Abstract
Post-transcriptional and post-translational modifications play a major role in Plasmodium life cycle regulation. Lysine methylation of histone proteins is well documented in several organisms, however in recent years lysine methylation of proteins outside histone code is emerging out as an important post-translational modification (PTM). In the present study we have performed global analysis of lysine methylation of proteins in asexual blood stages of Plasmodium falciparum development. We immunoprecipitated stage specific Plasmodium lysates using anti-methyl lysine specific antibodies that immunostained the asexual blood stage parasites. Using liquid chromatography and tandem mass spectrometry analysis, 570 lysine methylated proteins at three different blood stages were identified. Analysis of the peptide sequences identified 605 methylated sites within 422 proteins. Functional classification of the methylated proteins revealed that the proteins are mainly involved in nucleotide metabolic processes, chromatin organization, transport, homeostatic processes and protein folding. The motif analysis of the methylated lysine peptides reveals novel motifs. Many of the identified lysine methylated proteins are also interacting partners/substrates of PfSET domain proteins as revealed by STRING database analysis. Our findings suggest that the protein methylation at lysine residues is widespread in Plasmodium and plays an important regulatory role in diverse set of the parasite pathways.
Similar content being viewed by others
Introduction
Plasmodium falciparum, a protozoan parasite responsible for the severe form of human malaria, has a complex life cycle in two hosts, namely Anopheles mosquito and human. Parasites in these two hosts invade different cell types and propagate in distinct microenvironments. Although transcriptional regulation plays an important role in helping the parasite to adapt to distinct environments, however relatively few regulatory motifs and transcriptional regulators have been reported in Plasmodium so far1,2,3. Evidences are emerging to suggest that post-translational modifications (PTMs) play an important role in regulation of fundamental processes of Plasmodium growth and host invasion- including cell signaling and epigenetic control of gene regulation. Protein trafficking and interactions between various PTMs are the two very important processes that fine-tune the functions of several Plasmodium proteins4. Although several PTMs- such as phosphorylation, acetylation, palmotylation, ubiquitylation and lipidation have been identified in Plasmodium, however, only phosphorylation/dephosphorylation have been studied extensively5,6,7,8,9,10,11.
In the recent years, methylation of proteins has been ranked as the fourth common post-translational modification12 and is of common occurrence in human, Saccharomyces cerevisiae and Trypanosomes13,14,15,16. Protein methylation is mainly found on lysine and arginine residues, although there are reports of methylation of histidine and glutamic acid too17. Methylation, particularly lysine methylation is a well-studied phenomenon in histones, which involves addition of one to three methyl groups on the amino acid’s amine group to form mono, di or tri-methyllysine18. Histone lysine methylation is involved in transcriptional activation and silencing. The process is regulated by histone lysine methyltransferases (HKMTs) and histone lysine demethylases19. Recent proteome-wide lysine methylation studies indicate that the modifications also occur in non-histone proteins such as proteins linked to RNA processing, ribosome assembly, trafficking and signaling20,21.
Among the apicomplexan parasites, Plasmodium and Toxoplasma have orthologs of several chromatin remodeling proteins and enzymes responsible for protein methylation and acetylation22,23. In P. falciparum, the histone posttranslational modifications, mainly acetylation and methylation have been shown to play significant role(s) in red blood cell invasion and in virulence gene regulation24,25. Ten SET domain containing histone lysine methyltransferases (HKMTs), three histone-demethylase orthologs of lysine-specific demethylases (LSD1) and jumonji-C histone demethylases (jHDM) families have been described in Plasmodium. These proteins are the targets for novel drug development as the proteins show low sequence similarity to corresponding human counterparts22,26.
To understand the extent of lysine methylation in blood stage forms of Plasmodium falciparum, we analyzed the reactivity of anti-mono/dimethyl lysine and anti-trimethyl lysine antibodies with intact asexual blood stage Plasmodium parasites and further immunoprecipitated the Plasmodium lysates from the three blood stages, using these antibodies. Intriguingly, the LC-MS/MS analysis of the immunoprecipitates identified several non-histone methylated Plasmodium proteins linked with diverse functions such as transport, hemostatic processes and chromosome organization. These results suggest an important role of protein lysine methylation in regulation of various P. falciparum biological processes.
