Abstract
Expression of the TUSC2/FUS1 tumor suppressor gene in TUSC2 deficient EGFR wildtype lung cancer cells increased sensitivity to erlotinib. Microarray mRNA expression analysis of TUSC2 inducible lung cancer cells treated with erlotinib uncovered defects in the response to oxidative stress suggesting that increasing reactive oxygen species (ROS) would enhance therapeutic efficacy. Addition of the thioredoxin reductase 1 inhibitor (TXNRD1) auranofin (AF) to NSCLC cells treated with combination of TUSC2 forced expression with erlotinib increased tumor cell apoptosis and inhibited colony formation. TXNRD1 overexpression rescued tumors from AF-TUSC2-erlotinib induced apoptosis. Neutralizing ROS with nordihydroguaiaretic acid (NDGA) abrogated cell death induced by AF-TUSC2-erlotinib, indicating a regulatory role for ROS in the efficacy of the three drug combination. Isobologram-based statistical analysis of this combination demonstrated superior synergism, compared with each individual treatment at lower concentrations. In NSCLC tumor xenografts, tumor growth was markedly inhibited and animal survival was prolonged over controls by AF-TUSC2-erlotinib. Microarray mRNA expression analysis uncovered oxidative stress and DNA damage gene signatures significantly upregulated by AF-TUSC2-erlotinib compared to TUSC2-erlotinib. Pathway analysis showed the highest positive z-score for the NRF2-mediated oxidative stress response. Taken together these findings show that the combination of TUSC2-erlotinib induces additional novel vulnerabilities that can be targeted with AF.
Similar content being viewed by others
Introduction
Effective systemic delivery of tumor suppressor genes would have broad applicability in cancer therapeutics as tumor suppressor gene inactivation is the most common genetic abnormality in cancer. TUSC2 (tumor suppressor candidate 2, also known as FUS1) is a tumor-suppressor gene identified in the human chromosome 3p21.3 region, in which allele losses and genetic alterations occur early and frequently in many human cancers, including breast and lung1,2,3. Loss or reduction of TUSC2 expression has been detected in 100% of small-cell lung cancer and 82% of non-small cell lung cancer (NSCLC) cases. Restoration of TUSC2 expression significantly inhibits tumor growth and progression in mouse models4. These findings led to a Phase I clinical trial that showed safety and antitumor activity of TUSC2 nanovesicle-based systemic gene therapy administered intravenously in lung cancer patients5. TUSC2 nanovesicles combined with erlotinib, an inhibitor of activated epidermal growth factor (EGFR), synergistically inhibited tumor growth and metastases in NSCLC cells with wildtype EGFR by abrogating resistance pathways related to FGFR2 and mTOR activation6. We hypothesized that this combination would induce novel vulnerabilities in the cancer cell. One possibility was induction of vulnerabilities related to oxidative stress. A previous study of TUSC2 knock-out mice showed that TUSC2 is involved in mitochondrial function and plays a significant role in mediating stress-induced mitochondrial reactive oxygen species (ROS) in response to chronic oxidative stress7. An increase in ROS is associated with abnormal cancer cell growth and reflects a disruption of redox homeostasis due either to an elevation of ROS production or to a decline of ROS-scavenging capacity8,9,10.
Maintaining ROS homeostasis is critical for normal cell growth and survival. Cells control ROS levels by balancing ROS generation and elimination through tightly regulated ROS-scavenging systems. This includes the essential cellular antioxidant thioredoxin, a thiol-dependent electron donor system11. In addition to its role in DNA synthesis, thioredoxin directly catalyzes reduction of protein disulfides and regulates the cellular redox environment in a wide range of cellular activities12. Thioredoxin reductase 1 (TXNRD1) an NADPH-dependent oxidoreductase enzyme, best known for recycling thioredoxin to its reduced form, is overexpressed in a variety of human cancer cell lines and primary tumors, indicating its tumorigenic involvement13. Attention has indeed recently focused on developing specific inhibitors that target TXNRD114. Previous mechanistic characterization has shown that auranofin (AF), an oral, FDA-approved, lipophilic gold-containing compound prescribed for arthritis, inhibits thioredoxin reductase (TXNRD)15,16. AF interacts with the redox-active site of TXNRD thus preventing electron transfer.
Previously, we have reported a synthetic lethal interaction between the AKT pathway and TXNRD1 via the KEAP1/NRF2 antioxidant system. AF inhibits expression and phosphorylation of major effectors of the PI3K/AKT/mTOR pathway17,18. In this study, our data support a novel conceptual model in which we identify vulnerabilities in wild type EGFR NSCLC cells following treatment with the TUSC2-erlotinib combination which we found could be targeted with the repurposed drug AF.
Results
Elevated ROS level after TUSC2 and erlotinib treatment could be lethal for lung cancer cells
We recently reported that the TUSC2 gene delivered by nanovesicles combined with erlotinib inhibits lung cancer cell viability synergistically6. Array analysis was performed to identify potential vulnerabilities in lung cancer cells treated with a combination of TUSC2 nanovesicles and erlotinib. Lung cancer cell lines H157 and H1299 with doxycycline dose response inducible TUSC2 expression were developed using the Tet-On system. Measuring TUSC2 protein expression with various doses of Doxycycline treated cancer cells by western blotting indicated that TUSC2 proteins significantly increased upon Doxycycline treatments whilst β-actin remains unchanged. The TUSC2 expression was positively correlated to Doxycycline doses, shown in supplemental Fig. 1. Microarray mRNA expression analysis of TUSC2 inducible lung cancer cells treated with erlotinib, illustrated in Supplemental Figure 2A,B, uncovered defects in the response to oxidative stress including downregulation of HSPA6, IFNL2, PPP1R15A (GADD34), and GADD45B. This result suggested that increasing ROS would be lethal for the cancer cells thus enhancing therapeutic efficacy.
