, develops from the outer integument of ovule1. Botanically, the fruit is a drupe. Aril is sweet in taste, rich in calories, vitamin C, and is a good source of minerals2.
In angiosperms, seed development begins with double fertilization followed by the formation of embryo and endosperm. Endosperm development progresses through four phases: syncytial, cellularization, differentiation, and death3. In most dicots, initially the endosperm grows rapidly and is consumed at later stages. Thus, the cotyledon occupies most of the portion in a mature seed. Coordination in the growth of endosperm and embryo is instrumental in determining seed size in a fruit4. The size of seed is also limited by the expansion space permitted by seed coat. Some genes have been identified to play role in determining seed size in Arabidopsis thaliana5,6. However, no such studies have been reported on litchi.
Next generation sequencing technologies (NGS) and differential expression analyses have accelerated developmental studies in plants through the investigation of transcriptomes. In horticultural crops, only in a few cases, transcriptome sequence analysis has been used for the prediction of genes related to fruit development- as in case of Annona squamosa, Musa accuminata, Prunusavium, Pyrusbrets chneideri etc7,8,9,10. Comparative analysis of transcriptome in litchi has been used to speculate on the processes involved in the regulation of floral initiation by phytohormones, expression of flowering related genes11, genes related to shading stress12, fruit cracking13, reactive oxygen species (ROS) induced abortion of rudimentary leaves14, fruit abscission15, maturation, coloration16, abscission induced by carbohydrate stress17 and fruit ripening after cold storage18. To the best of our knowledge, there is no report on transcription profiling of early stage developing ovules in litchi.
Litchi seeds are very hard and bold, therefore not favourable to fruit juice processing industry and fresh fruit consumers. Small seed size or seedlessness is desirable in litchi, also because small-seeded fruits develop more pulp as compared to the bold-seeded genotypes19. Molecular characterization of seed development in ovule in genotypes with contrasting seed size may give leads to control seed development in litchi. In this study, we compared the transcriptional profiles in early-stage developing ovules of two litchi genotypes with contrast in seed size. The transcriptomes were assembled, annotated and analyzed for differential expression.
Results and Discussion
Sequence assembly and annotation
Transcriptome sequencing of litchi (bold and small seeded) ovule-specific libraries at 0, 6 and 14 DAA yielded 303,778,968 high-quality paired-end reads of average 101 nucleotides length. The reads were assembled into 1,19,939 transcripts (Trinity transcripts), with an average length of 865.38, median length of 489 and N50 length of 1,487. The 1,19,939 transcripts represented 87,072 unique components (unigenes) and their isoforms (Supplementary Table S1). Our study records the highest number of transcripts in litchi, reported till date.
The assembled transcripts were compared against the public databases (E-value ≤ 10−5). Out of the 1,19,939 transcripts, 48,968 (40.82%), 36,570 (30.49%) and 16,140 (13.45%) showed homology with the sequences in NCBI non-redundant (NR), Swiss-Prot and COG(Clusters of Orthologous Groups) databases, respectively. Comparative analysis was done with a few closely related species (Citrus sinensis, Populus trichocarpa, Ricinus communis, Glycine max, Fragaria vesca, Carica papaya and Vitis vinifera) and the model plant Arabidopsis thaliana. Highest homology was noticed with transcripts from Fragaria vesca (39.31%), followed by Citrus sinensis (38.90%), Populus trichocarpa (38.73%), Ricinus communis (38.25%) and Vitis vinifera (38.10%) (Supplementary Table S2). A total of 68,988 (57.51%) transcripts, including their possible splice variants, showed no homology (at an E-value of 10−5) with any of the eight species, NR and Swiss-Prot databases examined in the study. These transcripts could be specific to Litchi chinensis.
Functional categorization
The assembled data was classified into 25 groups by using the COG tool (Supplementary Fig. S1). This groups the sequences on the basis of evolutionary relationship among bacteria, algae and eukaryotes and provides a useful platform for protein classification of unsequenced organisms20. The analysis suggested that ovule development in litchi involved transcriptional, post transcriptional and metabolism related regulation. Further analysis was done by Gene Ontology (GO) tool that classifies genes by their molecular functions, biological processes in which they are involved, and the cellular location of the gene products. The 50,748 unique transcripts, showing best hits (E-value ≤ 10−5) in the aforementioned public databases, were examined with the GO tool. The transcripts, were categorized in 6,333 GO terms into biological processes, cellular components and molecular functions (Supplementary Fig. S2). In biological process category, the major representation was for metabolic processes (82.18%), cellular process (65.99%) and cellular metabolic processes (53.87%). Catalytic activity (69.30%) represented the most abundant sub-category in molecular function.
Differential gene expression in developing ovules between bold- and small-seeded litchi genotypes
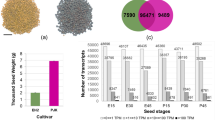
The expression patterns of transcripts between the early-stage ovules of bold (HC)- and small (HS)-seeded genotypes were investigated. The highest number of differentially expressed transcripts was noticed at 0DAA. A total of 7,946 transcripts expressed differentially (log2 fold change ≥ 2; P value ≤ 0.001) at 0DAA. Pair wise comparison of the developing ovules revealed common and exclusive differentially expressed transcripts at 0, 6 and 14 DAA between the two genotypes (Fig. 1A) (Supplementary Data 1 and 2). The 6,259 differentially expressed transcripts were exclusive to 0 DAA (Fig. 1A), suggesting a dominant role of maternal factors in the regulation of seed size in litchi. The maternal genes determine the development of seed coat exclusively, while also function in co-ordination with paternal genes in the development of endosperm and embryo. The role of maternal tissues in determining seed size has been emphasized earlier5,6,21.