Materials and Methods
Plasmodium falciparum culture
Plasmodium falciparum 3D7 was cultured in complete RPMI (1640 (Invitrogen Corporation, USA), 50 mg/L hypoxanthine (Sigma Aldrich Co., USA), 0.5 g/L Albumax I (Gibco, Thermofisher Scientific Inc., USA) and 2 g/L sodium bicarbonate (Sigma Aldrich Co., USA) using O+ human erythrocytes (4% haematocrit) under mixed gas (5% O2, 5% CO2 and 90% N2). Cultures were synchronized with 5% sorbitol for at least two successive cycles and harvested using saponin treatment.
Immunofluorescence assay
Thin smears were prepared from Plasmodium falciparum culture at ring, trophozoite and schizont stages. The glass slides were air dried and fixed with pre-chilled absolute methanol for 30 minutes. The smears were incubated with 4% BSA in PBS for 2 h at room temperature (RT) to block the non-specific binding. After washing with PBS, the smears were probed with anti-mono/dimethyl lysine polyclonal antibody (abcam; ab23366)(1:20) for overnight at 4 °C, followed by Alexa-Fluor 488 conjugated goat anti-rabbit IgG antibody (A11008; Thermofisher Scientific Inc. USA) (1:200) for 1 h at RT. The slides were washed and mounted with 4′,6-diamidino-2-phenylindoledihydrochloride (DAPI, Molecular Probes, USA) antifade solution (Molecular Probes, USA). Images were captured using a Nikon A1-R confocal microscope.
Preparation of parasite lysate, Immunoprecipitation and Western blot analysis
Immuno-precipitation was performed using Pierce® Crosslink Immunoprecipitation Kit (Thermofisher Scientific Inc., USA) according to manufacturer’s protocol. Briefly, parasite cell lysates from synchronized cultures of three asexual stages (ring, trophozoite and schizont) were prepared using IP lysis buffer. The stage specific cell lysates were incubated overnight at 4 °C with anti-mono/dimethyl (abcam; ab23366) or anti-trimethyl lysine (Immunechem Pharmaceutical Inc.; ICP0601) antibodies, cross-linked with Protein A/G PlusAgarose by DSS cross-linker. The antibody cross-linked resin was washed with TBS followed by two washes with lysis buffer. Finally, the resin was washed with conditioning buffer and antibody bound proteins were eluted with elution buffer.
Also, the peptides obtained after trypsin digestion of trophozoite stage parasite lysates were lyophilized, desalted and incubated overnight at 4 °C with anti-mono/dimethyl and anti-trimethyl lysine antibodies cross-linked with Protein A/G Plus agarose. The eluted peptides were lyophilized and separated into 12 fractions using hydrophilic liquid interaction chromatography (HILIC) over one hour. Each fraction was separately analyzed on LC-MS/MS.
For western blot analysis, equal amount (50 μg) of total protein from three stages were resolved on 10% SDS-PAGE and transferred onto PVDF membrane (Merck Millipore, Merck KGaA, USA) pre-activated with methanol. The membrane was blocked with 4% BSA and incubated with polyclonal anti-methyllysine antibodies (ImmuneChem Pharmaceuticals Inc, Canada, ICP0601; 1:50) overnight at 4 °C. After three washings with PBST, the membrane was incubated with HRP conjugated anti-rabbit IgG secondary antibody (Sigma Aldrich Pvt. Ltd., USA) for 1 h at RT. The membrane was developed and visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermofisher Scientific Incorporation, USA) as per manufacturer’s instructions.
For confirmation of proteins in the IP experiments, a fraction of IP eluates pulled using anti-methyllysine antibodies were boiled in 4X sample loading buffer, separated on 10% SDS-PAGE gels and transferred to a nitro-cellulose membrane. The membrane was blocked with blocking buffer (ODYSSEY infrared imaging systems; LI-COR) and probed with protein specific antisera (anti-PfP12; a 6-cysteine protein and anti-PfTSN; Tudor Staphylococcal Nuclease) using anti-mice IRDye 800CW secondary antibodies (LI-COR). Protein bands were imaged in ODYSSEY Infrared Imager (LI-COR).
