Abstract
Deranged metabolism is a hallmark of cancer, playing a significant role in driving the disease process. One such example is the induction of carcinogenesis by the oncometabolite D-2 hydroxyglutarate (D-2HG), which is produced by the mutated enzyme isocitrate dehydrogenase (IDH) occurring in subsets of leukaemias and brain tumours. The oncogenic property of D-2HG appears to stem from its ability to interfere with the activities of α-ketoglutarate-dependent dioxygenases, including the Jumonji family histone demethylases. Here, we find in colorectal cancer cells that even in the absence of IDH mutation, the levels of D-2HG and its enantiomer L-2HG were elevated through glutamine anaplerosis. D-2HG, but not L-2HG, increased the trimethylation of histone H3 lysine 4 of the promoter region of ZEB1, a master regulator of epithelial-mesenchymal transition (EMT), and increased the expression of the ZEB1 gene to directly induce EMT in colorectal cancer cells. EMT promotes the ability of cancer cells to invade the local tissue and enter into the bloodstream, leading to distant organ metastasis. D-2HG levels were elevated in colorectal cancer specimens, particularly in those associated with distant metastasis, supporting the observations in vitro and implicating the contribution of D-2HG in metastasis, the major cause of death in this disease.
Similar content being viewed by others
Introduction
Colorectal cancer is the third most common cancer and the fourth most common cause of cancer-related deaths worldwide, leading to approximately 700,000 deaths each year1. The frequency with which the disease presents at an advanced stage2, and therefore beyond the scope of curative surgical resection3, underlies the high number of deaths and emphasises the need for novel treatment strategies, underpinned by a better understanding of the disease.
Deranged metabolism is a hallmark of cancer, including in colorectal cancer4, and can encompass many of its components from the increased cellular uptake of glucose and other nutrients to the shift to anaerobic metabolism of glucose to lactate, allowing the rapidly proliferating malignant cells to meet their energetic and biosynthetic demands5. Moreover, we and others have found that deranged metabolism also drives oncogenesis6 and disease progression7,8, highlighting the significant role of cellular metabolism throughout the disease process and as a promising target for treatment.
Specific metabolites have been identified to be the crucial link between metabolism and oncogenesis, providing an insight into the molecular mechanisms behind this phenomenon9. Metabolites are the intermediate products of metabolism, and are the fuel, biomaterials and signalling molecules of the cells6,10. It appears to be the latter property of metabolites that endows them with the ability to alter gene expression in cancer cells, including through control of the epigenetic machinery6. One such metabolite is D-2-hydroxyglurate (D-2HG), the concentration of which is very low in healthy individuals, but high in subsets of leukaemias and brain tumours that harbour gain of function mutations of the enzymes isocitrate dehydrogenase 1 or 2 (IDH1 or 2)11. D-2HG induces leukemogenesis12 and malignant transformation of astrocytes13, but has no known physiological function, and is widely referred to as an “oncometabolite”. The oncogenic nature of D-2HG appears to stem from its ability to interfere with the activity of various α-ketoglutarate-dependent dioxygenases, including ten eleven translocation 5-methylcytosine hydroxylases (TET) involved in DNA demethylation and Jumonji family histone demethylases11,14, which can lead to widespread changes of DNA and histone methylation, resulting in the disruption of cellular proliferation and differentiation15,16.
The revelation that 2HG levels are elevated in breast cancer even with wild-type IDH, driven by glutamine anaplerosis (Fig. 1A)17,18,19, highlighted that 2HG levels could be elevated in a wider range of malignancies that do not necessarily harbour mutated IDH20. Also in the azoxymethane mouse model of intestinal cancer, 2HG levels were elevated in the tumours compared to normal tissues21, providing further support that the oncometabolite may be of importance in human colorectal cancer.
D- and L-2HG levels are elevated in colorectal cancer cells and is dependent on glutamine metabolism.
(A) The metabolic pathway of the production of D- and L-2HG from glutamine. Glutamine is metabolised to α-ketoglutarate (α-KG) in sequential steps, involving glutaminase (GLS1). α-KG can then be metabolised to D-2HG by IDH1 or 2, or to L-2HG by lactate dehydrogenase 1 (LDH1) or malate dehydrogenase (MDH). Furthermore, D-2HG is converted physiologically to α-KG by D-2HG dehydrogenase (D2HGDH), as is L-2HG to α-KG by L-2HG dehydrogenase (L2HGDH). (B) Baseline levels of D- and L-2HG were measured in three colorectal cancer cell lines (DLD1, HCT116, RKO) and a sarcoma cell line HT1080 known to harbour mutant IDH1 (R132C) by gas chromatography-mass spectrometry (GC-MS). Data are presented as means and standard deviations; **p < 0.01. (C) Sanger sequencing confirms the presence of IDH1 mutation (G97D) in the DLD1 cell line. IDH1 was wild-type in HCT116 and RKO cell lines. (D) D- and L-2HG levels were measured by GC-MS in DLD1 and RKO cell lines, after siRNA interference of GLS1. Data are presented as means and standard deviations; **p < 0.01.