Putative hormone related genes
The endogenous levels of different plant hormones such as auxins, gibberellins, cytokinins, abscisic acid, ethylene, and brassinosteroids play important roles in seed and fruit development. The BLASTx searches revealed the expression of a total of 2,331 trinity transcripts showing homology with hormone related genes in the developing ovules. The maximum representation was of the transcripts related to abscisic acid pathway followed by auxin in the ovule transcriptome data. A total of 430, 50 and 137 transcripts were differentially expressed (HC vs HS) at 0, 6 and 14 DAA, respectively. The transcripts related to auxin and brassinosteroid biosynthesis were down-regulated in small-seeded genotype at 0, 6 and14 DAA (Fig. 2A–C, Supplementary Data 3). The biosynthesis and transportation of auxin play important roles in cell division, differentiation, expansion, and determining apical to basal patterning of gynoecium22,23. Auxin accumulates at the tip of primordia from where it is transported to sub-epidermal layers through vascular route23. Majority of auxin transport related transcripts were down regulated at 0 and 6 DAA in the small-seeded genotype (Fig. 2A,B). Down-regulation of auxin transport in embryo sac may affect megagametogenesis and embryo patterning24. The putative transcripts for auxin efflux carrier family proteins such as PIN1 (AT1G73590; c40956_g1_i1&c45750_g1_i1), PIN6 (AT1G77110; c44908_g1_i1) and EIR1 (AT5G57090; c44908_g2_i2), involved in the maintenance of auxin gradient in embryo sac24, were down-regulated in the developing ovules of small-seeded litchi (Fig. 2A). The auxin transporter LAX2 (AT2G21050; c42412_g1_i1) regulates vascular development25. It was repressed in small-seeded ovules (Supplementary Table S3). Down-regulation of auxin transport in ovule may hamper megagametogenesis and embryo patterning24, and sometimes may induce parthenocarpy26. The suppression of gene expression for auxin transport could hamper proper seed development in litchi. Although we could not detect any transcript homologous to cytokinin biosynthesis pathway genes, in developing ovule of small-seeded litchi at 0 DAA, the candidate genes for cytokinin transport were up regulated. The higher level of cytokinin transport could make ovular tissues more sensitive to various stresses27. Reproductive tissues of flowering plants are sensitive to different stresses, which may influence ovule development28. Several transcripts which showed no expression in the ovule of bold-seeded genotype were significantly expressed in small-seeded litchi and were homologous to stress related genes at 0 DAA (Supplementary Table S4). This suggests that the small-seeded ovule experiences a metabolic state of stress, which subsequently hampers seed development.
Differentially expressed (log2 fold ≥ 2; P-value ≤ 0.001) hormone-related genes at early ovule developmental stages of bold-seeded (HC) vs small-seeded (HS) litchi genotypes.
Log2 (FPKM + 1) of putative auxin, brassinosteroid biosynthesis and auxin transport transcripts at (A) 0 DAA, (B) 6 DAA and (C) 14 DAA are shown.
Brassinosteroids play important role in initiation and formation of reproductive organs. These regulate stress tolerance, stomatal development and vascular differentiation29,30,31,32. Their role has been discussed in determining seed size by regulating endosperm growth and maternal imprinting33. Sterol methyltransferase 2 (c4669_g1_i1_AT1G20330) is involved in sterol biosynthesis and the loss of function leads to defective cotyledon growth34. DWARF1 (AT3G19820; c39853_g1_i1) is involved in the conversion of brassinosteroid precursor 24-methylenecholesterol to campesterol, and the loss of function induces dwarf phenotype, sterility and seed abortion35. We found relatively repressed expression of the putative genes for DWARF1 and sterol methyltransferase 2 in small-seeded litchi at 0DAA (Fig. 2A). The results agree with the up- and down-regulation of transcripts at 0 DAA for candidate genes identified as negative (IKU1_AT2G35230; c50429_g1_i1) and positive (KLU_ AT1G13710; c46160_g1_i3 & c46160_g1_i1 and ANT_AT4G37750; c46773_g2_i3) regulators of seed size in Arabidopsis5,36 (Supplementary Table S3). Putative transcript for brassinosteriod deactivation, sulphotransferase 1237 expressed at higher level at 14 DAA in the small-seeded genotype.
Putative transcription factor genes
Transcription factors (TFs) are key regulators of spatial, temporal and quantitative gene expression. The BLASTx analysis revealed a total of 2,155 trinity transcripts showing homology with TFs involved in early-stage ovule development in bold- and small-seeded litchi. The MYB family TFs were most abundant in the ovule transcriptome data of litchi. A total of 426, 33 and 125 TF related transcripts were differentially expressed (bold seeded vs small seeded) at 0, 6 and 14 DAA, respectively.
The expression of members of YABBY, WRKY, LSD, HD-ZIP and EIL family TFs was up-regulated in small-seeded litchi at 0 DAA (Fig. 3A, Supplementary Data 4). The YABBY family regulates polar differentiation of abaxial side of lateral organs, playing important role in organ morphogenesis38,39. Out of six YABBY genes known in Arabidopsis genome, four (FIL, YAB2, YAB3 and YAB5) are expressed in leaf and leaf derived organs, and two (INO and CRC) are restricted to floral organs39. The putative genes for FIL (c35059_g1_i1, c35059_g2_i1, c38598_g1_i1&c38598_g1_i2; AT2G45190), YAB2 (c36528_g2_i1_AT1G08465), YAB5 (c32907_g1_i1_AT2G26580), and CRC (c30105_g1_i1_AT1G69180) showed higher expression in the ovules of small-seeded genotype (Fig. 3A). CRC is expressed in placental tissue for proper carpel identity and nectar development39,40. In contrast, putative INO (c42867_g1_i1_AT1G23420) was down-regulated in the ovules of small-seeded genotypes (Fig. 3B). INO plays important role in the development of outer integuments, and its down-regulation leads to seedless phenotype in Arabidopsis41 and Annona squamosa42.
Differentially expressed (log2 fold ≥ 2; P-value ≤ 0.001) putative transcription factor genes in early ovule developmental stages of bold-seeded (HC) vs small-seeded (HS) litchi genotypes.
Log2 (FPKM + 1) of the genes (A) upregulated in small-seeded litchi genotype at 0DAA; (B) down-regulated in small-seeded litchi genotype at 0 DAA; (C) NAC and MYB transcription factors at 6 DAA and (D) AP2 family transcription factors at 14 DAA.