Tryptic digestion and LC-MS/MS analysis
Eluted proteins from immunoprecipitation were subjected to in-solution digestion. Samples were subjected to subsequent reduction and alkylation of disulfide bonds with 10 mM dithiothreitol (DTT) and 40 mM iodoacetamide for 1 hr. at room temperature. Digestion was performed using trypsin (1:50, enzyme: total protein) (Promega corporation, USA) for overnight at 37 °C and stopped by adding 0.1% trifluoroacetic acid. The samples were cleared by centrifugation at 10000 rpm for 10 min. The digested proteins were concentrated using Speed Vac (Thermofisher Scientific Inc., USA) and analyzed on Orbitrap Velos Pro mass spectrometer coupled with nano-LC 1000 (Thermofisher Scientific Inc., USA). The peptide mixtures were loaded on to a reverse phase C-18 pre-column (Acclaim PepMap, 75 μm × 2 cm, 3 μm, 100 A°, Thermofisher Scientific Inc., USA), in line with an analytical column (Acclaim PepMap, 50 μm × 15 cm, 2 μm, 100 A°). The peptides were separated using a gradient of 5% to 50% of solvent B (0.1% formic acid in 95/5 acetonitrile/water) in 180 min for immunoprecipitates and 120 min for HILIC fractions. The peptides were analyzed in data dependent mode where the precursors were acquired (MS) in Orbitrap at a resolution of 60000 and a minimum of 1000 counts were needed to trigger the MS/MS. Top 20 precursors were allowed to fragment using CID (collision induced dissociation) in Ion trap with collision energy of 35 for the immunoprecipitates. For HILIC fractions, precursors were fragmented using high-energy collision dissociation (HCD) and detected in Orbitrap at a resolution of 7500. Charge state screening of precursors and monoisotopic precursor selection was enabled. Unassigned and singly charged ions were rejected. The parent ions once fragmented were excluded for next 50 s with exclusion mass width of ±10 ppm. The lock mass (m/z 445.120024) enabled the accurate measurement in MS for immunoprecipitated samples and in both MS as well as MS/MS for HILIC fractions. The acquired spectra were analyzed using SEQUEST algorithm in Proteome Discoverer (PD; version 1.4) software, with a precursor tolerance of 20 ppm and tolerance of 0.6 Da for MS/MS for CID and 0.1 Da for HCD against P. falciparum database version 10 downloaded from PlasmoDB27. Carbamidomethyl (C), Deamidation (NQ) and mono-methyl, di-methyl and trimethyl (K) were set as variable modifications and five missed cleavages were allowed. The resultant identified peptides were validated using Percolator at 5% False Discovery Rate (FDR) (q value < 0.05), which uses PEP (Posterior Error Probability) and q value for validations.
Lysine methylation motif analysis
The motif search included six amino acid residues N-terminal and C-terminal to each methylation sites. Putative lysine methylation sites were analyzed, based on previously known methyl lysine associated motifs in other organisms, for example LK and MK. To determine the sequence motifs, we developed a PERL script to fetch six amino acids upstream as well as downstream of a central lysine. The frequency of amino acids near lysine residue was also analyzed using WebLogo28. To compare neighboring residues in the methylated and non methylated lysines in the proteomics identified proteins, we used Two Sample Logo29.
Gene Ontology (GO) analysis
GO enrichment was performed for functional classification and cellular localization of the methylated proteins using PlasmoDB (version 13.0).
Interaction analysis using STRING
We downloaded the STRING30 Protein-Protein Interaction (PPI) network for P. falciparum and converted it into R object using igraph package31. Next, we removed the Edges with no experimental evidence of interaction (Experimental < 0). However, the network contained redundant edges and loops which were removed too, using simplify() function from igraph package. SET domain containing proteins along with its top 10 nearest neighbors were retrieved and the network was visualized using Cytoscape32.
Results
Anti-methyl lysine specific antibodies recognize Plasmodium proteins at asexual blood stages
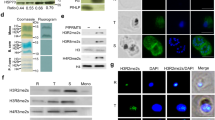
Plasmodium genome sequencing analysis has revealed existence of a number of methyltransferases, particularly SET-domain-containing proteins and demethylases that are responsible for lysine methylation in the genome22,33. To assess the extent of lysine methylation in Plasmodium proteins in asexual blood stages, we tested the reactivity of commercially available anti-mono/di methyl lysine and anti-trimethyl lysine antibodies with the parasite lysate and with intact blood stage P. falciparum parasites by western blot and immunofluorescence analysis respectively. As shown in Fig. 1A, anti-methyl lysine specific antibodies recognized specific bands at ring, trophozoite and schizont stages, although the extent of reactivity appeared more at the schizont stage. The results were further confirmed by immunofluorescence assay where a considerable staining was observed at all the three parasite blood stages, thereby indicating the extensive methylation of Plasmodium proteins at lysine residues. Staining was mainly observed in the periphery of nucleus as blue DAPI stain overlapped with the anti-lysine antibody immunostains, however some staining was also seen in parasite cytoplasm (Fig. 1B).
