Abstract
The extraordinary hypolipidemic effects of polyphenolic compounds from tea have been confirmed in our previous study. To gain compounds with more potent activities, using the conformations of the most active compound revealed by molecular docking, a 3D-QSAR pancreatic lipase inhibitor model with good predictive ability was established and validated by CoMFA and CoMISA methods. With good statistical significance in CoMFA (r2cv = 0.622, r2 = 0.956, F = 261.463, SEE = 0.096) and CoMISA (r2cv = 0.631, r2 = 0.932, F = 75.408, SEE = 0.212) model, we summarized the structure-activity relationship between polyphenolic compounds and pancreatic lipase inhibitory activities and find the bulky substituents in R2, R4 and R5, hydrophilic substituents in R1 and electron withdrawing groups in R2 are the key factors to enhance the lipase inhibitory activities. Under the guidance of the 3D-QSAR results, (2R,3R,2′R,3′R)-desgalloyloolongtheanin-3,3′-O-digallate (DOTD), a potent lipase inhibitor with an IC50 of 0.08 μg/ml, was obtained from EGCG oxidative polymerization catalyzed by crude polyphenol oxidase. Furthermore, DOTD was found to inhibit lipid absorption in olive oil-loaded rats, which was related with inhibiting the activities of lipase in the intestinal mucosa and contents.
Similar content being viewed by others
Introduction
Tea (Camellia sinensis) has been widely consumed as healthy beverages in the world1. People worldwide have acknowledged that tea possesses many beneficial effects on their health, particularly the prevention of obesity and the improvement of lipid metabolism2. Epidemiological data showed that habitual tea consumption can lower the body fat and regulate the plasma lipoprotein constructions3,4. Our previous clinical studies confirmed that the polyphenolic compounds of tea with extraordinary hypolipidemic effects5. Apart from polyphenols, caffeine is also an important component in tea to promote lipid metabolism. However, the side effects of central nervous system limit the use of caffeine’s thermogenic and fat oxidizing properties. Thus, the lipase inhibitory effect of polyphenols has attracted great attention6.
Polyphenols of tea are predominantly catechins and tannins7. It is also well-known that heat epimerization produces catechin derivatives during processing and extracting such as (−)-epigallocatechin (EGC), (−)-epicatechin-3-gallate (ECG) and (−)-epigallocatechin-3-gallate (EGCG)8. It has been reported that these catechins and their derivatives possess antioxidant, antiglycemic, anticancer and antilipidemic properties7. Researchers also reported that theaflavins and the polymerized polyphenols can alter lipid metabolism through inhibiting fatty acid synthesis and enhancing fatty acid oxidation9. The study of Nakai et al. showed theaflavins had stronger inhibitory activities than tea polyphenols. They assumed that the polymerization of flavan-3-ols and/or the galloyl moieties in their structures might be needed for their enhancement of inhibition of pancreatic lipase10. However, this hypothesis needs more validation. In our study, the docking and 3D-QSAR models were performed to depict the structure-activity relationship between polyphenolic compounds and pancreatic lipase inhibition activities, which might indicate some key information for optimizing hypolipidemic activity of the compounds. The polyphenolic compounds identified the common core structure of EGCG. The models were of satisfactory statistical significance and predictive ability and we were intrigued by the QSAR and those polymers which gave excellent lipid lowering effect. Using the conformations of the most active compound revealed by molecular docking, a 3D-QSAR pancreatic lipase inhibitor model with good predictive ability was also established and validated by CoMFA and CoMISA methods to further understand the structure-activity relationship between polyphenolic compounds and pancreatic lipase inhibitory activities. In order to get compounds with better activities, the polymerized polyphenols were catalyzed by crude polyphenol oxidase according to the QSAR results which indicate better activity of EGCG’s dimer with specific orientation.
Results
Docking analysis of tea flowers polyphenols
The Fig. 1 depicts the binding modes between the binding pocket of lipase and compound 8, as well as the alignment of compounds based on the conformation retrieved by docking. The docking conformation of template molecule, compound 8, is shown in Fig. 1a. Based on the benzopyrone ring as the common substructure, we can see all molecules overlap well as in Fig. 1b. The hydroxyl groups at pyrogallol in R1 position form a H-bond as an acceptor of hydrogen-bond with the -NH group in the Leu153 and Phe77 residues of lipase, while the other two hydroxyls form H-bonds with Ser152 and Phe77 residues to serve as a donor of hydrogen bond. These observations from Fig. 1c precisely match the acceptor contour maps and the donor of hydrogen bond in the corresponding CoMSIA models.