AF-TUSC2-erlotinib enhance tumor cell death synergistically
We hypothesized that inhibition of TXNRD1 by AF should increase cellular ROS thus increasing lethality in the presence of TUSC2-erlotinib. We next tested the effect of AF on cells treated with TUSC2-erlotinib. Cell viability was evaluated in three wild type EGFR TUSC2-deficient NSCLC cell lines, Calu-3, Calu-6 and H522. Cells were transiently transfected with TUSC2 cDNA and treated with clinically achievable concentrations of AF and erlotinib. Treatment groups were: (1) 0.5–0.6 μM AF; (2) 1 μM erlotinib; (3) 0.5–0.6 μM AF plus 1 μM erlotinib. Untreated cells and cells treated with AF, erlotinib, or AF and erlotinib were used as controls. AF alone had a minimum inhibitory effect on Calu-3 and Calu-6, but reduced viability of H522 by 50% (Fig. 1). In all three cell lines, TUSC2 forced expression reduced viability more effectively than either AF or erlotinib. TUSC2 transient expression combined with AF and erlotinib reduced viability of Calu-6, Calu-3, and H522 by 78%, 64%, and 75% respectively, which was more effective than any other tested treatments.
AF-TUSC2-erlotinib enhance tumor cell death significantly.
Wild type EGFR NSCLC Calu-3, Calu-6, and H522 were transiently transfected with 4 μg TUSC2 cDNA, treated with 0.5–0.6 μM auranofin and/or 1 μM erlotinib and assayed for viability using XTT assay. Data represent the mean of three independent experiments each in quadruplicates, and was statistically analyzed using a three-way ANOVA and two-tailed t test. After adjusting for multiple comparisons, a significant inhibitory effect was found for the TUSC2-erlotinib-AF combination compared to any other treatment. Data shown represent the mean ± SE of three independent experiments.
Next, we evaluated the effect of TUSC2-erlotinib-AF combination on two wildtype EGFR Tet -inducible TUSC2 NSCLC cell clones H157 and H1299. Doxycycline treatment was used to induce TUSC2 expression which was confirmed by western blot in every experiment. Consistent with the above results, cell death by TUSC2-erlotinib-AF was higher than TUSC2-AF, TUSC2-erlotinib, or AF-erlotinib. Viability of H1299 and H157 was reduced by over 70% and 80% respectively (Fig. 2A,B). Turkey’s multiple comparisons test was performed for pairwise differences between different treatment groups and found that TUSC2-erlotinib-AF had the highest probability of a cooperative effect (>98%).
Synergistic effect of AF-TUSC2-erlotinib combination.
(A,B) Wild type EGFR TUSC2 Tet-On stable clones H157 and H1299 were treated with 1 μg/ml doxycycline (TUSC2 expression); 2.3 μM erlotinib; 0.5 μM auranofin; 1 μg/ml doxycycline/2.3 μM erlotinib; 1 μg/ml doxycycline/0.5 μM Auranofin; 2.3 μM erlotinib/0.5 μM Auranofin; and 1 μg/ml doxycycline/1 μg/ml doxycycline and 0.5 μM auranofin for 72 hours. Untreated cells were used as control. Cell viability was assessed with XTT assay. After adjusting for multiple comparisons, a significant inhibitory effect was found for the TUSC2-erlotinib-AF combination compared to any other treatment using three-way ANOVA and two-tailed t test. Data shown represent the mean ± SE of three independent experiments. (C,D) Loewe additivity model was used at 95% confidence intervals (CI); II < 1, II = 1, and II > 1 correspond to the drug interactions’ being synergistic, additive, and antagonistic, respectively. Blue needles represent synergy; Grey represents combination of the three drugs that had interaction index (II) less than one without synergy; Black represents combination of the three drugs with an interaction index (II) above one without antagonism; Red and green surface represent antagonistic and additive effects respectively.
To further characterize the interaction between AF, TUSC2, and erlotinib, and determine its magnitude, we assessed viability at different individual dose-levels. Five cell viability measurements were taken at each different combination: Doxycycline (0, 0.4, 0.6, 0.8, 1.0, and μg/ml), Erlotinib (0, 0.6, 0.9, 1.2, 1.8, and 2.3 μM), and AF (0, 0.5, 0.75, 1, 2, and 3 μM). After subtracting background, measurements were divided by average viability at AF = 0, doxycycline = 0, and erlotinib = 0. We used the Loewe additivity model at 95% confidence intervals. Isobolograms was used to generate Interaction Index (II). II < 1, II = 1, and II >1 correspond to the drug interactions’ being synergistic, additive, and antagonistic, respectively. For the H1299 clone, the optimum synergy was observed at 0.25 μg/ml doxycycline, 0.5 μM erlotinib, and 0.5 μM AF; 0.25 μg/ml doxycycline, 0.75 μM erlotinib, and 0.5 μM AF; 0.5 μg/ml doxycycline, 0.75 μM erlotinib, and 0.5 μM AF; 0.75 μg/ml doxycycline, 0.5 μM erlotinib, and 0.75 μM AF. Similarly, for the H157 clone, optimum synergy was observed at 1 μg/ml doxycycline, 0.6 μM erlotinib, 0.5 μM AF; and at 1.2 μg/ml doxycycline, 0.6 μM erlotinib, and 0.5 μM AF. Synergy was also observed at auranofin and erlotinib concentrations between 1–3 μM. Figure 2C,D shows results represented in isobolograms for H1299 and H157 respectively. These results indicate that AF-TUSC2-erlotinib combination synergistically induces cell death of wild type EGFR Dox-inducible TUSC2 H1299 and H157 clones at nanomolar concentrations.