Extensive lysine methylation occurs at asexual blood stages of P. falciparum.
Anti-methyl-lysine specific antibodies recognize the proteins from three different blood stages of P. falciparum. (A) Representative western blot showing extent of lysine methylation at ring, trophozoite and schizont stages when the parasite lysates were probed with anti-methyl-lysine specific antibody. (B) The same antibodies also immuno-stained the asexual blood stages of the parasite as examined through immuno-fluorescence assay. The representative confocal microscopy images are shown above. DIC- bright field, DAPI-stained nucleus (blue), immunofluorescent cells labeled with anti-Methyl lysine antibody (green), and merged images.
Identification of P. falciparum lysine methylated proteins
Next, we carried-out global proteome analysis of lysine methylated Plasmodium proteins at asexual blood stages by immunoprecipitating the proteins from Plasmodium lysates prepared from the three asexual blood stages; ring, trophozoite and schizont using either anti-mono/di methyl lysine and anti-trimethyl lysine antibodies. Mass spectroscopic analysis of immunoprecipitated proteins identified a total of 570 putative lysine methylated proteins in P. falciparum asexual blood stages (Fig. 2). The two antibodies pulled-down approximately the same number of proteins at each stage, with considerable overlap. The one hundred and thirty two proteins identified in the proteome analysis were common to all the three stages (Fig. 2, Supplementary Table 1). To determine whether these putative methylated proteins have methylated lysine residues, we analyzed the spectra of the peptides generated in mass spectrometric analysis. As shown in Table 1, we could identify 364 K-methylated sites on 266 peptides corresponding to Plasmodium proteins. The representative spectra of few of the methylated peptides corresponding to Plasmodium proteins are shown in Supplementary Fig. 1.
Lysine methylated proteins of Plasmodiun falciparum.
Anti-methyl-lysine antibodies were used to immunoprecipitate stage specific lysine methylated proteins from P. falciparum lysates. N = number of proteins, MK, K = Proteins immunoprecipitated by anti-monomethyl lysine antibody, TMK, TK = Proteins immunoprecipitated by anti-trimethyl lysine antibody.
A number of reports have shown that the pre-fractionation of tryptic peptides either by SCX or HILIC provides a sort of enrichment of methylated peptides that can be better captured on LC-MS/MS for site identification. Between the two methods, HILIC has provided more number of methylation sites as compared to SCX in S. cerevasiae34. We performed HILIC chromatography analysis on trypsin digested parasite lysate generated from trophozoite stage. Twelve fractions were collected in two replicates and each of the fractions was separately analyzed on LC-MS/MS. As shown in Table 1, we could identify 247 sites in 236 peptides corresponding to Plasmodium proteins. Representative spectra of few of these K-methylated peptides are shown in the Supplementary Fig. 2. In total, 605 methylated lysine sites in 502 peptides were identified corresponding to 422 Plasmodium proteins. A couple of previous reports in human/mouse cell lines and Saccharomyces cerevisiae have shown that many of the lysine methylated sites are associated with EK, LK and MK motifs. We searched the motifs in 570 putative lysine methylated proteins identified by immunoprecipitation of parasite lysates. We could observe LK and EK motifs in many Plasmodium proteins (Fig. 3). The “Two Sample Logo” visualization of the residues surrounding methylated and non methylated lysines amongst the proteomics identified proteins reveal that the residues surrounding methylated lysines are different from those surrounding the non methylated lysines (Supplementary Fig. 4). Among the Plasmodium methylated peptides, we identified 152 monomethyl, 249 dimethylated and 210 trimethylated sites in the mass spectrometry analysis corresponding to these proteins (Fig. 3).
Analysis of identified lysine-methylated proteins for the presence of known motifs.
(A) Motif representation of methylated lysine sites along with a consensus sequence logo in P. falciparum. All the 605 confirmed sites were examined to know the presence of conserved motifs. (B) Motif representation of previously reported sites in other organisms.