WRKY family of TF are involved in various stress responses and plant developmental processes43. Putative transcripts of LSD one like 1 gene (c44752_g1_i2 &c44752_g1_i3; AT1G32540) was up-regulated in small-seeded genotype (Fig. 3A). LSD one like 1 is known to cause stress induced cell death in Arabidopsis thaliana44. HD-ZIP is involved in various developmental processes in plants45. HB-5, a member of HD-ZIP (c35680_g1_i1_AT4G36740), induced during ovule abortion46, was highly expressed in small-seeded genotype. EIL family members regulate various genes involved in developmental processes and stress response47. Putative transcripts for ethylene-insensitive3-like3 (c46629_g2_i1&c46629_g1_i1; AT1G73730) and ethylene insensitive 3 (c49072_g1_i1 & c50367_g2_i1; AT3G20770) involved in ethylene signaling were up-regulated in small-seeded genotype (Fig. 3A). In Arabidopsis thaliana, ethylene insensitive 3 is potent to activate ethylene pathway in absence of ethylene48. Ethylene responsive genes are found to be up-regulated in stress induced ovule abortion47.
The TF families, ARF, CPP, Dof, E2F and GRF, were down regulated in small-seeded genotype at 0DAA (Fig. 3B, Supplementary Data 4). ARFs activate or repress genes in response to auxins49 CPP family plays important role in regulating development of reproductive tissues and cell division50. TSO 1 (c45960_g1_i1, c45960_g1_i2, c45960_g1_i3, c49343_g1_i1, c50381_g2_i1 & c50381_g2_i4; AT3G22780), TSO1-like CXC2 (c50381_g2_i2, c50381_g2_i3, c50381_g2_i5, c50381_g2_i6 & c51075_g1_i4; AT4G14770) and TSO1-like CXC domain containing proteins (c50381_g2_i7_AT3G04850) of CPP family were down regulated in small-seeded genotype (Fig. 3B). TSO 1 and TSO1-like CXC are essential for development of reproductive tissue and cell cycle control51,52. E2F proteins regulate transcription of genes expressed in a quiescent stage53. Putative transcripts of E2F transcription factor family E2F3 (c47924_g2_i1&c47924_g2_i2; AT2G36010), E2F-like2 (c49929_g1_i11& c49929_g1_i12; AT3G01330), E2F-like3 (c44582_g1_i1_ AT3G48160) and E2F1 (c47924_g3_i1&c47924_g3_i3; AT5G22220) are key components of cell cycle regulation54. These were down-regulated in small-seeded genotype (Fig. 3B). GRF family members regulate cell expansion and its mutants develop smaller cotyledon size55. The results indicate up-regulation of several stress related TFs (Fig. 3A) in the ovules of small-seeded genotype.
The candidate genes for NAC TFs were up-regulated in small-seeded litchi at 6 DAA (Fig. 3C, Supplementary Data 4). NAC transcription factors participate in the regulation of seed development and stress responses56,57. The putative transcripts for MYB family of TF were down-regulated in small-seeded genotype (Fig. 3C). MYB transcription factors regulate biosynthesis of flavonoids in grapes, required for proper pollination and embryo development in plants58. The A-class homeotic gene APETALA2 (AP2), a floral homeotic TF, is known to negatively regulate seed size but promote integument growth59. AP2 was up-regulated in small-seeded genotype at 14 DAA (Fig. 3D). This is in agreement with the development of fruits with under developed seed and well-developed arils of integument origin (Fig. 4), in the small seeded litchi.
Ovule identity determining genes
Based on homology with the genes that have been attributed role in ovule development and embryogenesis in Arabidopsis thaliana, we mapped the transcripts, expressed differentially between the bold- and small seeded genotypes, on a model ovule developmental pathway. Higher expression of NGATHA (AT2G46870; c41974_g2_i1 & c41974_g4_i2), BEL1-like (AT2G35940; c45509_g1_i1, c45509_g1_i2, c45509_g1_i3, c39557_g3_i1 & c39557_g3_i2), HB-5 like (AT4G36740; c35680_g1_i1), LOB domain containing protein (AT2G28500; c36314_g1_i1) and HB-7 like gene (AT2G46680; c44555_g1_i1 & c44555_g1_i2) was noticed in the small-seeded litchi at 0DAA (Fig. 2A, Supplementary Table S3). BEL1-like homeodomain 1 protein controls ovule patterning through auxin signaling60. Its ectopic expression in embryo sac leads to defects in nuclear migration, cellularization and embryogenesis61. Ectopic expression of LOB domain containing proteins in flowering tissues causes alteration of flower size and shape and may lead to seed abortion62. The Arabidopsis mutants, lacking differentiated gametophyte, exhibit higher level of expression of HB-763. NGATHA regulates auxin-mediated development of female reproductive structures in plants. Its overexpression has been reported to reduce organ growth e.g. leaves, flowers and cotyledons64.
Cell cycle is tightly co-regulated with cell patterning, and morphogenesis65. The candidate genes involved in the cell cycle regulation, cyclin D1 (AT1G70210; c47767_g1_i1 & c47767_g3_i1), cyclin A3 (AT1G47230; c46681_g1_i1) cyclin D4 (AT5G65420; c47767_g1_i2), dUTP pyrophosphatase (AT3G46940; c26922_g1_i1 & c38696_g1_i1), E2F target gene 1 (AT2G40550;c46496_g1_i1), minichromosome maintenance chromosome 5 (MCM5) (AT2G07690; c46324_g1_i1), ribonucleoside-diphosphatereductase small chain C/TSO2 (AT3G27060; c39450_g1_i1), DNA primase/POLA3 (AT5G41880; c41508_g1_i1,c41508_g1_i2 & c41508_g2_i1) and RPA70B (AT5G08020; c37107_g1_i1) were down-regulated in litchi small-seeded genotype at 0 DAA (Supplementary Table S3). E2F target gene 1 controls cell cycle progression by cohesion of sister chromatid and DNA repair66. Minichromosome maintenance 5 is involved in initiation of DNA replication in cell67. No reads were detected for fructose-bisphosphatealdolase 2 (AT4G38970; c43203_g3_i1), required for biomass accumulation68 in the developing ovules (0, 6, and 14 DAA) of small-seeded genotype. At hormone level, the events of cell division, differentiation and expansion in plant tissues are regulated by auxin23. The transcriptional pattern in the developing ovules of small seeded genotypes showed suppression of auxin transport.