AF-TUSC2-erlotinib reduce tumor cell colony formation synergistically
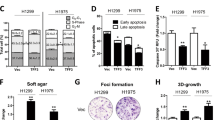
To further evaluate the biological activity of the TUSC2-erlotinib-AF combination, we tested its effect on tumor cell colony formation. In dose titration experiments at nanomolar concentrations, we found that the ability of the wild type EGFR Dox-inducible TUSC2 H1299 clone to form colonies was reduced to almost 100% (Fig. 3A). AF, TUSC2, and erlotinib, reduced colonies by 30%, 25%, and 10%, respectively. AF-erlotinib or TUSC2-erlotinb reduced colonies by 50%, whereas AF-TUSC2 treatment resulted in 80% inhibition. AF-TUSC2-erlotinib inhibited colony formation to the greatest extent (>95%) which is consistent with the observed cell viability results.
Auranofin/TUSC2/erlotinib reduce tumor cell colony formation and enhance apoptosis synergistically.
(A) H1299 Tet-inducible TUSC2 clone was grown in RPMI-1640 medium containing 400 μg/ml G418 prior to treatment with: (1) 0.4 μg/ml Doxycycline; (2) 0.2 μM Auranofin; (3) 0.8 μM erlotinib; (4) 0.4 μg/ml Doxycycline/0.2 μM Auranofin; (5) 0.2 μM Auranofin/0.8 μM erlotinib; and (6) 0.4 μg/ml Doxycycline/0.2 μM Auranofin/0.8 μM erlotinib. Survived colonies were fixed with 3.7% formalin, stained with 1% crystal violet, counted with a stereomicroscope, and analyzed with Image-J software. Values represent the mean of three independent experiments. The statistical significance of differences between treatments was calculated by using three-way ANOVA and two-tailed t test; P < 0.05 was considered significant. The cooperative effect of DC-TUSC2 and erlotinib combination was assessed using a Bayesian parametric bootstrapping approach. The Statistical software S-PLUS 8.0 was used for all analyses. (B) Following treatment of H1299 Tet-inducible TUSC2 clone with 1 μg/ml Doxycycline, 0.8 μM Auranofin, and 0.8 μM Erlotinib, cells were harvested, washed with PBS, and stained with Annexin-V (Promega, Madison, WI). Apoptotic activity was measured by flow cytometry using FlowJo software (Tree Star Corp, Ashland, OR). Values represent the mean of three independent experiments, each in quadruplicate.
AF-TUSC2-erlotinib induces apoptosis
We analyzed whether the observed synergy between AF, TUSC2, and erlotinib involves apoptosis. AF-TUSC2-erlotinib resulted in a seven fold increase in apoptosis compared to the control which was greater than each individual agent or the two agent combinations (Fig. 3B). These results suggest that apoptosis contributes to the synergistic tumor cell death by AF-TUSC2-erlotinib.
AF-TUSC2-erlotinib inhibits tumor growth and prolongs survival of xenograft mice
To investigate the efficacy of AF-TUSC2-erlotinib in vivo, eight wild type EGFR H1299-derived xenograft mice were allocated to each treatment group: DOTAP: cholesterol (DC)-empty vector; AF; DC-TUSC2 nanoparticle complex-erlotinib; and DC-TUSC2-AF-erlotinib. The DC-TUSC2-AF-erlotinib combination inhibited tumor growth more effectively than AF or TUSC2-erlotinib (Fig. 4A). The mean tumor volumes for control, AF, TUSC2-erlotinib, and AF-TUSC2-erlotinib were 600.29 ± 311.17; 353.30 ± 172.28 mm3; 456.94 ± 172.28 mm3, and 222.03 ± 98.14 mm3, respectively (p < 0.005). The posterior probability of a cooperative effect between AF, TUSC2 and erlotinib was greater than 99%, which means that there were less than 1 in 100 chance that the effect of this combination was not cooperative. DC-TUSC2-AF-erlotinib had no apparent effect on weight or change in food or water intake (Fig. 4B). DC-TUSC2-AF-erlotinib prolonged animal survival (median 44 days) compared with AF (median 32 days) or erlotinib/TUSC2 (median 36 days), shown in Fig. 4C.
Auranofin/TUSC2/erlotinib inhibits tumor growth and prolong animal survival significantly.
A subcutaneous mouse model of the human NSCLC using H1299 cells was adopted to evaluate effect the trinary combination: Auranofin plus systemic delivery of DC-based TUSC2 nanoparticles, and erlotinib. (A) Tumor volumes were calculated taking length of the longest diameter across the tumor and with width of the corresponding perpendicular diameter using the formula: length x width2 × 0.52. The rate of tumor growth inhibition was calculated as 100% × (treated tumor size/control tumor size). (B) Mice were weighed throughout the experiments. (C) Overall survival of animals with various treatments were estimated by Kaplan-Meier method. Log-rank (Mantel-Cox) test was performed to compare overall survival between TUSC2/erlotinib treated mice and TUSC2/erlotinib/Auranofin treated mice (P < 0.0001).