To confirm the identity of non-histone lysine methylated proteins, we subjected the immunoprecipitated Plasmodium lysate to western blot analysis using anti-PfP12 or anti-PfTSN antisera. As shown in Fig. 4, PfP12 and PfTSN are detected in IP lysate, thus confirming the lysine methylation of the proteins. Altogether, several mass spectrometric analysis runs as well as western blot analysis indicate extensive lysine methylation of Plasmodium proteins at asexual blood stages.
Classification of lysine methylated proteins
Mass spectrometry analysis of the anti-methyl lysine antibody immuno-precipitated proteins from the Plasmodium lysates suggested that over 10% of the proteins encoded in the P. falciparum genome are modified by lysine methylation. To get insight into the role of lysine methylation in the parasite growth and development, we utilized the fully annotated genome database (PlasmoDB) to describe correlations for lysine methylated proteins, their families, subcellular localization and biological function. The identified lysine methylated proteins were classified based on their subcellular localization with GO term analysis using PlasmoDB. As seen in Fig. 5A, we were able to obtain the GO assigned localization of only 78% of the lysine methylated proteins, since a large number of the query proteins were hypothetical. The percentage of proteins with known or predicted subcellular localization was highest for cytoplasmic proteins (Fig. 5A). Many of the identified lysine methylated proteins appeared to be part of protein-protein or protein-DNA complexes associated with chromosome or ribosome.
Classification of lysine methylated proteins in P. falciparum.
(A) based on cellular components and (B) based on function and (C) on the basis of conserved domains. The 570 identified lysine methylated proteins were categorized based on their known or likely functions and cellular localization. Proteins with no annotations in PlasmoDB were categorized as unknown proteins. A pie chart shows the distribution of the proteins based on domain super families.
The diverse localization profile of lysine-methylated proteins suggests that this modification regulates a wide range of cellular functions. To define the functional classes of lysine methylated proteins, we data mined the P. falciparum annotated genome database for all identified methylated proteins. Similar to the localization GO terms, we were able to predict functional classes for only 62% of the identified lysine methylated proteins (Fig. 5B). Many of these annotated proteins seem to be associated with the transport/trafficking processes, chromosomal organization and translation regulation (Fig. 5B).
To get insights into the role(s) of lysine methylated Plasmodium proteins identified by mass spectrometry analysis, we sorted the proteins based on other conserved domains. As shown in Fig. 5C, several of these proteins belong to major super families such as protein kinase C (PKc), ATS, P-loop_ NTPase, PHD_SF and AdoMet_MTases_SF. The set also included 21 PfEMPs and 17 rifin proteins, indicating a role of lysine methylation in antigenic diversity in P. falciparum.
Lysine methyltransferase-substrate interactome networks and cross talks between PTMs
For further exploration of the breadth of Plasmodium protein lysine methylation and associated lysine methyltransferases (KMTs), we generated KMT-substrate networks using STRING database and published literature. These networks are depicted in Supplementary Fig. 3 and are described in Supplementary Table 2. All the Plasmodium SET domain proteins show a number of associated partners and many of them are non-histone proteins such as endoplasmin putative (GRP94), bromodomain protein 1 (BP1), proliferating cell nuclear antigen1, guanylyl cyclase (GCalpha). Importantly, some of the lysine-methylated proteins identified in our proteome analysis are also part of this KMTs-substrate network (Supplementary Table 2). For example endoplasmin putative (GRP94) and proliferating cell nuclear antigen 1 (PCNA1) proteins are identified in STRING as well as in our mass spectrometry analysis. We also observed associations among the set domain proteins. For example SET3 protein shows association with SET8 and SET4 proteins in the STRING analysis (Supplementary Table 2).
Extensive cross talk has been reported and predicted between different PTMs in different organisms, including Plasmodium. We compared the methylated lysine containing proteins with the previously published studies for phosphorylation and acetylation in P. falciparum5,6,8,35. This comparison showed that 209 lysine-methylated proteins were phosphorylated too. Further, 173 Plasmodium proteins were acetylated as well as lysine methylated and 113 proteins possess all three modifications (Supplementary Table 3). Several proteins from the STRING analysis were either acetylated or phosphorylated (Supplementary Table 2), thus indicating extensive cross talks between various PTMs.