Embryogenesis related genes
In litchi, the preliminary zygotic cell division happens during 3 to 7 DAA, and the embryo enters globular stage from 14 to 21 DAA69. We investigated expression pattern of several embryogenesis related genes in the two contrasting litchi genotypes. Ole e 1 allergen and extensin family protein (AT2G34700; c38654_g1_i1), involved in pollen tube guidance during fertilization70, was down-regulated in developing ovules of the small-seeded litchi (Supplementary Table S3). The flavin binding monooxygenase (AT5G25620; c50631_g2_i1 & c41400_g1_i3) that provides ROS tolerance71 during pollination was down regulated in the small seeded genotype (Fig. 2A). Asymmetric cell division of zygote is very important for embryo development and auxin plays an important role in the establishment of polarity72. Expression of Topless/TPR2 (AT3G16830; c51185_g3_i1, c51185_g3_i2, c51185_g3_i3 and c51185_g3_i4) was suppressed in small-seeded litchi genotype (Supplementary Table S3). In the loss of function mutant of this gene, axis establishment is severely affected in Arabidopsis73. The putative gene was down–regulated in the small-seeded genotype. EP3 chitinase (AT3G54420; c45503_g1_i1 & c45503_g1_i2) and phytosulfokine 2/PSK2 (AT2G22860; c79596_g1_i1) hamper embryo development by inducing somatic embryogenesis74. These genes showed higher expression in small-seeded litchi genotype at 0 DAA (Supplementary Table S3).
Embryogenesis begins with the fertilization of egg, forming zygote. It undergoes asymmetrical division, followed by defined pattern of embryo growth to subsequently form cotyledon75. Essential genes for seed development have been identified in Arabidopsis thaliana by analyzing the mutants that severely affect embryo developmental stages76. A total of 440 seed development related Arabidopsis – like transcripts were identified in the litchi transcriptome data. Such litchi transcripts, that appear to effect embryo development at pre-globular (AT3G01610, AT1G01370, AT2G32950, AT1G12260, AT5G49010, AT5G15920, AT5G13690, AT5G27740 and, AT1G08840), globular (AT5G07280, AT1G67320, AT3G06350, AT2G44190 andAT5G1870), transition (AT3G54720) and cotyledon stage (AT5G48600, AT2G01190, AT3G50870, AT4G02060, AT2G34650, AT1G08560, AT5G13480, AT1G44900, AT1G76620, AT1G66520, AT5G62410, AT5G49160, AT1G71720, AT3G54650, AT1G78580 and AT1G67730) were down-regulated at 0DAA in small-seeded litchi genotype (Fig. 5, Supplementary Table S5). Replication factor C subunit 3 (AT5G27740; c50794_g10_i1, c50794_g10_i2, c50794_g10_i3, c50794_g10_i4 and c50794_g10_i6) is a clamp loader during replication. Its mutagenesis causes severe defects in embryo development at pre-globular stage in Arabidopsis thaliana77. COP 1 (AT2G32950; c44060_g2_i2 & c44060_g2_i5) regulates seed development by suppressing expression of photomorphogenic pathway and enhancing expression of dark induced seed development pathway78. N-acetyl–glucosaminidase catalyzes the breakdown of sugars in proteoglycans, and mutation in the gene may induce embryo defects at pre-globular stage79. N-acetyl–glucosaminidase/CYL 1 (AT5G13690; c48465_g2_i1 & c48465_g2_i2) was down-regulated at 0 and 6 DAA in small-seeded genotype. Histone H3 variant (AT1G01370; c43813_g1_i1) is important for kinetochore assembly and chromosome segregation, its knock out causes small seed with reduced fertility80. Structural maintenance of chromosome 5 (AT5G15920; c48305_g2_i1) is involved in DNA double strand repair81. Endosperm defective 1 (AT2G44190; c44482_g1_i1) is a microtubule-associated protein, essential for cell division of embryo and endosperm82. RUNKEL (AT5G18700; c50689_g1_i1) is responsible for phragmoplast organization and cell plate expansion83. ALTERED MERISTEM PROGRAM 1 (AT3G54720; c47383_g5_i1 & c48054_g2_i2) regulates pollen tube attraction by synergid cells84. Structural maintenance of chromosomes protein 4 (AT5G48600; c19609_g1_i1 & c19609_g1_i2) plays important role in condensation of chromatin during mitosis85. Trehalose-6-phosphate synthase 1 (AT1G78580; c48632_g2_i1, c48632_g2_i2, c48632_g2_i3, c48632_g2_i4 & c48632_g2_i5) regulates trehalose synthesis, and is essential for seed maturation86. MMC 2 (AT1G44900; c47148_g1_i1 & c47148_g1_i2) is a cell cycle regulator that plays important role in stem cell maintenance and differentiation87. F BOX-LIKE 17 (AT3G54650; c48319_g1_i1) positively regulates cell proliferation88. At 6 DAA, transcripts causing embryo defects leading to terminal phenotypes at pre-globular (AT5G13690; c48465_g2_i1 & c48465_g2_i2), globular (AT2G45690; c43594_g1_i10) and transition stage (AT5G67570; c50526_g1_i5) were down-regulated in the small-seeded litchi cultivar (Fig. 5). Mutation in the gene encoding Shrunken Seed protein (AT2G45690), which is involved in peroxisome assembly and protein trafficking, leads to shrunken seeds89. Pentatricopeptide repeat containing protein, delayed greening 1 (AT5G67570; c50526_g1_i5) is a regulator of early chloroplast development and gene expression90. The homologues of transcripts causing embryo development at pre-globular (AT3G20070; c50859_g5_i6 and AT3G17300; c51543_g1_i2) and globular stage (AT5G16715; c51777_g1_i10 and AT1G64790; c38034_g1_i2 & c49435_g2_i3) were down-regulated in the small-seeded litchi genotype at 14 DAA (Fig. 5). However, the master regulatory genes, if any, that may lead to differential expression of the genes downstream have not yet been identified. Several genes that expressed differentially between HC and HS could not be functionally annotated due to lack of homology with the database. A total of 3,413 differentially expressed transcripts could not match with any known protein in the database. These unknown transcripts could be important for the development of seeds in litchi.