TRNXD1 inhibition renders cells more sensitive to TUSC2-erlotinib treatment
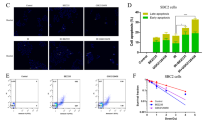
We analyzed the role of TXNRD1, the primary target of auranofin, in the observed AF-TUSC2-erlotinib synergy. TXNRD1 enzymatic activity was measured after exposure of the wild type EGFR Tet-inducible TUSC2 H1299 clone to AF-TUSC2-erlotinib. AF reduced TXNRD1 activity by 35%, which is consistent with previous reports (Fig. 5A). Doxycycline, erlotinib, and doxycycline-erlotinib slightly reduced TXNRD1 enzymatic activity. Doxycycline-AF and AF-erlotinib reduced TXNRD1 to the extent of auranofin alone whereas AF-Dox-Erlotinib treatment resulted in a 50% inhibition. TXNRD1 overexpression abrogated the ability of AF to enhance sensitization to doxycycline-TUSC2 (Fig. 5B). These results suggest that TXNRD1 inhibition renders cells more sensitive to TUSC2-erlotinib.
TRNXD1 inhibition renders cells more sensitive to TUSC2 and erlotinib treatment and involves increase in ROS levels.
(A) Thioredoxin Reductase 1 (TXNRD1) activity was measured using Thioredoxin Reductase 1 (TXNRD1) Activity Assay Kit with TXNRD1 antibody. Following treatments, Tet-inducible TUSC2 H1299cell lysates were incubated in the absence or presence of DTT (10 mM) at 37 °C in TE buffer. TXNRD1 was purified using Chroma Spin TE-10 columns and incubated with increasing concentrations of NAPQI (0.1–100 μM) or DMSO. TXNRD1/insulin mixture (50 nM purified rat liver TXNRD1, 0.5 mM NADPH, and 170 μM insulin in TE buffer) was added and changes in absorbance at 340 nm were recorded. TXNRD1 activity was calculated as the linear change in absorbance per min and expressed as a percentage of the enzyme activity of DMSO-treated control samples. (B) H1299 cells were transfected with 1 μg TXNRD1 cDNA and treated as indicated before assayed for survival with XTT assay. (C) H1299 Tet-inducible TUSC2 cells were cultured in a black 96-well culture plates and washed with HBSS before incubation in 100 μl of 1X cell-permeable fluorogenic probe 2’, 7’-Dichlorodihydrofluorescin diacetate (DCFH-DA) at 37 °C for 60 minutes. After TUSC2 induction with 1 μg doxycycline for 24 hours, cells were treated with 0.7 μM auranofin and/or 0.9 μM erlotinib for two hours. All treatment were in triplicates. Fluorescence was analyzed with a fluorometric plate reader at 480/530 nm. Hydrogen peroxide, H2O2 at 1000 μM was used a positive control. DCFH-DA treatment was 60 minutes. (D) H1299 cells were treated with 5 μmol/L NDGA before exposure to the indicated treatments and assayed for cell survival with XTT assay. TUSC2 induction by doxycycline was confirmed by western blot in all experiments. Data shown represent the mean ± SE of three independent experiments.
AF-TUSC2-erlotinib synergy is mediated through increased ROS production
We have previously reported that AF alters cellular ROS balance17. Therefore we analyzed whether AF-TUSC2-erlotinib interaction is regulated by ROS. ROS expression levels were assessed in wild type EGFR Tet-inducible TUSC2 H1299 clone after exposure to the AF-TUSC2-erlotinib combination. AF, erlotinib, doxycycline, AF/erlotinib, and doxycycline-erlotinib had a minimal effect (Fig. 5C). In contrast, compared to control, doxycycline-AF and doxycycline-AF-erlotinib enhanced ROS levels by 80% and almost 100%, respectively. To determine whether ROS is required for synergy by AF-doxycycline-erlotinib, cells we treated with ROS blocker nordihydroguaiaretic acid (NDGA; 5 μM) before exposure to the combination. As Fig. 5D shown, NDGA reversed cell growth inhibition, which indicates that AF-doxycycline-erlotinib efficacy occurs through ROS.
Gene expression profile and pathway analysis
To identify differentially expressed genes and specific pathways involved in AF-TUSC2-erlotinib synergy, we used Illumina Human HT-12V4 expression bead chip platform across the TUSC2-inducible H1299 clone. The dataset for each of the control and seven treatment groups (AF, erlotinib, doxycycline, AF-erlotinib, AF-TUSC2, erlotinib-TUSC2, and AF-TUSC2-erlotinib) were analyzed individually to generate specific sets of genes for which expression levels were up or downregulated between erlotinib-TUSC2 and AF-TUSC2-erlotinib treatments. Volcano plots and Heatmap were used to visualize the results (Fig. 6A,B). The resulting aggregate scores were used for pathway analysis based on the selected genes at a false detection rate (FDR) of 0.0001. The results show a gene profile that differentiates between erlotinib-TUSC2 and AF-TUSC2-erlotinib (Fig. 6B). Genes that are upregulated all encode proteins for oxidative stress and DNA damage (Fig. 6C). Their expression was activated by 6.389 to 82.222 fold with HMOX1, HSPA6 and FTH1 being the highest. Ingenuity pathway analysis showed the highest positive z-score for the NRF2-mediated oxidative stress response pathway (Fig. 6D). These results support an important role for antioxidant and DNA damage response gene networks in AF-TUSC2-erlotinib synergy.
Gene expression profile and pathway analysis.
Illumina array Human HT-12V4 expression bead chip platform was used across the Tet-inducible TUSC2 H1299 clone treated with auranofin-doxycycline-erlotinib vs doxycycline-erlotinib. Each treatment group was in triplicates. Beta-Uniform Mixture models were used to adjust for multiple comparisons. Significant genes were identified by One Way ANOVA, and Tukey’s HSD tests were used to analyze pairwise comparisons. Fold change larger than 2 or smaller than −2 were considered as significant at p < 0.05. (A) Volcano plot: 164 genes marked in red at FDR of 0.0001 were selected. (B) Heatmap: Genes with FDR of 0.0001 and fold change >2 were considered as significant genes. doxycycline-erlotinib: gray, auranofin-doxycycline-erlotinib: yellow. (C) List of top genes significantly upregulated between binary and trinary combination treatment. (D) Ingenuity pathway analysis Pathway analysis based on statistical significance and strength of association with extracts revealed the highest positive z-score for NRF2-mediated oxidative stress response.