Discussion
Processes related to intraerythrocytic development of malaria parasite that contributes to malaria associated morbidity and mortality are tightly regulated at transcription as well as translation levels36,37,38. Genome sequencing has shown that basal transcription and translational machineries are conserved in Plasmodium parasite, although few recognizable transcription factors have been identified so far39,40. Like in other eukaryotes, PTMs such as phosphorylation, ubiquitination, sumoylation, acetylation and methylation play an important role in regulating the functions of several Plasmodium proteins4. In recent times, methylation of proteins has emerged as one of the most prevalent post-translational modifications41, in the present study we have examined the lysine methylproteome of P. falciparum.
Plasmodium genome encodes nine SET domain and two JmjC-domain genes, indicating the presence of protein lysine methylation in malaria parasites22. Our current understanding of protein lysine methylation in Plasmodium is mainly restricted to histones and its role(s) in regulation of var gene expression. Tri-methylated histone 3 lysine 9 (H3K9me3) has been linked to exclusive expression of certain var genes42, while H3K36me3 has been shown to be involved in the repression of these genes25. To know the extent of lysine methylation in Plasmodium parasites in particularly the non-histone lysine methylation, we performed immunoprecipiataion of Plasmodium asexual blood stage lysates with two Methyl lysine specific antibodies and identified lysine methylated proteins by LC-MS/MS analysis. We further validated many of these lysine methylated proteins and also identified additional lysine methylated proteins by HILIC fractionation followed by LC-MS/MS analysis at trophozoite stage. Similar approaches have been previously applied to identify Saccharomyces cerevisiae and human lysine methylated proteins43,44. Based on spectral analysis, we could identify 364 sites in IP analysis and 247 sites in HILIC analysis. In total, we have identified 605 lysine methylated sites in 422 proteins.
A number of previous studies in human and yeast have shown that methylated lysine amino acid residues are part of a motif that has either leucine or methionine at – 1 position45,46. We could indeed identify leucine residue at -1 position in a number of Plasmodium specific methylated peptides identified in our study too. To get functional insights into the roles of methylated lysine containing proteins, especially the non-histone proteins, Gene Ontology (GO) analysis of these Plasmodium proteins was performed as described earlier44,46. Besides histones, we identified a large number proteins involved in transport, protein folding, translational elongation and other important biological processes. For example, lysine-methylated peptides corresponding to a number of ribosome-associated proteins, translation elongation factors, HSPs and DNA mismatch repair proteins were identified in our study. We also identified lysine methylation in several helicases, hydrolases, histone deacetylase complex proteins, kinases and phosphatases. Many of these proteins are methylated in other organisms too44,47. Remarkably, a number of Plasmodium specific proteins are also identified in the present study. These include many surface/secretory proteins; RON3, ROP14, 6-cysteine protein (p12), trophozoite excretory protein (TEX1) Rifin and PfEMP1. Interestingly, two inner membrane complex proteins; 1g and 1c (IMC 1g and 1c) are heavily methylated with 16 and 15 lysine methylation sites, respectively. Intriguingly, many of the lysine methylated proteins, especially the Plasmodium surface proteins and proteins involved in gliding motility of merozoites during invasion, have also been shown to be phosphorylated5. Summarily, the results suggest an important role of PTMs and their cross talks in the invasion of RBC by merozoites. It is important to mention here that in our analysis we could identify many methylated proteins belonging to PfEMP and Stevor family, whose transcription and antigenic variations have been linked with chromatin/epigenetic memory that includes methylation of histones25,42,48.
To validate our lysine methylome analysis, we used the STRING protein interaction database49 to understand the protein interaction networks for PfSET domain proteins and compared the results with the data generated in our methylome analysis. A few common non-histone proteins were identified in the two datasets. In addition, we also examined the proteins identified in STRING analysis for acetylation and phosphorylation. A number of Plasmodium proteins showed two or more PTMs, thereby suggesting cross talks among various PTMs. Such cross talks among PTMs such as between methylation and acetylation or between methylation and phosphorylation or between two methylation sites on the same proteins are reported in other organisms too41,44. Such cross talks probably fine tune the function of individual proteins and elucidation of such cross talks between several PTMs may shed new lights on system biology of this human pathogen. Finally, the data presented here shows that protein lysine methylation is quite wide spread in P. falciparum which may be an important gene regulatory processes. However, in order to gain deeper understanding of the role of lysine methylation in Plasmodium development and sustenance, a conditional knockdown of each of the nine PfSET domain proteins followed by a quantitative lysine methylome analysis will be required. Additionally, it will be important to experimentally explore each of the nine PfSET domain proteins that may also be important for development of novel chemotherapeutics for malaria. Thus the parasite lysine methylome analysis performed by us is a step forward in elucidating the complex nature of the gene regulatory processes in Plasmodium, where only a few transcription factors have been functionally characterized.