In renal cell carcinoma, L-2HG, the opposite enantiomer of D-2HG, is elevated secondary to decreased expression of L-2HG dehydrogenase, the enzyme responsible for the metabolisation of L-2HG22. L-2HG is also the product of glutamine anaplerosis, but in contrast to D-2HG, it is produced from the reduction of α-ketoglutarate by lactate dehydrogenase A or malate dehydrogenase (Fig. 1A)23. L-2HG is thought also to be an oncometabolite, but it is distinct from D-2HG in its bioactivity, given that it does not cause leukemogenesis or astrocyte transformation12,13.
Here we explore the role of D- and L-2HG in colorectal cancer, and find the levels of these metabolites to be elevated in colorectal cancer cells. D-2HG, but not L-2HG, directly induced epithelial-mesenchymal transition (EMT) in colorectal cancer cells, which is associated with cancer cells acquiring the ability to invade the surrounding tissues and enter the bloodstream, leading to distant organ metastasis24. D-2HG also induced histone modifications associated with increased gene expression at the promoter region of the ZEB1 gene, which encodes a master regulator of EMT. Furthermore, clinical specimens with higher levels of D-2HG were more likely to be associated with distant organ metastasis, supporting the significant contribution of the metabolite in cancer metastasis.
Results
D- and L-2HG Levels are Elevated in Colorectal Cancer Cells and is Dependent on Glutamine Metabolism
To explore the potential of D- or L-2HG playing a role in colorectal cancer, we began by measuring the levels of the metabolites in colorectal cancer cell lines by gas chromatography-mass spectrometry (GC-MS). In two of the three cell lines analysed (DLD1 and RKO), both D- and L-2HG were elevated at levels comparable to or exceeding the concentrations measured in the sarcoma cell line HT1080, harbouring mutant IDH1 (Fig. 1B). Given that mutations of IDH1 and IDH2 are the widely known cause of elevated levels of D-2HG, these genes were sequenced, confirming the presence of mutated IDH1 (G97D) in the DLD1 cell line, previously revealed to be present in the same cell line25 and to lead to the production D-2HG26 (Fig. 1C). However the IDH1 and 2 enzymes were wild-type in RKO cells, leading to the speculation that D-2HG could be elevated in this cell line instead from glutamine anaplerosis, which would also explain the levels of L-2HG being elevated18,23 (Fig. 1A). In support of glutamine anaplerosis fuelling the production of D- and L-2HG, the concentration of both metabolites were reduced in the colorectal cancer cells when glutaminase expression was knocked down, an enzyme responsible for the first step of several in the catabolism of glutamine to α-ketoglutarate (Fig. 1D, Supplementary Fig. S1A). Furthermore, the baseline expressions of glutaminase and other enzymes involved in the catabolism of glutamine to α-ketoglutarate, aspartate aminotransferase 2 and glutamate dehydrogenase 1, were elevated in the DLD1 and RKO cell lines relative to the HCT116 cell line (Supplementary Fig. S1B and C). Given gene expression is one of the determinants of metabolic flux, these results suggest that the differences in the expression of enzymes involved in glutamine metabolism may explain the variation in the levels of D- and L-2HG across the colorectal cancer cell lines.
In diffuse large B-cell lymphoma, D-2HG levels have been found to be elevated secondary to mutation of the enzyme D-2HG dehydrogenase (D2HGDH)27, which has a physiological role of converting D-2HG to α-ketoglutarate. We therefore examined for mutations of D2HGDH, and its counterpart for L-2HG, L-2HG dehydrogenase (Fig. 1A) in DLD1, HCT116 and RKO cells through sequencing, and found both enzymes to be wild-type in the three colorectal cancer cell lines. Furthermore, the expression of D2HGDH and L2HGDH were not consistently lower in DLD1 and RKO cells in comparison to HCT116 cells, as might have been expected if the decreased metabolisation of 2HG was to explain the higher levels of these metabolites in DLD1 and RKO cells (Fig. S1D). These results add further weight to the notion that it is the production of 2HG rather than its metabolisation that governs the levels of D- and L-2HG in colorectal cancer cell lines.
D-2HG Induces EMT in Colorectal Cancer Cells
To examine the potential role of D- and L-2HG in colorectal cancer, these metabolites were administered as octylester derivatives to the HCT116 cell line, in which neither of the metabolites were elevated at baseline. 2HG octylesters are commonly used in place of 2HG given they can permeate across the cell membrane, to be converted by intracellular esterases to 2HG19,22. After the administration of D-2HG octylester over 20 passages, we found that the colorectal cancer cells transformed into spindle-shaped cells, which can signify the acquisition of the mesenchymal phenotype. In contrast, no obvious changes in cellular morphology were observed with the administration of L-2HG octylester (Fig. 2A). The acquisition of the mesenchymal phenotype would be of clinical significance, since this phenotype is associated with the promotion of the cancer cells’ ability to invade the local tissues and enter the bloodstream, leading to distant organ metastasis28. Indeed, HCT116 cells treated with D-2HG octylester acquired increased ability for migration (Fig. 2B) and invasion (Fig. 2C), demonstrating that D-2HG is sufficient to induce some of the cardinal features of the mesenchymal phenotype. Consistent with the above, the basal cellular migration was higher in DLD1 and RKO cells, the cells with elevated D-2HG levels in comparison to HCT116 (Fig. 2D). Cellular migration was inhibited when 2HG levels were reduced through knockdown of glutaminase, but was partially rescued by the administration of D-2HG octylester (Fig. 2E), signifying that cellular migration is also partially dependent on D-2HG.