Functional mapping of transcripts that express differentially between the small (HS) and bold (HC) seeded litchi genotypes.
The function has been assigned based on the phenotypes reported in Arabidopsis thaliana following loss of function mutations in homologous genes. Heatmaps in different columns represent the expression levels in ovule at 0, 6 and 14 DAA.
A distinct expression pattern of the genes in early-stage ovule development might lead to under-developed embryos (Fig. 6), resulting into the development of small seed size in litchi (Fig. 4).
Shift in gene expression during ovule development
As shown in Fig. 1B, during transition from 0 to 6 DAA, a total of 618 differentially expressed transcripts were common between bold and small-seeded ovules; whereas, 1,569 and 2,184 differentially expressed transcripts were exclusively present in the ovules of bold- and small-seeded genotypes, respectively. During transition from 6 to 14 DAA a total of 277 differentially expressed transcripts were common between bold and small seeded ovules; whereas, 1,761 and 393 differentially expressed transcripts were exclusive to bold and small-seeded ovules, respectively (Fig. 1B). In developing ovules, zygotic division is a metabolically active process at which apical-basal body of the polarized zygote is established by asymmetric divisions. The Homeobox proteins, WRKY and receptor kinases are known to express at high level during zygotic division91,92,93. The homologues of these genes were up-regulated in large-seeded genotype at 6 DAA as compared to 0 DAA ovules, in contrast to the small-seeded ovules, where these were down regulated. Expression of auxin, gibberellic acids and brassinosteriods was up-regulated at 6 DAA and then decreased at 14 DAA in bold-seeded genotype, which is in accordance with the pattern of expression in an earlier report on litchi94. No such trend was observed in small-seeded genotype, with little change in expression pattern between 6 to 14 DAA (Supplementary Fig. S3).
The expression differences between the developmental stages of ovules may lead to partial or complete abortion of seed in litchi.
Validation of ovule-specific differentially expressed genes obtained from RNA-seq libraries in two genotypes with contrasting seed size
To validate the differential expression profiles identified in the digital data obtained by RNA-seq, we performed in real time PCR on 4 representative transcripts selected for different levels of differential expression at 6, 8 at 14 DAA (Fig. 7). The representative transcripts were selected from among both annotated and non-annotated entries and with contrasting levels of expression in the two genotypes (Supplementary Table S6). The expression pattern obtained in real time assay was comparable with that analyzed from RNA-seq data. The high and significant Pearson correlation coefficient (0.808) of RNA-seq with real time results suggests high confidence in the expression values obtained from the transcriptome data. Hence, the RNA-seq data can be used reliably for relative expression analysis of genes in seed development pathway in litchi.
In conclusion, we report the first transcriptome information on early-stage ovule development in litchi genotypes with contrasting seed size. We identified various differentially expressed genes that showed homology to the genes reported in database. Our analysis shows that the genes involved in endosperm development as well as embryo development were associated with small seed size in the small-seeded genotype of litchi. Such genes were related to hormone biosynthesis pathways, transcription factors, ovule identity determinants, and embryogenesis. The developing ovules of small seeded genotype showed higher expression of several stress related transcripts, suggesting the association of metabolic stress with improper seed development in litchi. A number of genes expressed differentially at 0 DAA, suggesting that some factors resident in maternal tissue may set in the process that leads to the reduced seed size in the small-seeded litchi genotypes.
Materials and Methods
Plant materials and sample collection
Plant materials were collected from Fruit Research Station, Punjab Agriculture University, Hoshiarpur, Punjab, India. Two litchi genotypes of contrasting seed size - Calcuttia (bold seeded) and Seedless Late (small seeded), were selected for the study. In our previous study, these two genotypes were reported to be closely genomically related, with the Nei 72 genetic distance of 0.3695. The hermaphrodite female flowers, with two lobed styles and sticky stigma, were identified on the panicle as highly receptive flower stage and marked as 0 DAA (days after anthesis). Developing fruits at 0, 6, and 14 DAA were harvested randomly from three healthy trees each, of the two genotypes. The samples were surface sterilized with absolute ethanol before harvesting, as described previously7. The harvested samples were immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
RNA isolation and library construction for transcriptome analysis
Ovules were excised from 20–30 developing fruits at each developmental stage, in chilled (−20 °C) ethanol, under Leica M205 C stereo microscope in RNase free condition (Supplementary Figs S4 and S5). Total RNA was extracted by using Spectrum Plant Total RNA kit (Sigma Aldrich, St. Louis, MO, USA), following manufacture user’s protocol. DNase I (Sigma Aldrich St. Louis, MO, USA) treatment was given to remove genomic DNA contamination. The RNA integrity was established, using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The equivalent quantity of total RNA extracted from the ovules of three biological replicates (plants) was pooled for transcriptome sequencing.
Transcriptome sequencing and de novo assembly
Paired-end Illumina mRNA libraries were generated from total RNA samples from ovules at 0, 6, and 14 DAA for the two genotypes in accordance with the manufacturer’s instructions (Illumina Inc., USA). Transcriptome sequencing was performed on IlluminaHiSeq2000 platform (Sandor, Hyderabad, India). The raw reads were pre-processed by removing adaptor sequences, and discarding empty reads and low-quality sequences. The high quality paired end 101 bp reads of each sample were then used for transcriptome de novo assembly using Trinity pipeline (release 2015-02-14) at default parameters. The sequences assembled by Trinity (Trinity transcripts) are referred to as transcripts in the manuscript.
Comparative analysis
A comparative analysis was performed by using the litchi transcripts as queries (BLASTx; E value ≤ 10−5) against the protein databases of crops such as, Arabidopsis thaliana (ftp://ftp.psb.ugent.be/pub/plaza/plaza_ public_dicots_ 03/Fasta/proteome. ath.tfa.gz), Citrus sinensis (ftp://ftp.psb.ugent.be/pub/plaza/plaza_public_dicots_03/Fasta/proteome.csi. tfa.gz). Ricinus communis (ftp://ftp.psb. ugent. be/pub/plaza/plaza_public_ dicots_03/Fasta/proteome. rco.tfa.gz), Populus trichocarpa (ftp://ftp.psb. ugent.be/pub/plaza/plaza_public_ dicots_03/Fasta/proteome.ptr.tfa.gz), Fragaria vesca (p.t. be/pub/plaza/plaza_public_ dicots_03/Fasta/proteome. fve.tfa.gz), Carica papaya (ftp://ftp.psb.ugent.be/pubt./plaza/plaza_ public_dicots_03/Fasta/proteome.cpa.tfa.gz) and Glycine max (a_public_dicots_03/Fasta/proteome.gma. tfa.gz).