Additional Information
How to cite this article: Kaur, I. et al. Widespread occurrence of lysine methylation in Plasmodium falciparum proteins at asexual blood stages. Sci. Rep. 6, 35432; doi: 10.1038/srep35432 (2016).
References
Guerra, C. A. et al. The limits and intensity of Plasmodium falciparum transmission: implications for malaria control and elimination worldwide. PLoS medicine 5, e38, 10.1371/journal.pmed.0050038 (2008).
Le Roch, K. G. et al. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics 9, 513, 10.1186/1471-2164-9-513 (2008).
Hoffman, M. D., Sniatynski, M. J. & Kast, J. Current approaches for global post-translational modification discovery and mass spectrometric analysis. Analytica chimica acta 627, 50–61, 10.1016/j.aca.2008.03.032 (2008).
Doerig, C., Rayner, J. C., Scherf, A. & Tobin, A. B. Post-translational protein modifications in malaria parasites. Nat Rev Microbiol 13, 160–172, 10.1038/nrmicro3402 (2015).
Solyakov, L. et al. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat Commun 2, 565 (2011).
Treeck, M., Sanders, J. L., Elias, J. E. & Boothroyd, J. C. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell host & microbe 10, 410–419, 10.1016/j.chom.2011.09.004 (2011).
Lasonder, E. et al. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. Journal of proteome research 11, 5323–5337, 10.1021/pr300557m (2012).
Pease, B. N. et al. Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. J Proteome Res 12, 4028–4045, 10.1021/pr400394g (2013).
Jones, M. L., Collins, M. O., Goulding, D., Choudhary, J. S. & Rayner, J. C. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell host & microbe 12, 246–258, 10.1016/j.chom.2012.06.005 (2012).
Miao, J. et al. Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Molecular microbiology 89, 660–675, 10.1111/mmi.12303 (2013).
WHO. World Malaria Report. http://www.who.int/malaria/publications/world_malaria_report_2014/en/ (2014).
Khoury, G. A., Baliban, R. C. & Floudas, C. A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 1 (2011).
Boisvert, F. M., Cote, J., Boulanger, M. C. & Richard, S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics 2, 1319–1330 (2003).
Low, J. K., Hart-Smith, G., Erce, M. A. & Wilkins, M. R. Analysis of the proteome of Saccharomyces cerevisiae for methylarginine. Journal of proteome research 12, 3884–3899 (2013).
Lott, K. et al. Global proteomic analysis in trypanosomes reveals unique proteins and conserved cellular processes impacted by arginine methylation. J Proteomics 91, 210–225 (2013).
Lake, A. N. & Bedford, M. T. Protein methylation and DNA repair. Mutation research 618, 91–101, 10.1016/j.mrfmmm.2006.09.010 (2007).
Grillo, M. A. & Colombatto, S. S-adenosylmethionine and protein methylation. Amino acids 28, 357–362, 10.1007/s00726-005-0197-6 (2005).
Hart-Smith, G. et al. Stoichiometry of Saccharomyces cerevisiae lysine methylation: insights into non-histone protein lysine methyltransferase activity. J Proteome Res 13, 1744–1756, 10.1021/pr401251k (2014).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719, 10.1016/j.cell.2007.01.015 (2007).
Cao, X. J., Arnaudo, A. M. & Garcia, B. A. Large-scale global identification of protein lysine methylation in vivo. Epigenetics 8, 477–485, 10.4161/epi.24547 (2013).
Bremang, M. et al. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Molecular bioSystems 9, 2231–2247, 10.1039/c3mb00009e (2013).
Cui, L., Fan, Q., Cui, L. & Miao, J. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. International journal for parasitology 38, 1083–1097, 10.1016/j.ijpara.2008.01.002 (2008).
Sullivan, W. J. Jr., Naguleswaran, A. & Angel, S. O. Histones and histone modifications in protozoan parasites. Cellular microbiology 8, 1850–1861, 10.1111/j.1462-5822.2006.00818.x (2006).
Freitas-Junior, L. H. et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36, 10.1016/j.cell.2005.01.037 (2005).