D-2HG is sufficient and necessary to induce a mesenchymal phenotype in colorectal cancer cells.
(A) The morphology of HCT116 cells are pictured after treatment with D-2HG (250 μM) or L-2HG (250 μM) over 20 passages. (B) The migration of HCT116 cells treated with D- or L-2HG (250 μM over 20 passages) was assessed by wound healing assay at 48 hours. Scale bar: 200 μm. (C) The invasion of HCT116 cells treated with D- or L-2HG (250 μM over 20 passages) were assessed with the matrigel invasion assay. Scale bar: 400 μm. (D) The migration of colorectal cancer cell lines (HCT116, DLD1, RKO) were assessed by wound healing assay after 96 hours. (E) The migration of the DLD1 cell line by wound healing assay after siRNA interference of GLS1 and/or treatment with D-2HG octylester. Measurements were taken at 48 hours. Data are presented as means and standard deviations of at least three independent experiments; **p < 0.01.
The reduced expression of the epithelial marker E-cadherin (CDH1), with the concurrent increase in the expression of the mesenchymal markers fibronectin (FN1) or vimentin (VIM) in the HCT116 cell line, as well as in two other colorectal cancer cell lines Caco2 and HT29 (Fig. 3A–C, Supplementary Fig. S2A and B) confirmed the induction of epithelial-mesenchymal transition by D-2HG across colorectal cancer cells with different genetic background (Supplementary Table S1). These observations were not accompanied by any consistent alteration in the rate of cellular proliferation across the colorectal cancer cell lines treated by 2HG octylesters (Supplementary Fig. S2C).
D-2HG induces epithelial-mesenchymal transition in colorectal cancer cells.
(A) qPCR of epithelial marker (CDH1) and mesenchymal marker (FN1) in HCT116 cells treated with D- or L-2HG (250 μM) over 20 passages. (B) qPCR of epithelial marker (CDH1) and mesenchymal marker (VIM) in Caco-2 cells treated with D- or L-2HG (250 μM) over 4 passages. (C) qPCR of epithelial marker (CDH1) and mesenchymal marker (VIM) in HT29 cells treated with D- or L-2HG (250 μM) over 2 passages. Data are presented as means and standard deviations of at least three independent experiments; *p < 0.05, **p < 0.01.
D-2HG Directly Induces EMT in Colorectal Cancer Cells
Given that D-2HG can be metabolised to α-ketoglutarate by D2HGDH, it remained to be clarified whether the induction of EMT is a direct effect of D-2HG, or through its conversion to another metabolite (Fig. 4A). In order to resolve this ambiguity, D2HGDH was knocked down by RNA interference (Fig. 4B) to reduce the metabolisation of D-2HG and to enhance the direct effect of this metabolite. Knockdown of D2HGDH resulted in the enhancement of the induction of migration and invasion, in association with the increased expression of the mesenchymal marker in response to D-2HG octylester (Fig. 4C–E). Furthermore, knockdown of D2HGDH facilitated the reduction in the expression of the epithelial marker in response to D-2HG octylester (Fig. 4F). These findings suggest that D-2HG is directly responsible for the induction of EMT in colorectal cancer cells.
D-2HG is a direct inducer of EMT in colorectal cancer cells.
(A) D-2HG is metabolised to α-KG by D-2HG dehydrogenase (D2HGDH). (B) qPCR confirming knockdown of D2HGDH by shRNA. (C) Migration of HCT116 cells with knockdown of D2HGDH and D-2HG treatment (250 μM) over 2 passages. (D) Invasion of HCT116 cells with knockdown of D2HGDH and D-2HG treatment (250 μM) over 2 passages. (E) FN1 expression of HCT116 cells with knockdown of D2HGDH and D-2HG treatment (250 μM) over 2 passages. (F) CDH1 expression of HCT116 cells with knockdown of D2HGDH and D-2HG treatment (250 μM) over 2 passages. Data are presented as means and standard deviations of at least three independent experiments; *p < 0.05, **p < 0.01.