Discussion
Molecular vulnerabilities in lung cancer are frequently not targetable by existing drugs. We identified a gene delivery strategy that creates vulnerabilities in pathways susceptible to existing drugs. Restoration of the TUSC2/FUS1 tumor suppressor gene, which also functions as a multikinase inhibitor, enhances killing of EGFR wild type human lung cancer cells by the EGFR tyrosine kinase inhibitor erlotinib4,6,19. TUSC2/erlotinib is currently in Phase II clinical trials in stage 4 NSCLC patients who are EGFR wildtype and have tumor progression on other treatments5. In this report, we found that addition of thioredoxin reductase 1 (TXNRD1) inhibitor auranofin to the TUSC2/erlotinib combination synergistically enhanced cancer cell death by induction of apoptosis through an increase in ROS. We tested several wild type EGFR NSCLC lines with different histologies and genetic backgrounds to determine whether the observed effects were specific to certain cells or genomic subtypes. These cell lines have low to absent levels of TUSC2 expression. Restoration of TUSC2, both transient and stable, sensitized these cell lines to erlotinib, an effect that was clearly enhanced after AF addition at clinically achievable concentrations. The efficacy of AF-TUSC2-erlotinib in vitro was validated in vivo in mice bearing NSCLC tumor xenografts.
A quantitative isobologram statistical model was used to characterize the interactions among AF, TUSC2, and erlotinib. We combined dose-response curves for each combination with one, two, and three drugs, and compared multiple concentrations in the nanomolar range. AF, TUSC2, or erlotinib each produced similar tumor cell killing effects, although that of TUSC2 was slightly better. When combined at low doses, efficacy was statistically superior for AF-TUSC2-erlotinib, compared to that of the individual effects indicating a departure from additivity, which indicates synergy.
TXNRD1 is the only enzyme known to catalyze the reduction of thioredoxin and hence is a central component in regulation of reactive oxygen species11,12. Expression and activity of TXNRD1 has been reported to be upregulated in various tumors, and interconnection between its inactivation and cell growth inhibition and apoptosis has been shown13,14. AF, alone, or combined with TUSC2-forced expression or erlotinib reduced TRND1 enzymatic activity at different potency levels, however the three agent combination was the most effective. TXNRD1 overexpression rescued cells from AF/TUSC2/erlotinib induced cell death, which indicates that this enzyme plays a functional role in the combined efficacy. Reducing ROS with NDGA abrogated the effect of AF on TUSC2-erlotinib. Thus, it is conceivable that AF improves sensitivity of wild type EGFR NSCLC to TUSC2-erlotinib by mediating increased ROS via inhibition of TXNRD1 in cells rendered less responsive to oxidative stress.
Microarray mRNA expression analysis revealed an oxidative stress and DNA damage gene signature differentially regulated between TUSC2-erlotinib and auranofin-TUSC2-erlotinib combination treatments. Ten genes in encoding proteins of iron and oxygen metabolism were significantly upregulated: HMOX1, HSPA6, FTH1, FTH1P8, FTH1P3, FTH1P11, OSGIN1, FTH1P12, PLK2, and FTH1P2. Pathway analysis showed the highest positive z-score for NRF2-mediated oxidative stress response. Nrf2 is a transcriptional regulator that targets heme oxygenase-1 (HMOX1), ferritin (FTH) genes, oxidative Stress induced growth inhibitor 1 (OSGIN1), and TXRD1. These genes function to protect cells from oxidative damage during stress and are implicated in cancer initiation, progression and inhibition. In NSCLC patients, NRF2 is frequently deregulated and may effect tumor initiation and progresson19,20. These results are consistent with our recent report of synthetic lethality between the TXNRD1 and AKT pathways which occurred through the KEAP1/NRF2 cellular antioxidant axis17. NRF2 controls expression and activity of TXNRD1 and is activated by the ROS and AMP-activated protein kinase (AMPK) signaling system, which has emerged in recent years as a regulator of the redox-state of the cell21,22. AMPK activation was proposed to sensitize NSCLC cells to erlotinib23. It is worth noting that TUSC2 directly activates AMPK allowing AMPK to drive cell dearth more effectively5,24.
In this paper we show that the combination of TUSC2 and erlotinib induces additional novel vulnerabilities that can be targeted with an approved drug, auranofin. Thus interrogation of genetically modified lung cancer cells may reveal novel vulnerabilities targetable by existing drugs. A clinical trial showed that the TUSC2 gene can be delivered intravenously in patients using nanovesicles and achieves sufficient TUSC2 protein expression levels in tumors to mediate response5. Thus the combination of these drugs is feasible in future clinical trials for patients with wild type EGFR.
Materials and Methods
Cell Culture
Wild type EGFR human non-small cell lung adenocarcinoma cell lines with low or absent TUSC2 expression, H1299, H522, and H157 were provided by Drs. John V. Heymach (MD Anderson Cancer Center), Adi Gazdar and John D. Minna (The University of Texas Southwestern Medical Center at Dallas). Calu-3 and 6 cells were purchased from ATCC I (Manassas, VA). Cells were maintained in RPMI-1640 medium (Manassas, VA), supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA).