Functional annotation
Functional annotation of the transcripts was performed by BLASTx (E-value ≤ 10−5) against databases: NCBI non-redundant (nr) database (http://www.ncbi.nlm.nih.gov), Swiss-Prot protein database (http://www.expasy.ch/ sprot), and Clusters of Orthologous Groups of proteins (http://www.ncbi. nlm.nih.gov/COG). The process was accelerated using WImpiBLAST tool96 on institute’s high performance computing cluster.
The litchi transcripts showing homology with the protein database of different crops (Arabidopsis thaliana, Citrus sinensis, Populus trichocarpa, Ricinus communis, Glycine max, Fragaria vesca, Carica papaya and Vitis vinifera) were used for GO term assignment.
Gene ontology (GO) analysis
GO terms for the litchi transcripts were obtained from PLAZA (http://bioinformatics.psb.ugent.be/plaza/versions/plaza_v3_dicots/download/index), using the corresponding IDs of aforementioned eight crops. The GO terms were used as inputs in the agriGO analysis tool (http://bioinfo.cau.edu.cn/agriGO/analysis.php)97 for GO categorization (P value ≤ 0.05) with customized annotation option. The GO (P value ≤ 0.05) was given as input in the REVIGO tool (http://revigo.irb.hr/)98, results of REVIGO were obtained in.csv format and used for histogram preparation.
Transcript, abundance estimation and identification of differentially expressed transcripts
The de novo assembled transcripts were used as a reference to map the individual Illumina reads, using script ‘align_and_estimate_abundance.pl’ bundled with Trinity suite. Abundance estimation was done with the RSEM version 1.2.999, at default parameter using BOWTIE2 version 2.1.0100 mapping. Relative measure of transcript abundance was obtained as TPM (Transcripts per million) and FPKM (Fragments per Kilobase of transcripts per Million mapped reads). Expression levels of different transcripts were compared among the samples by edgeRBioconductor101, using script run_DE_analysis.pl at default parameter. EdgeR performs an additional TMM (Trimmed Mean of M-Values) scaling normalization to examine differences in total RNA production across the samples102.
EdgeR Bioconductor uses negative binomial distribution as statistical model for identification of possible differentially expressed transcripts in the absence of biological replicates103.
Quantitative real-time PCR analysis
Differentially expressed transcripts obtained by high throughput sequencing were validated by quantitative real-time PCR. Total RNA were isolated from ovules at 6 and 14 DAA, and cDNA was synthesized from total RNA (500 ng) using oligo (dT) primers and M-MLV reverse transcriptase, according to the manufacturer’s instructions (Invitrogen, USA) in a 20 μl volume. Between the two libraries (HC and HS), four and eight differentially expressed genes at 6 DAA and 14 DAA respectively, were selected to observe their expression patterns between the two cultivars (Supplementary Data 5). Transcript levels were analyzed by quantitative real-time PCR using the fast SYBR green master mix (Applied Biosystems, USA) and an ABI 7500 Real-Time PCR System (Applied Biosystems, USA) according to the manufacturers’ instructions. All biological replicates were analyzed in duplicate. Real-time PCR reactions were normalized to the Ct values for litchi LcActin (HQ615689). The relative expression levels of the target genes were calculated using the formula 2−ΔΔCT.
Data Availability
Raw sequencing data is available through NCBI Sequence Read Archive, accession number: http://www.ncbi.nlm.nih.gov/sra?term=SRP076141. All samples were sequenced as 101-nucleotide paired-end reads on an Illumina HiSeq2000 sequencer.
Additional Information
How to cite this article: Pathak, A. K. et al. Transcriptional changes during ovule development in two genotypes of litchi (Litchi chinensis Sonn.) with contrast in seed size. Sci. Rep. 6, 36304; doi: 10.1038/srep36304 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Ye, X. L., Wang, F. X. & Qian, N. F. Embryological studies of Litchi chinensis. Acta Metallurgica Sinica 14, 1–3 (1992).
Gangehei, L. et al. Oligonol a low molecular weight polyphenol of lychee fruit extract inhibits proliferation of influenza virus by blocking reactive oxygen species-dependent ERK phosphorylation. Phytomedicine 17, 1047–1056 (2010).
Berger, F. Endosperm development. Curr.Opin. Plant Bio. 2, 28–32 (1999).
Chaudhury, A. M. et al. Control of early seed development. Annu. Rev. Cell Dev. Biol. 17, 677–699 (2001).
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M. & Lenhard, M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106, 20115–20120 (2009).
Xia, T. et al. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25, 3347–3359 (2013).
Gupta, Y. et al. De novo assembly and characterization of transcriptomes of early-stage fruit from two genotypes of Annona squamosa L. with contrast in seed number. BMC Genomics 16, 86 (2015).
Asif, M. H. et al. Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biology 14, 316 (2014).
Alkio, M., Jonas, U., Declercq, M., Nocker, S. V. & Knoche, M. Transcriptional dynamics of the developing sweet cherry (Prunus avium L.) fruit: sequencing, annotation and expression profiling of exocarp-associated genes. Horticulture Research 1, 11 (2014).
Xie, M. et al. Transcriptome profiling of fruit development and maturation in Chinese white pear (Pyrusbrets chneideri Rehd). BMC Genomics 14, 823 (2013).
Zhang, H. N. et al. Transcriptomic analysis of floral initiation in litchi (Litchi chinensis Sonn.) based on de novo RNA sequencing. Plant Cell Rep. 33, 1723–1735 (2014).
Li, C. et al. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genomics 14, 552 (2013).