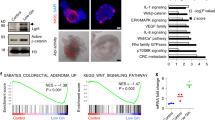
D-2HG Induces EMT by Increasing the Expression of ZEB1 and Promotes the Trimethylation of Histone 3 Lysine 4 at the Promoter Region of the ZEB1 Gene
The induction of EMT is orchestrated by transcription factors that switch on the expression of the mesenchymal genes and switch off the expression of the epithelial genes. Consistent with this, the administration of D-2HG octylester increased the expression of the ZEB1 gene, which encodes one of the key transcription factors that regulate the induction of EMT (Fig. 5A). The expression of other transcription factors that regulate EMT did not increase in response to the administration of D-2HG octylester (Supplementary Fig. S3A). Furthermore, the induction of EMT by D-2HG was demonstrated to be dependent on increased ZEB1 expression, through the complete abrogation of the alteration in CDH1 and FN1 expression when siRNA knockdown of ZEB1 was performed (Fig. 5B).
D-2HG increases the expression of ZEB1 and the trimethylation of H3K4 at the promoter region of this gene.
(A) qPCR of ZEB1 in HCT116 cells treated by D- or L-2HG (250 μM) over 20 passages. (B) qPCR of ZEB1, FN1 and CDH1 in HCT116 cells treated by D-2HG (250 μM) and siRNA knockdown of ZEB1; *p < 0.05, **p < 0.01. (C) ChIP qPCR of trimethylated H3K4 at the promoter region of ZEB1 in HCT116 cells treated by D- or L-2HG (250 μM) over 20 passages. Data are presented as means and standard deviations of at least three independent experiments; **p < 0.01.
Previous reports highlight the alteration of gene expression by D-2HG through inhibition of Jumonji histone demethylase activity, resulting in the increased levels of histone methylation at the gene promoter regions11,14. ChIP-qPCR confirmed the increase in trimethylation of histone H3 lysine 4 (H3K4) at the promoter region of the ZEB1 gene when D-2HG octylester was administered, which is a histone modification associated with increased gene expression (Fig. 5C). Alongside this, the acetylation of histone H3 lysine 9 (H3K9) and histone H3 lysine 27 (H3K27), both histone modifications associated with activation of gene expression, were also increased in the promoter region of the ZEB1 gene in response to the administration of D-2HG octylester (Supplementary Fig. S3B). In contrast, there was no obvious change in the global levels of trimethylated H3K4 or acetylated histones (Supplementary Fig. S3C). These results signify the selective nature of histone modification by D-2HG, which in this case includes the promoter region of the ZEB1 gene.
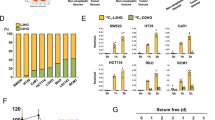
D-2HG Level is Elevated in Colorectal Cancer, and Colorectal Cancer with Higher Levels of D-2HG are Associated with Distant Metastasis
In order to examine the clinical relevance of the above findings, D-2HG levels were measured in human colorectal cancer specimens (n = 28). This demonstrated D-2HG levels to be elevated in human colorectal cancer tissues compared to non-cancerous tissues, in the absence of mutations of IDH1 or IDH2 (Fig. 6A, Supplementary Table S2). Also when the colorectal cancer specimens were divided into two groups according to D-2HG levels at the median value, colorectal cancers with higher levels of D-2HG were associated with an increased frequency of distant metastasis, as well as an increased trend for a higher T-stage (Fig. 6B, Supplementary Table S2). These findings support the observations from in vitro and suggest the significant activity of D-2HG in inducing EMT and distant metastasis in human colorectal cancer.
D-2HG is increased in colorectal cancer compared to non-cancerous tissue, and elevated D-2HG levels are associated with distant organ metastasis.
(A) D-2HG were measured in fresh-frozen colorectal cancer and paired non-cancerous tissues (n = 28) by an enzymatic assay. (B) The colorectal cancer cases were divided according to whether the D-2HG levels were above or below the median value and assessed for frequencies of distant organ metastasis and T-stage. *p < 0.05, **p < 0.01.
Discussion
In this study, we demonstrate that D- and L-2HG are elevated in some of the colorectal cancer cells, and that the D- enantiomer is directly responsible for inducing EMT through increased expression of ZEB1, a master regulator of EMT. Our data is consistent with the notion that D-2HG, an inhibitor of Jumonji-family histone demethylase11,14, induces EMT by increasing the trimethylation of H3K4 at the promoter region of the ZEB1 gene, to increase the expression of this master regulator of EMT. We also found D-2HG to be elevated in colorectal cancer, particularly in the cases associated with distant metastasis, supporting a significant role of the oncometabolite in the human disease.
Previous metabolome analyses have shown that the concentrations of a number of metabolites to be deranged in malignancies29, including in colorectal cancer30. However, it has been challenging to demonstrate whether the altered metabolite levels are merely the consequence of the disease or whether they have an active role in driving the disease process. In this study, we provide further support that deranged metabolism drives the disease process in cancer, and that this is mediated by metabolites8,31. Our findings are also consistent with an earlier report that IDH mutation can induce EMT through the production of D-2HG and the increased expression of ZEB132. Here we demonstrate the relevance and the significance of D-2HG in colorectal cancer, showing that endogenous D-2HG levels can be elevated by glutamine anaplerosis in the absence of IDH mutation, and that D-2HG levels are associated with distant metastasis.