Reagents
Erlotinib was obtained from the clinical pharmacy at the University of Texas MD Anderson Cancer Center (MDACC, Houston, TX). Auranofin, NDGA, and Doxycycline, were purchased from Sigma-Aldrich Corporation (Saint Louis, MO). TXNRD1 activity assay kit was purchased from abcam, (Cambridge MA). TUSC2 polyclonal antibody was developed in Bethyl Laboratories, Montgomery, TX. ROS kit was purchased from Cell Biolabs Inc (San Diego, CA). Lipofectamine 2000 was purchased from Invitrogen Corporation (Carlsbad, CA). DOTAP: Cholesterol (DC)–TUSC2 complexes were made as previously described4.
Stable Tet-inducible TUSC2 Cell Lines
Tet-inducible TUSC2 expressing cells were produced using Lenti-X Tet-On advanced inducible expression system (Clontech, Mountain View, CA) according to the manufacturer’s instructions. Briefly, H1299, and H157 cells were infected with lentivirus generated with the pLVX-Tet-On advanced vector to constitutively express the tetracycline-controlled transactivator. After G418 selection, surviving colonies were expanded and infected with pLVX-Tight-Puro-Luc. New colonies were selected and expanded under G418 and puromycin. Surviving clones were selected after exposure to 2 μg/ml doxycycline for 48 hours, and assessed for TUSC2 expression by Western blotting.
Cell viability assay
We used 2,3-bis(2-methoxy-4-nitro-5-sulfophenly)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) assay to evaluate overall treatment effect on cell viability. Calu3, Calu 6, and H522 were transfected with 4 μg TUSC2 cDNA before treatment with 0.8 μM erlotinib and/or 0.5 μM AF. Combined treatments for TUSC2 stable clones H1299 and H157 were: 1 μg/ml doxycycline plus 1 μM erlotinib; 1 μg/ml doxycycline and 0.5 μM AF; 1 μM erlotinib and 0.5 μM AF; and 1 μg/ml doxycycline and 0.5 μM AF and 1 μM erlotinib for 72 hours. Several other concentrations at nanomolar ranges of AF, doxycycline, erlotinib were used to test for synergy. Untreated cells and cells treated with AF, doxycycline, or erlotinib served as controls. The statistical significance of differences between treatments was calculated using three-way ANOVA and two-tailed t test; P < 0.05 was considered significant. The percentage of viable cells was determined by the ratio of absorbance of treatment and control groups: ODT/ODC × 100% from three independent experiments, each in triplicates. Average of three cell viability measurements were calculated after subtracting background and divided by average cell viability at AF = 0, doxycycline = 0, and erlotinib = 0.
Colony Formation Assay
H1299 Tet-inducible TUSC2 clone was grown in RPMI-1640 medium containing 400 μg/ml G418 prior to treatment with: (1) 0.4 μg/ml doxycycline; (2) 0.2 μM AF; (3) 0.8 μM erlotinib; (4) 0.4 μg/ml doxycycline/0.2 μM AF; (5) 0.2 μM AF/0.8 μM erlotinib; and (6) 0.4 μg/ml doxycycline/0.2 μM AF/0.8 μM erlotinib. Survived colonies were fixed with 3.7% formalin, stained with 1% crystal violet, counted with a stereomicroscope, and analyzed with Image-J software. Values represent the mean of three independent experiments.
Flow Cytometry/apoptosis assays
Following treatment of Tet-inducible TUSC2 H1299 clone with 1 μg/ml doxycycline, 0.8 μM AF, and 0.8 μM erlotinib, cells were harvested, washed with PBS, and stained with Annexin-V (Promega, Madison, WI). Apoptotic activity was measured by flow cytometry using FlowJo software (Tree Star Corp, Ashland, OR). Values represent the mean of three independent experiments, each in quadruplicate.
Animal experiments and analysis of tumor growth
Protocols were approved by the Ethics of Animal Experiments committee, The University of Texas MD Anderson Cancer Center, in accordance with NIH Guidelines for the Care and Use of Laboratory Animals. Six to eight week old Balb/c athymic nude mice female were purchased from Harlan Sprague Dawley (Indianapolis, IN). Animals were inoculated subcutaneously with wild type EGFR TUSC2-deficient human H1299 cells (3 × 106). When tumors reached an average volume of 0.1 cm3, mice were randomly divided into four treatment groups (n = 8 animals per group): (1) PBS control; (2) AF (10 mg/kg) delivered once daily five times per week via i.p. injection for two weeks; (3) Erlotinib (30 mg/kg) orally feed daily with a total of 8 times, and intravenous injections with DC-TUSC2 complexes at a dose of 25 μg of plasmid DNA, 10 nmol DC in 100 μL of 5% dextrose in water every 48 hours for a total of three injections; and (4) AF-TUSC2-Erlotinib combination. Mice were sacrificed when tumors reached a size of 1.5-cm diameter. Evaluation of tumor size was made without knowledge of the treatment groups. Tumor volumes were analyzed at day 0, 3, 8, 14, 16, and 19, by measuring the longest diameter across the tumor and its corresponding perpendicular diameter (Length × Width2 × 0.52). Data was analyzed independently by the biostatistician.