Li, W. C. et al. De novo assembly and characterization of pericarp transcriptome and identification of candidate genes mediating fruit cracking in Litchi chinensis Sonn. Int. J. Mol. Sci. 15, 17667–177685 (2014).
Lu, X. et al. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC Genomics 15, 805 (2014).
Li, C., Wang, Y., Ying, P., Ma, W. & Li, J. Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front. Plant Sci. 6, 502 (2015).
Lai, B. et al. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genomics 16, 225 (2015).
Li, C. et al. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 6, 439 (2015).
Yun, Z. et al. Comparative transcriptome and metabolome provides new insights into the regulatory mechanisms of accelerated senescence in litchi fruit after cold storage. Sci. Rep. 6, 19356 (2016).
Varoquaux, F., Blanvillain, R., Delseny, M. & Gallois, P. Less is better: new approaches for seedless fruit production. Trend. Biotechnol. 18, 233–242 (2000).
Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 (2000).
Ohto, M. A., Fischer, R. L., Goldberg, R. B., Nakamura, K. & Harada, J. Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102, 3123–3128 (2005).
Nemhauser, J. L., Feldman, L. J. & Zambryski, P. C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888 (2000).
Perrot-Rechenmann, C., Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2, a001446 (2010).
Ceccato, L. et al. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. Plos One 8, e66148 (2013).
Paret, B. et al. AUX/LAX Genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24, 2874–2885 (2012).
Dorcey, E., Urbez, C., Blázquez, M. A., Carbonell, J. & Perez-Amador, M. A. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 58, 318–332 (2009).
Traub, M. et al. The fluorouridine insensitive 1 (fur1) mutant is defective in equilibrative nucleoside transporter 3 (ENT3), and thus represents an important pyrimidine nucleoside uptake system in Arabidopsis thaliana. Plant J. 49, 855–864 (2007).
Zinn, K. E., Tunc-Ozdemir, M. & Harper, J. F. Temperature stress and plant sexual reproduction: uncovering the weakest links. J. Exp. Bot. 61, 1959–1968 (2010).
Clouse, S. D. & Sasse, J. M. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 49, 427–451 (1998).
Gudesblat, G. E. et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell. Biol. 14, 548–554 (2012).
Kim, T. W., Michniewicz, M., Bergmann, D. C. & Wang, Z. Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422 (2012).
Wang, Z. Y., Bai, M. Y., Oh, E. & Zhu, J. Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724 (2012).
Sun, X., Shantharaj, D., Kang, X. & Ni, M. Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant. Biol. 13, 611–620 (2010).
Carland, F., Fujioka, S. & Nelson, T. The Sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153, 741–756 (2010).
Klahre, U. et al. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690 (1998).
Li, N. & Li, Y. Maternal control of seed size in plants. J. Exp. Bot. 66, 1087–1097 (2015).
Smith, D. W. et al. Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana. Proteins 57, 854–857 (2004).
Sawa, S. et al. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes & Dev. 13, 1079–1088 (1999).
Siegfried, K. R. et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128 (1999).
Baum, S. F., Eshed, Y. & Bowman, J. L. The Arabidopsis nectary is an ABC-independent floral structure. Development 128, 4657–4667 (2001).
Baker, S. C., Robinson-Beers, K., Villanueva, J. M., Gaiser, J. C. & Gasser, C. S. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145, 1109–1124 (1997).
Lora, J., Hormaza, J. I., Herrero, M. & Gasser, C. S. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 108, 5461–5465 (2011).
Rushton, P. J., Somssich, I. E., Ringler, P. & Shen, Q. J. WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010).
Epple, P., Mack, A. A., Morris, V. R. F. & Dangl, J. L. Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 100, 6831–6836 (2003).
Ruberti, I., Sessa, G., Lucchetti, S. & Morelli, G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 10, 1787–1791 (1991).
Sun, K., Cui, Y. & Hauser, B. A. Environmental stress alters genes expression and induces ovule abortion: reactive oxygen species appear as ovules commit to abort. Planta 222, 632–642 (2005).
Yamasaki, K. et al. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J. Mol. Biol. 348, 253–264 (2005).
Chao, Q. et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 (1997).
Tiwari, S. B., Hagen, G. & Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543 (2003).
Yang, Z. et al. Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and Rice. J. Mol. Evol. 67, 266–277 (2008).
Hauser, B. A., He, J. Q., Park, S. O. & Gasser, C. S. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 127, 2219–2226 (2000).
Andersen, S. U. J. et al. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J. Exp. Bot. 58, 3657–3670 (2007).
van den Heuvel, S. & Dyson, N. J. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell. Biol. 9, 713–724 (2008).
Gutierrez, C. The Arabidopsis Cell Division Cycle. The Arabidopsis Book/American Society of Plant Biologists 7, e0120 (2009).
Kim, J. H., Choi, D. & Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104 (2003).
Kim, S. G., Kim, S. Y. & Park, C. M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226, 647–654 (2007).
Nuruzzaman, M., Sharoni, A. M. & Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front.Microbiol. 4, 248 (2013).
Czemmel, S., Hepple, S. C. & Bogs, J. R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249, 109–118 (2012).
Krizek, B. A., Prost, V. & Macias, A. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12, 1357–1366 (2000).
Bencivenga, S., Simonini, S., Benkova, E. & Colombo, L. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897 (2012).
Pagnussat, G. C., Yu, H. J. & Sundaresan, V. Cell-fate switch of synergid to egg cell in Arabidopsiseostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592 (2007).
Shuai, B., Reynaga-Pena, C. G. & Springer, P. S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761 (2002).
Liu, L. et al.Auxin in Plant Growth and Stress Responses. In Phytohormones: A window to metabolism, signaling and biotechnological applications (eds Phan, L. S. T. & Sikander, P. ) 1–35 (Springer, 2014).
Kwon, S. H. et al. Overexpression of a Brassica rapa NGATHA gene in Arabidopsis thaliana negatively affects cell proliferation during lateral organ and root growth. Plant Cell Physiol. 50, 2162–2173 (2009).
Jenik, P. D., Jurkuta, R. E. J. & Barton, M. K. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA Polymerase ɛ. Plant Cell 17, 3362–3377 (2005).