The D-2HG concentrations measured in colorectal cancers were modest relative to cancers that harbour mutant IDH, such as leukaemias and brain tumours11,33,34,35. However our findings are consistent with the modest elevation of D-2HG reported in renal cell carcinoma, in which IDH mutation are also rare22. Importantly, colorectal cancers with higher D-2HG levels were more frequently associated with distant metastasis, consistent with the fact that even the modest increase in D-2HG is of clinical significance24.
Our study also highlights the discordant activity of the enantiomers D- and L-2HG in colorectal cancer, consistent with previous reports from other malignancies12,13. Despite L-2HG also being inhibitors of Tet2 and histone demethylases36, only the D- enantiomer of 2HG has been implicated in leukaemogenesis and astrocyte transformation12,13. This discordant activity has so far been explained by the ability of D-2HG, but not L-2HG, to stimulate the prolyl 4-hydroxylases activity of enzymes encoded by Egl nine homolog 1 (EGLN1) gene, and enhance the degradation of hypoxia inducible factor (HIF) transcription factor12,13. However, an earlier report of EMT being induced by HIF-1α in colorectal cancer37, suggested the induction of EMT by D-2HG does not occur through an axis involving HIF-1α. Indeed, the increase in the trimethylation of H3K4 being demonstrated at the promoter region of ZEB1 in response to D-2HG provided an alternative explanation. Why the histone modification occurred only in response to D-2HG and not L-2HG is open to speculation, with one explanation being that there is a yet to be identified H3K4 demethylase that is inhibited only by D-2HG, and not L-2HG, active at the promoter regions of specific genes including ZEB1.
In addition to the trimethylation of H3K4, we also observed the acetylation of H3K9 and H3K27 at the promoter region of the ZEB1 gene in response to D-2HG octylester. The acetylation of these lysine residues on histones is also associated with increased gene expression. The mechanism by which D-2HG causes histone acetylation remains to be elucidated, but may involve hitherto unreported interactions with histone acetyltransferases or histone deacetylases, or may be the knock-on effect of the H3K4 trimethylation, resulting in the recruitment of protein complexes that can alter the acetylation of neighbouring lysine residues. It would be worthwhile exploring the precise mechanisms by which D-2HG contributes to the disease process, given that epigenetic alterations can potentially be corrected in cancer, presenting with therapeutic opportunity38.
Reports from leukaemia demonstrate its dependence on elevated levels of D-2HG for oncogenesis and disease maintenance12,39, and early phase clinical trials of inhibitors against mutant IDH in leukaemia patients demonstrate provisional yet promising results40. The significance of D-2HG in colorectal cancer brought to light by the current study adds further weight to the importance of also investigating the role of IDH mutation occurring in a small subset of colorectal cancer. The same R132 IDH1 mutation that are found in leukaemia also occur in some cases of colorectal cancer41,42, but the significance of this IDH1 mutation in colorectal cancer is not yet known. We were limited in our study to using the DLD1 cell line that harbours an atypical mutation of IDH1 (G97D), because to the best of our knowledge of the literature there are no colorectal cancer cell lines that harbour the IDH1 mutation at R132, and justified in part by it being associated with high levels of 2HG nevertheless.
Cancer treatment targeting D-2HG has the potential to be both effective and safe, including for colorectal cancer. Further work is required to investigate the wider role of 2HG in colorectal cancer, including in oncogenesis. Interestingly, a recent report of 2HG being one of the very few metabolites whose levels are reduced in individuals administered aspirin43, currently the most effective drug available for chemoprevention of colorectal cancer44, tentatively suggests that D-2HG could be involved with oncogenesis of this disease. Furthermore, treatments targeting D-2HG can be envisaged to be safe, since there are no known physiological role of this metabolite45, which is normally maintained at low levels in health46.
In summary, we demonstrate D-2HG to be elevated in colorectal cancer and that it directly induces EMT in colorectal cancer cells, a phenotype associated with cellular invasion. The findings from human colorectal cancer specimens also support the significant role of D-2HG in promoting distant organ metastasis. Treatment strategies centred around reducing the levels of D-2HG or inhibiting its downstream effects in colorectal cancer could be effective.
Experimental Procedures
Cell Lines, Culture
The human colorectal cancer cell lines Caco2, DLD1, HCT116, HT29 and RKO were obtained from ATCC (VA) and the sarcoma cell line HT1080 was obtained from the Japan Cancer Research Resources Bank (Tokyo, Japan). The cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM D6046; Sigma-Aldrich, MO), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, MA), and maintained at 37 °C and 5% CO2 in a humidified incubator. Cells were reseeded every 3–4 days, prior to reaching confluency.
Reagents and Antibodies
2HG octylesters were purchased as separate enantiomers, D-2HG octylester (16366, Cayman Chemical Company, MI) or L-2HG octylester (16367, Cayman Chemical Company, MI). The antibodies used for Western blot analysis were glutaminase antibody (ab93434, Abcam, 1:000 dilution) and E-cadherin antibody (#3195, Cell Signaling Technology, 1:1000 dilution).