Illumina gene expression
Total RNA was extracted from Tet-inducible TUSC2 H1299 clone in treatments groups using RNeasy plus micro kits (Qiagen). Quality was assessed with a 2100 Bioanalyzer (Agilent). RNA (300 ng) was reversed transcribed to generate amplified biotinylated cRNA according to Eberwine procedure using Illumina TotalPrep RNA Amplification kit. cRNA (750 ng) was hybridized overnight to Illumina HT-12 Bead Arrays, washed and stained with streptavidin-Cy3 (Amersham-Pharmacia Biotech) according to the Illumina protocol. Arrays were scanned on a Bead Array Reader (Illumina). Raw measurements of the intensity of each bead were captured directly and processed as “bead-level”. All measurements were processed as “probe-level” data by GenomeStudio software (Illumina). One way ANOVA was used to identify the differential expressed genes among treatment groups. Beta-Uniform Mixture models were used to adjust for multiple comparisons. For the significant genes identified by One Way ANOVA, Tukey’s HSD Tests were used to do the post hoc analysis for pairwise comparisons. Each group had three replicates. Treatment groups were compared with controls and each binary combination was compared with AF alone. Finally, combinations of AF-TUSC2-erlotinib and TUSC2-erlotinib were compared to each other. Genes with p < 0.05 (by Tukey’s HSD tests) and fold change larger than 2 or smaller than −2 were considered as differentially expressed. Volcano plots and Heatmap for each comparison was generated using the R gplots program. Ingenuity pathway analysis based on statistical significance and strength of association with extracts was used for z-score.
TXNRD1 assay
Thioredoxin Reductase 1 (TXNRD1) activity was measured using Thioredoxin Reductase 1 (TXNRD1) Activity Assay Kit according to the instructions (abcam, Cambridge MA). Briefly, the assay uses a 96-well plate with an antibody specific to TXNRD1 to isolate the enzyme pre-coated onto the wells. Following treatments, Tet-inducible TUSC2 H1299 cell lysates were prepared and incubated in the absence or presence of DTT (10 mM) at 37 °C in TE buffer for 15 min. Then, TXNRD1 was purified using Chroma Spin TE-10 columns to remove DTT. Aliquots of purified TXNRD1 (final concentration 1 μM) were incubated with increasing concentrations of NAPQI (0.1–100 μM) or DMSO in a final volume of 100 μl of TE buffer at room temperature. After 30 min, 100 μl of a TXNRD1/insulin mixture (50 nM purified rat liver TXNRD1, 0.5 mM NADPH, and 170 μM insulin in TE buffer) was added and changes in absorbance at 340 nm were recorded. TXNRD1 activity was calculated as the linear change in absorbance per min and expressed as a percentage of the enzyme activity of DMSO-treated control samples.
ROS Intracellular Activity
Tet-inducible TUSC2 H1299 cells were cultured in a black 96-well culture plate for 24 hours and washed with HBSS before incubation in 100 μl of 1X cell-permeable fluorogenic probe 2’, 7’-Dichlorodihydrofluorescin diacetate (DCFH-DA) at 37 °C for 60 minutes. Cells were treated with 1 μg/ml doxycycline for 24 hours to induce TUSC2 and exposed to 0.7 μM auranofin and/or 0.9 μM erlotinib for two hours. All treatments were in triplicates. Fluorescence was analyzed with a fluorometric plate reader at 480/530 nm. Hydrogen peroxide, H2O2, at 1000 μM was used a positive control.
Statistical analysis
Statistical analysis was performed with SAS 9.4 (SAS Institute Inc., Cary, North Carolina, USA). All data are presented as mean ± SD. The statistical significance of differences between treatments was tested by using three-way ANOVA and two-tailed t test; P < 0.05 was considered significant. The magnitude of drug interaction being synergistic, additive, or antagonistic was assessed by The Loewe additivity model25. CalcuSyn was used to generate median effect plot and 3D isobologram. S-plus 8.2 (TIBCO Software Inc., Palto Alto, CA) was used to calculate Interaction Index (II) and its 95% confidence interval. The cooperative effect of DC-TUSC2 and erlotinib and auranofin combination was assessed using a Bayesian parametric bootstrapping approach. The Statistical software S-PLUS 8.2 and SAS 9.4 were used for all analyses. Treatment effect in animal model was measured by the mean tumor volume on a log-scale in each treatment arm. The treatment effects of 4 groups were compared by ANOVA and pairwise mean comparisons were assessed with and without Turkey’s adjustment. Overall survival of animals with various treatments was estimated by Kaplan-Meier method. Log-rank (Mantel-Cox) test was performed to compare overall survival between different treatments.
Additional Information
How to cite this article: Xiaobo, C. et al. TUSC2(FUS1)-erlotinib Induced Vulnerabilities in Epidermal Growth Factor Receptor(EGFR) Wildtype Non-small Cell Lung Cancer(NSCLC) Targeted by the Repurposed Drug Auranofin. Sci. Rep. 6, 35741; doi: 10.1038/srep35741 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Wistuba, I. I. et al. High Resolution Chromosome 3p Allelotyping of Human Lung Cancer and Preneoplastic/Preinvasive Bronchial Epithelium Reveals Multiple, Discontinuous Sites of 3p Allele Loss and Three Regions of Frequent Breakpoints. Cancer Research 60, 1949–1960 (2000).
Ji, L., Minna, J. D. & Roth, J. A. 3p21.3 tumor suppressor cluster: prospects for translational applications. Future Oncology 1, 79–92 (2005).
Lerman, M. I. & Minna, J. D. The 630-kb Lung Cancer Homozygous Deletion Region on Human Chromosome 3p21.3: Identification and Evaluation of the Resident Candidate Tumor Suppressor Genes. Cancer Research 60, 6116–6133 (2000).
Ito, I. et al. Liposomal vector mediated delivery of the 3p FUS1 gene demonstrates potent antitumor activity against human lung cancer in vivo. Cancer Gene Ther 11, 733–739 (2004).
Lu, C. et al. Phase I Clinical Trial of Systemically Administered TUSC2(FUS1) Nanoparticles Mediating Functional Gene Transfer in Humans. Plos One 7, e34833 (2012).