Takahashi, N. et al. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. Plos Genet. 6, e1000817 (2010).
Shultz, R. W., Lee, T. J., Allen, G. C., Thompson, W. F. & Hanley-Bowdoin, L. Dynamic localization of the DNA replication proteins MCM5 and MCM7 in plants. Plant Physiol. 150, 658–669 (2009).
Uematsu, K., Suzuki, N., Iwamae, T., Inui, M. & Yakawa, H. Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 63, 3001–3009 (2012).
Liang, Y. S., Teng, Y. S., Wang, C. H. & Ke, L. S., Types of aborted seed and quality evaluation of ‘Wuheli’ litchi (Litchi chinensis Sonn.). Afr. J. of Agri. Res. 7, 2910–2917 (2012).
de Dios Alche, J., M’rani-Alaoui, M., Castro, A. J. & Rodriguez-Garcia, M. I. Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant and Cell Physiol. 45, 1149–1157 (2004).
Cheol, P. H., Cha, J. Y. & Yun, D. J. Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signaling & Behav. 8, e24495 (2013).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Long, J. A., Ohno, C., Smith, Z. R. & Meyerowitz, E. M. TOPLESS Regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006).
Matthys-Rochon, E. Secreted molecules and their role in embryo formation in plants: a min-review. Acta. Biol.Cracov. 47, 23 (2005).
Mayer, U. & Jurgens, G. Pattern formation in plant embryogenesis: A reassessment. Sem. Cell Dev. Biol. 9, 187–193 (1998).
Tzafrir, I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220 (2004).
Reynolds, N., Fantes, P. A. & MacNeill, S. A. A key role for replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 27, 462–469 (1999).
Deng, X. W., Caspar, T. & Quail, P. H. Cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes & Dev. 5, 1172–1182 (1991).
Ronceret, A., Gadea-Vacas, J., Guilleminot, J. & Devic, M. The alpha-N-acetyl-glucosaminidase gene is transcriptionally activated in male and female gametes prior to fertilization and is essential for seed development in Arabidopsis. J. Exp. Bot. 59, 3649–3659 (2008).
Lermontova, I. et al. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 68, 40–50 (2011).
Watanabe, K. et al. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688–2699 (2009).
Pignocchi, C. et al. ENDOSPERM DEFECTIVE1 Is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21, 90–105 (2009).
Krupnova, T. et al. Microtubule-associated kinase-like protein RUNKEL needed for cell plate expansion in Arabidopsis cytokinesis. Curr. Biol. 19, 518–523 (2009).
Kong, J., Lau, S. & Jargens, G. Twin plants from supernumerary egg cells in Arabidopsis. Curr. Biol. 25, 225–230 (2014).
Siddiqui, N. U., Rusyniak, S., Hasenkampf, C. A. & Riggs, C. D. Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta 223, 990–997 (2006).
Gamez, L. D., Baud, S. & Graham, I. A. The role of trehalose-6-phosphate synthase in Arabidopsis embryo development. Biochem. Soc. Trans. 33, 280–282 (2005).
Ni, D. A. et al. The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol. 184, 311–322 (2009).
Noir, S. et al. The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-Box Protein FBL17. Plant Cell 27, 1461–1476 (2015).
Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A. & Goodman, H. M. The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis Seeds. Science 284, 328–330 (1999).
Chi, W. et al. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584 (2008).
Bosca, S., Knauer, S. & Laux, T. Embryonic development in Arabidopsis thaliana: from the zygote division to the shoot meristem. Front. Plant. Sci. 2, 93 (2011).
Petricka, J. J., Van Norman, J. M. & Benfey, P. N. Symmetry breaking in plants: molecular mechanisms regulating asymmetric cell divisions in Arabidopsis. Cold Spring Harb. Perspect. Biol. 1, a000497 (2009).
Souter, M. & Lindsey, K. Polarity and signalling in plant embryogenesis. J. Exp. Bot. 51, 971–983 (2000).
Wei, C. & Liuxin, L. U. Relationship between embryonic development and changes of endogenous hormones in Litchi (Litchi chinensis Sonn.) ovules. Chin. j. appl. environ. biol. 6, 419–422 (2000).
Pathak, A. K., Singh, S. P. & Tuli, R. Amplified fragment length polymorphism fingerprinting to identify genetic relatedness among Lychee cultivars and markers associated with small-seeded cultivars. J. Am. Soc. Hortic. Sci. 139, 657–668 (2014).
Sharma, P. & Mantri, S. S. WImpiBLAST: Web interface for mpiBLAST to help biologists perform large-scale annotation using high performance computing. Plos One 9, e101144 (2014).
Du, Z., Zhou, X., Ling, Y., Zhang, Z. & Su, Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38 (Web Server issue), W64 (2010).
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. Plos One 6, e21800 (2011).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M. & Gilad, Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517 (2008).
Acknowledgements
This work was supported by Department of Biotechnology (DBT), Govt. of India. A.K.P. acknowledges DBT fellowship. R.T. acknowledges DST for J C Bose Fellowship. Authors acknowledge the Director, Fruit Research Station (PAU, Hoshiarpur, Punjab) for permitting sample collection and Kiran Bankar (Bionivid) for help in execution of Trinity pipeline.
Author information
Authors and Affiliations
Contributions
R.T. proposed and designed the study. S.P.S. guided the experiments and data analysis. A.K.P. performed the experiments, and analyzed data. S.S.M. supervised assembly and annotation. Y.G. and A.K.S.G. helped in detailed annotations. A.K.P. wrote the manuscript, S.P.S. and R.T. improved presentation. All authors read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pathak, A., Singh, S., Gupta, Y. et al. Transcriptional changes during ovule development in two genotypes of litchi (Litchi chinensis Sonn.) with contrast in seed size. Sci Rep 6, 36304 (2016). https://doi.org/10.1038/srep36304
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36304
This article is cited by
-
Transcriptome analysis at mid-stage seed development in litchi with contrasting seed size
3 Biotech (2022)
-
Metabolomic and transcriptomic profiling of three types of litchi pericarps reveals that changes in the hormone balance constitute the molecular basis of the fruit cracking susceptibility of Litchi chinensis cv. Baitangying
Molecular Biology Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.