Cell Proliferation Assay
Cells were seeded at 1,000 cells per well in 96-well plates with 200 μL of culture medium. At the indicated time points, the cells were fixed with 25% glutaraldehyde and stained with 0.05% crystal violet/20% methanol solution. After washing away the excess crystal violet, 0.05 M NaH2PO4/50% Et-OH was added and colometric quantification was conducted with the absorbance set at 595 nm.
Small RNA Interference and Transfection
Small interfering RNA (siRNA) targeting GLS1 (NM_001256310.1, NM_014905.4) or ZEB1 (NM_001128128.2) was used, alongside nonspecific siRNA as negative control (Thermo Fisher Scientific, MA). At 50% confluency, the siRNA was transfected with Lipofectamine RNA iMax (Thermo Fisher Scientific, MA).
shRNA Construct and Lentivirus Production
Lentiviral shRNA construct, TRCN0000064034 targeting human D2HGDH was obtained from the MISSION TRC-Hs1.0 library (Sigma-Aldrich, MO). Control cells were transfected using the same procedure but with an empty control vector. HCT116 cells were selected using 1 μg/ml of puromycin over 2 weeks to establish stable cell lines.
Quantitative PCR
Total RNA was extracted from cultured cells or clinical specimens using phenol followed by precipitation (TRIzol kit, Thermo Fisher Scientific, MA). cDNA was synthesized by reverse transcription using ReverTra Ace (TOYOBO, Osaka, Japan). Quantitative PCRs was performed with the LightCycler (Roche Applied Science, Penzberg, Germany) and Thunderbird (TOYOBO, Osaka, Japan). The primer sequences used in this study are summarized in Supplementary Table S3.
Sanger Sequencing
Genomic DNA was extracted from cultured cells using a commercially available kit (QIAGEN, Hilden, Germany). Sequencing of IDH1 used primers covering amino acid residues 41–138, and sequencing of IDH2 used primers covering amino acid residues 125–226. The coding region and the intron/exon boundaries of D2HGDH and L2HGDH were also sequenced. The primers used for sequencing are detailed in Supplementary Table S4. Cycle sequencing was carried out using the BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific, MA).
Western Blot
After washing the cells in ice-cold phosphate buffer solution (PBS), cells were pelleted and total protein extracts were made by lysing in radio-immunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, MA). Cell lysates were measured for their protein concentrations using the Bradford method (BioRad, CA), after which they were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, then transferred to polyvinylidene difluoride membranes using iBlot (Life Technologies, CA). The membranes were incubated in primary antibodies overnight at 4 °C, then incubated in HRP-linked secondary antibodies (GE Healthcare Life Sciences, Little Chalfont, UK) for 1 hour at room temperature. Antigen-antibody complexes were visualized with chemiluminescence (BioRad, CA).
Flow Cytometry
E-cadherin expression was determined by flow cytometry. Cells were detached with versene and suspended in ice cold PBS with 5 μL E-cadherin-AlexaFluor 488 (#324110, BioLegend). After 1 hour of incubation, each sample was analysed using the BD FACSAriaTM II instrument (BD Bioscience).
Wound Healing Assay
Cells were seeded in 6 well plates in triplicates in numbers sufficient to become confluent within 1–2 days. A wound was then introduced using the tip of a P200 pipette, after which the cells were washed in PBS and cultured in DMEM with 1% FBS. The width of the wounds was assessed at the indicated time points, taking the mean of the narrowest and the widest points in a microscopic field.
Invasion Assay
5 × 104 HCT116 cells were seeded into the upper chamber of the 24-well BioCoat Matrigel Invasion Assay (BD Biosciences, San Jose) in triplicates in DMEM without FBS. DMEM containing 10% FBS was added to the lower chamber to induce cell invasion by chemotaxis. After 48 hours, the non-invaded cells remaining above the membrane were removed with cotton swabs, whilst the cells that had invaded through the membrane were stained using the Diff Quik kit (Sysmex, Kobe, Japan). The number of cells that had invaded through the matrigel in three random microscopic fields were counted in each chamber.
Gas Chromatography-Mass Spectrometry
Derivatisation was conducted in order to measure D- and L-2HG separately as (D and L)-0-acetyl-2-HG acid di-(D)-2-butyl esters, using GC-MS as previously described47. Disodium D-2HG (H8378, Sigma-Aldrich), Disodium L-2HG (90790, Sigma-Aldrich), and Disodium(RS)-2-hydroxy-1,5-pentanedioate-2,3,3-d3 CDN, (D-7496, C/D/N Isotopes, Pointe-Claire, Canada) were used as standards and for final quantification of the metabolites. Briefly, cell pellets stored at −80 °C were lysed in water and acidified to pH1-2 with HCl. After extraction with ethyl acetate and drying under nitrogen, tri-TMS derivatives of 2HG were made, before butylation and then acetylation. After evaporation and dissolving the extracts in chloroform, samples were injected to an Agilent 7890A GC equipped with an Agilent 5975C mass selective detector operating in splitless mode, using electron impact ionization at ionizing voltage of 70 eV and electron multiplier set to 1060 V. Helium was used for chamber gas at a flow rate of 30 cm/sec. GC temperature started at 120 °C for 3 minutes, ramped to 230 °C at 4 °C/min and held for 5 minutes, then ramped to 300 °C at 20 °C/min and held for 10 minutes. Mass range of 100–600 m/z was recorded at 2.71 scans/second.