Jiang, L. et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227, 10.1038/nature12361 (2013).
Malmquist, N. A., Moss, T. A., Mecheri, S., Scherf, A. & Fuchter, M. J. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc Natl Acad Sci USA 109, 16708–16713, 10.1073/pnas.1205414109 (2012).
Bahl, A. et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic acids research 31, 212–215 (2003).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 (2004).
Vacic, V., Iakoucheva, L. M. & Radivojac, P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 22, 1536–1537, 10.1093/bioinformatics/btl151 (2006).
Szklarczyk, D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research 39, D561–568, 10.1093/nar/gkq973 (2011).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal, Complex Systems 1695, 1–9 (2006).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, 10.1101/gr.1239303 (2003).
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511, 10.1038/nature01097 (2002).
Uhlmann, T. et al. A method for large-scale identification of protein arginine methylation. Mol Cell Proteomics 11, 1489–1499, 10.1074/mcp.M112.020743 (2012).
Cobbold, S. A., Santos, J. M., Ochoa, A., Perlman, D. H. & Llinas, M. Proteome-wide analysis reveals widespread lysine acetylation of major protein complexes in the malaria parasite. Scientific reports 6, 19722, 10.1038/srep19722 (2016).
Foth, B. J. et al. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 10, M110 006411, 10.1074/mcp.M110.006411 (2011).
Le Roch, K. G. et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508, 10.1126/science.1087025 (2003).
Luah, Y. H., Chaal, B. K., Ong, E. Z. & Bozdech, Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One 5, e10252, 10.1371/journal.pone.0010252 (2010).
Aravind, L., Iyer, L. M., Wellems, T. E. & Miller, L. H. Plasmodium biology: genomic gleanings. Cell 115, 771–785 (2003).
Coulson, R. M., Hall, N. & Ouzounis, C. A. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res 14, 1548–1554, 10.1101/gr.2218604 (2004).
Biggar, K. K. & Li, S. S. Non-histone protein methylation as a regulator of cellular signalling and function. Nature reviews. Molecular cell biology 16, 5–17, 10.1038/nrm3915 (2015).
Perez-Toledo, K. et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res 37, 2596–2606, 10.1093/nar/gkp115 (2009).
Couttas, T. A., Raftery, M. J., Padula, M. P., Herbert, B. R. & Wilkins, M. R. Methylation of translation-associated proteins in Saccharomyces cerevisiae: Identification of methylated lysines and their methyltransferases. Proteomics 12, 960–972, 10.1002/pmic.201100570 (2012).
Wu, Z. et al. A chemical proteomics approach for global analysis of lysine monomethylome profiling. Mol Cell Proteomics 14, 329–339, 10.1074/mcp.M114.044255 (2015).
Pang, C. N., Gasteiger, E. & Wilkins, M. R. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics 11, 92, 10.1186/1471-2164-11-92 (2010).
Guo, A. et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 13, 372–387, 10.1074/mcp.O113.027870 (2014).
Moore, K. E. & Gozani, O. An unexpected journey: lysine methylation across the proteome. Biochimica et biophysica acta 1839, 1395–1403, 10.1016/j.bbagrm.2014.02.008 (2014).
Chookajorn, T. et al. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA 104, 899–902, 10.1073/pnas.0609084103 (2007).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–D452, 10.1093/nar/gku1003 (2015).
Acknowledgements
This work was financially supported by Department of Biotechnology, Govt. of India (BT/PR6963/BID/7/427/2012 and BT/BI/25/066/2012 awarded to DG, and BT/01/CEIB/11/V/01 awarded to PM). ES is recipient of the Junior Research Fellowship of the Department of Biotechnology, Government of India; and AK is a recipient of Senior Research Fellowship of the CSIR, Government of India.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments D.G. and P.M. Contributed reagents/materials D.G., P.M. and A.M. Performed the experiments I.K., M.Z. and E.S. Performed analysis of experiments P.M., I.K., M.Z., A.M. and D.G. Performed computational analysis M.Z., I.K., D.G. and A.K. Wrote the main manuscript text D.G., P.M. and I.K. All the authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kaur, I., Zeeshan, M., Saini, E. et al. Widespread occurrence of lysine methylation in Plasmodium falciparum proteins at asexual blood stages. Sci Rep 6, 35432 (2016). https://doi.org/10.1038/srep35432
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35432
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.