Dai, B. et al. Exogenous Restoration of TUSC2 Expression Induces Responsiveness to Erlotinib in Wildtype Epidermal Growth Factor Receptor (EGFR) Lung Cancer Cells through Context Specific Pathways Resulting in Enhanced Therapeutic Efficacy. Plos One 10, e0123967 (2015).
Yazlovitskaya, E. M., Voziyan, P. A., Manavalan, T., Yarbrough, W. G. & Ivanova, A. V. Cellular oxidative stress response mediates radiosensitivity in Fus1-deficient mice. Cell Death Dis 6, e1652 (2015).
Ray, P. D., Huang, B.-W. & Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cellular Signalling 24, 981–990 (2012).
Gupta, S. C. et al. Upsides and Downsides of Reactive Oxygen Species for Cancer: The Roles of Reactive Oxygen Species in Tumorigenesis, Prevention, and Therapy. Antioxidants & Redox Signaling 16, 1295–1322 (2011).
Storz, P. Oxidative Stress in Cancer. In Oxidative Stress and Redox Regulation (eds Jakob, U. & Reichmann, D. ) 427–447 (Springer Netherlands, Dordrecht, 2013).
Arnér, E. S. J. & Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. European Journal of Biochemistry 267, 6102–6109 (2000).
Lu, J. & Holmgren, A. The thioredoxin antioxidant system. Free Radical Biology and Medicine 66, 75–87 (2014).
Karlenius, T. C. & Tonissen, K. F. Thioredoxin and Cancer: A Role for Thioredoxin in all States of Tumor Oxygenation. Cancers 2, 209 (2010).
Fulvio, S. et al. Thioredoxin Reductase and its Inhibitors. Current Protein & Peptide Science 15, 621–646 (2014).
Zou, T., Lum, C. T., Lok, C.-N., Zhang, J.-J. & Che, C.-M. Chemical biology of anticancer gold(iii) and gold(i) complexes. Chemical Society Reviews 44, 8786–8801 (2015).
Furst, D. E. Mechanism of Action, Pharmacology, Clinical Efficacy and Side Effects of Auranofin: An Orally Administered Organic Gold Compound for the Treatment of Rheumatoid Arthritis. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 3, 284–296 (1983).
Dai, B. et al. KEAP1-Dependent Synthetic Lethality Induced by AKT and TXNRD1 Inhibitors in Lung Cancer. Cancer Research 73, 5532–5543 (2013).
Li, H. et al. Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells. (2015).
Tian, Y. et al. Emerging roles of Nrf2 signal in non-small cell lung cancer. Journal of Hematology & Oncology 9, 14 (2016).
Jaramillo, M. C. & Zhang, D. D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes & Development 27, 2179–2191 (2013).
Gorrini, C., Harris, I. S. & Mak, T. W. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12, 931–947 (2013).
Hayes, J. D. & Dinkova-Kostova, A. T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences 39, 199–218 (2014).
Praveen, P., Hülsmann, H., Sültmann, H., Kuner, R. & Fröhlich, H. Cross-talk between AMPK and EGFR dependent Signaling in Non-Small Cell Lung Cancer. Scientific Reports 6, 27514 (2016).
Meng, J. et al. The Tumor Suppressor Gene TUSC2 (FUS1) Sensitizes NSCLC to the AKT Inhibitor MK2206 in LKB1-dependent Manner. Plos One 8, e77067 (2013).
Yadav, B., Wennerberg, K., Aittokallio, T. & Tang, J. Searching for Drug Synergy in Complex Dose–Response Landscapes Using an Interaction Potency Model. Computational and Structural Biotechnology Journal 13, 504–513 (2015).
Acknowledgements
This work was supported in part by the National Institutes of Health/National Cancer Institute through The University of Texas MD Anderson Cancer Center’s Cancer Center Support Grant CA-016672 - Lung Program and Shared Core Facilities, Specialized Program of Research Excellence (SPORE) Grant CA-070907 (J. Minna and J. A. Roth), Lung Cancer Moon Shot Program, The University of Texas MD Anderson Cancer Center, and by a sponsored research agreement from Genprex, Inc. (R. Mehran).
Author information
Authors and Affiliations
Contributions
X.C., M.F. and R.S. carried out experiments, data collection and analysis; M.M. Scientific supervision and review; J.S. and V.B. statistics analysis of in vitro and in vivo data; Y.Z. and W.J. bioinformatics analysis; J.A.R. is PI; R.M., B.F. and L.J. collaborators in experimental design; J.A.R. and M.M. performed data analysis and co-wrote the manuscript.
Ethics declarations
Competing interests
Jack A. Roth is a consultant, stock owner (including pending patent) in Genprex, Inc., and Reza Mehran has a sponsored research agreement with Genprex, Inc. All other authors have declared that no competing interests exist. This does not alter the authors’ adherence to all policies of the Scientific Reports-Nature on sharing data and materials.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xiaobo, C., Majidi, M., Feng, M. et al. TUSC2(FUS1)-erlotinib Induced Vulnerabilities in Epidermal Growth Factor Receptor(EGFR) Wildtype Non-small Cell Lung Cancer(NSCLC) Targeted by the Repurposed Drug Auranofin. Sci Rep 6, 35741 (2016). https://doi.org/10.1038/srep35741
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35741
This article is cited by
-
Mitochondrial Fus1/Tusc2 and cellular Ca2+ homeostasis: tumor suppressor, anti-inflammatory and anti-aging implications
Cancer Gene Therapy (2022)
-
The gold complex auranofin: new perspectives for cancer therapy
Discover Oncology (2021)
-
miR-125b-5p inhibits cell proliferation, migration, and invasion in hepatocellular carcinoma via targeting TXNRD1
Cancer Cell International (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.