Enzymatic Assay for D-2HG
The enzymatic assay for D-2HG (K213-100, BioVision, CA) was used according to the manufacturer’s instructions. Briefly, 10–20 mg of tissues stored at −80 °C were homogenized in the supplied buffer solution using the TaKaRa BioMasher (9791A, Clontech Laboratories, CA). The sample solution was deproteinised using a kit (K808-200, BioVision, CA) according to the manufacturer’s protocol, mixed with D-2HG substrate mix and D2HGDH enzyme and incubated at 37 °C for 1 hour, after which colometric assessment was made at OD450nm.
Clinical Tissue Samples
Colorectal cancer specimens were harvested at the time of surgical resection in the Department of Gastroenterological Surgery, Osaka University during 2011. Written informed consent was obtained from all patients regarding the use of the resected specimens in this study, and all experimental methods involving human tissues were carried out in accordance with the guidelines and regulations approved by the Ethics Committee at the Graduate School of Medicine, Osaka University (Approval Number 15222-2). None of the patients underwent neoadjuvant chemo- or radiotherapy. Samples were immediately frozen at −80 °C for storage. The specimens were analysed under the approval of the Ethics Committee at the Graduate School of Medicine, Osaka University (Approval Number 15222-2).
Statistical analysis
Data is presented as mean ± standard deviation. Students’ t-test and Fisher’s exact test were used where appropriate to determine whether differences were statistically significant using JMP Pro 12 software (SAS Institute, Cary, NC). P < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Colvin, H. et al. Oncometabolite D-2-Hydroxyglurate Directly Induces Epithelial-Mesenchymal Transition and is Associated with Distant Metastasis in Colorectal Cancer. Sci. Rep. 6, 36289; doi: 10.1038/srep36289 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Torre, L. A. et al. Global cancer statistics, 2012. CA. Cancer J. Clin. 65, 87–108 (2015).
Kim, Y. W. & Kim, I. Y. The Role of Surgery for Asymptomatic Primary Tumors in Unresectable Stage IV Colorectal Cancer. Ann. Coloproctol. 29, 44–54 (2013).
Anwar, S., Peter, M. B., Dent, J. & Scott, N. A. Palliative excisional surgery for primary colorectal cancer in patients with incurable metastatic disease. Is there a survival benefit? A systematic review. Color. Dis. 14, 920–930 (2012).
Christofk, H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011).
Pavlova, N. N. & Thompson, C. B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23, 27–47 (2016).
Hamabe, A. et al. Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA. 111, 15526–15531 (2014).
Miyo, M. et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer 113, 492–499 (2015).
Yang, M., Soga, T. & Pollard, P. J. Oncometabolites: linking altered metabolism with cancer. J. Clin. Invest. 123, 3652–3658 (2013).
Haas, R. et al. Intermediates of Metabolism: From Bystanders to Signalling Molecules. Trends Biochem. Sci. 41, 460–471 (2016).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Losman, J.-A. et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 (2013).
Koivunen, P. et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488 (2012).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Lu, C. et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012).
Wise, D. R. et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 108, 19611–19616 (2011).
Mullen, A. R. et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 7, 1679–1690 (2014).
Terunuma, A. et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest. 124, 398–412 (2014).
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 105, 18782–18787 (2008).
Montrose, D. C. et al. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev. Res. (Phila). 5, 1358–1367 (2012).
Shim, E.-H. et al. L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 4, 1290–1298 (2014).
Intlekofer, A. M. et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 22, 304–311 (2015).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation 119, 1420–1428 (2009).
Bleeker, F. E. et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 30, 7–11 (2009).
Ward, P. S. et al. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene 31, 2491–2498 (2012).
Lin, A.-P. et al. D2HGDH regulates alpha-ketoglutarate levels and dioxygenase function by modulating IDH2. Nat. Commun. 6, 7768 (2015).
Gupta, G. P. & Massagué, J. Cancer metastasis: building a framework. Cell 127, 679–695 (2006).
Kobayashi, T. et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 22, 571–579 (2013).
Nishiumi, S. et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One 7, e40459 (2012).
Shaul, Y. D. et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell 158, 1094–1109 (2014).
Grassian, A. R. et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT). J. Biol. Chem. 287, 42180–42194 (2012).
DiNardo, C. D. et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood 121, 4917–4924 (2013).
Fathi, A. T. et al. Prospective serial evaluation of 2-hydroxyglutarate, during treatment of newly diagnosed acute myeloid leukemia, to assess disease activity and therapeutic response. Blood 120, 4649–4652 (2012).
Gross, S. et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344 (2010).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Zhang, W. et al. HIF-1α Promotes Epithelial-Mesenchymal Transition and Metastasis through Direct Regulation of ZEB1 in Colorectal Cancer. PLoS One 10, e0129603 (2015).
Herman, J. G. & Baylin, S. B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 349, 2042–2054 (2003).
Kats, L. M. et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell 14, 329–341 (2014).
IDH1 inhibitor shows promising early results. Cancer Discov. 5, 4 (2015).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2, 401–404 (2012).
Liesenfeld, D. B. et al. Aspirin Reduces Plasma Concentrations of the Oncometabolite 2-Hydroxyglutarate: Results of a Randomized, Double-Blind, Crossover Trial. Cancer Epidemiol. Biomarkers Prev. 25, 180–187 (2016).
Cooper, K. et al. Chemoprevention of colorectal cancer: systematic review and economic evaluation. Health Technol. Assess. 14, 1–206 (2010).
Losman, J.-A. & Kaelin, W. G. J. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013).
Struys, E. A. et al. Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause D-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 76, 358–360 (2005).
Gibson, K. M. et al. Stable-isotope dilution analysis of D- and L-2-hydroxyglutaric acid: application to the detection and prenatal diagnosis of D- and L-2-hydroxyglutaric acidemias. Pediatr. Res. 34, 277–280 (1993).
Acknowledgements
We thank Dr. Ofusa from Idea Consultants, Inc. (Osaka, Japan) for assistance with measurement of 2HG by GC-MS. We thank the members of our laboratories for their fruitful discussions. Institutional endowments were received partially from Taiho Pharmaceutical Co., Ltd., Evidence Based Medical Research Center, Yakult Honsha Co., Ltd., Chugai Co., Ltd., and Merck Co., Ltd. [M.K., J.K., A.A., T.S., Y.D., M.M., H.I.]. This work was also supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (http://www.mext.go.jp/english/; #22130005, #25112708, #25134711, #24390315, #26670604; M.M., H.I.); a Grant-in-Aid from the Ministry of Health, Labour and Welfare (http://www.mhlw.go.jp/english/; #H23-003; M.M., H.I.); a grant from P-DIRECT (http://p-direct.jfcr.or.jp/english/; #04; H.I.); a grant from the National Institute of Biomedical Innovation (http://www.nibio.go.jp/english/index.html; #12-4;M.M., H.I.); and a grant from Osaka University Drug Discovery Funds (http://www.osaka-u.ac.jp/en/index.html; M.M., H.I.). The study was supported in part by Takeda Science and Medical Research Foundation (http://www.takeda-sci.or.jp/index.html; M.M., H.I.), Princess Takamatsu Cancer Research Fund (http://www.ptcrf.or.jp/english; M.M., H.I.), Suzuken Memorial Foundation (http://www.suzukenzaidan.or.jp; M.K.), Yasuda Medical Foundation (http://www.yasuda-mf.or.jp; N.N.), Pancreas Research Foundation (http://www.jprf.or.jp/shoreisho.html; K.K.), Nakatani Foundation (http://www.nakatani-foundation.jp; H.I.), and Nakatomi Foundation of Japan (https://www.nakatomi.or.jp/en/index.html; M.K.). H.C. received generous support from the Interdisciplinary Program for Biomedical Sciences, Osaka University and from the Japan Society for the Promotion of Science Research Fellowship.
Author information
Authors and Affiliations
Contributions
H.C., N.N., M.K., H.T., J.N., T.H., K.K, T.M., T.S., Y.D., M.M. and H.I. designed the research project; H.C., N.N. and H.I. conducted the experiments; H.C., N.N., N.H., A.A., K.T., J.K., M.M. and H.I. analysed the data; and H.C., N.N., N.H., A.A., K.T., J.K., M.M. and H.I. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Colvin, H., Nishida, N., Konno, M. et al. Oncometabolite D-2-Hydroxyglurate Directly Induces Epithelial-Mesenchymal Transition and is Associated with Distant Metastasis in Colorectal Cancer. Sci Rep 6, 36289 (2016). https://doi.org/10.1038/srep36289
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36289
This article is cited by
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
L-2hydroxyglutaric acid rewires amino acid metabolism in colorectal cancer via the mTOR-ATF4 axis
Oncogene (2023)
-
Long-term 1,2-dimethylhydrazine triggers pathological remodeling of colon mucosa through repression of sestrin2, nuclear factor (erythroid-derived 2)-like 2, and sirtuin4 stimulating mitochondrial stress and metabolic reprogramming
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Defining the landscape of metabolic dysregulations in cancer metastasis
Clinical & Experimental Metastasis (2022)
-
Gliosarcoma vs. glioblastoma: a retrospective case series using molecular profiling
BMC Neurology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.