CoMFA and CoMSIA analysis
Both of the predicted and actual pIC50 values of the training and test set compounds are displayed in Table 1. The data from CoMSIA and CoMFA models are depicted graphically as scatter diagrams in Fig. 2a,b, respectively. The two diagrams demonstrate an outstanding match of actual and predicted activities. Table 2 shows the parameters of PLS analysis. The model of CoMFA presents satisfactory leave-one-out cross-validation (q2 value = 0.622), non-cross-validation (r2 value = 0.956), standard errors of estimate (SEE) of 0.069 and F value of 261.463 with an optimal component of 3. The steric field and electrostatic field descriptors explain 44.6% and 55.4% of the variance, respectively. Furthermore, CoMSIA models constitute of descriptors containing steric, hydrophobic and H-bond donors were of a high q2 value (0.631) and r2 value (0.932). Other parameters including the optimal component, SEE and F value are 4, 0.122 and 75.408, respectively. Contributions of three selected descriptors are 0.232 for steric, 0.345 for hydrophobic and 0.423 for hydrogen bond donor, respectively.
External validation
The external validation is a vital part of the model verification and its r2pred values calculated based on both training and test set are respectively 0.918 and 0.773 for CoMFA and CoMSIA model (Table 2). The favorable external validation parameters indicate that our models are of high accommodating capacities in predicting the pIC50 values of new derivatives.
3D Contour maps of QSAR
In this study, compound 8 was used as the template molecule in QSAR models. For CoMFA steric field (Fig. 3a), the green contours (80% contribution) mean these regions are of favorable steric tolerance and the yellow contours mean 20% contribution for disfavored region. The green contours located around the aromatic ring indicate that this region with bulky substituents would increase the biological activity. That’s why the compounds 3, 5 and 16 where hydroxyl is induced in R2 show poorer activity than other compounds with bulky substituents. The red and blue contours in Fig. 3b respectively show the favorable locations for the electronegative and electropositive groups. A red contours encircling the carboxyl of benzoyl ring characterize where positively charged substituents are unfavorable. The higher activities of compounds 1, 2, 4, 8 and 11 than other compounds agree with the conclusion of electrostatic fields map.
Contours of CoMFA and CoMSIA analysis with compound 8 as template molecule.
(a) The steric fields of CoMFA. Green contours refer to the regions in which bulky groups elevate activities. Yellow contours suggest the regions in which bulky groups reduce activities; (b) The electrostatic fields of CoMFA. The contours in blue and red respectively represent favored regions of electron-donating groups and electron-withdrawing groups; (c) The contours in green and yellow respectively indicate bulky and minor groups favored regions; (d) Hydrophobic fields of CoMSIA. The contours in blue and red respectively locate the favored hydrophilic and hydrophobic regions; (e) Hydrogen-bond donor fields of CoMSIA. The contours in cyan and purple respectively locate the regions in which hydrogen-bond donor groups are favorable and unfavorable.
As to the CoMSIA contours plots, the steric, hydrophobic, hydrogen bond donor fields are dipicted in Fig. 3c–e. The steric contour maps of CoMSIA are alike to those of CoMFA. In hydrophobic counter maps, red (with 80% contribution) and blue (with 20% contribution) contours indicate hydrophobic and hydrophilic favored sites, respectively. A large blue contour can be observed around the R1 position, indicating that hydrophilic substituents induced here can enhance the activity. This confirm that compounds with phenol possessed better bioactivity. The red region near the side chain of R2 emphasizes a structural preference of hydrophobic substituents. For instance, compounds 1, 8, 10 and 11 whose substituents in R2 are ester groups show better activities. Hydrogen bond donor fields are depicted in purple (20%) and cyan (80%) contours for unfavorable and favorable regions, respectively. Cyan contour located near R2 indicates hydrogen bond donor induced here can increase the activity of compounds.
Structure-activity relationship
According to the results of docking and 3D-QSAR, the structure-activity relationship of these polyphenols are summarized in Fig. 4a. Briefly, R1 site favors the hydrophilic, hydrogen-bond acceptor groups; the bulky, electron withdrawing groups at R2 site are likely to improve the activity; the minor and bulky substituent at R3 and R4 positions would increase the activity, respectively. Bulky groups at R5 position are favored to the activity.
The oxidized tea polyphenol and its inhibitory activity of pancreatic lipase
The structure-activity relationship indicates useful information of rationally designing. Under the guidance of the 3D-QSAR results, a compound named (2R,3R,2′R,3′R)-desgalloyloolongtheanin-3,3′-O-digallate (DOTD) was obtained from EGCG oxidative polymerization catalyzed by crude polyphenol oxidase. The structure of DOTD is shown in Fig. 4b. The effect of DOTD on the pancreatic lipase in vitro was determined and results showed that DOTD showed strong lipase inhibition, with an IC50 of 0.08 μg/ml.
Activities of DOTD on TG level in olive oil-loaded rats
Based on the results from molecular docking, we inferred that DOTD might affect lipid absorption by inhibiting pancreatic lipase, thus converts TG to monoglycerides and free fatty acids. Thus, lipid tolerance test was conducted to determine the influence of DOTD on TG level. Figure 5a shows the chronological changes of TG in blood after oral administration of olive oil to rats. In the control group, the TG level in blood reached the peak at 6 h after administration of olive oil. Comparing to the control group, DOTD (50 and 100 mg/kg) significantly decreased the TG level at the time point of 2, 4, 6 and 8 h. DOTD (50 and 100 mg/kg) showed better effects on decreasing TG level in olive oil-loaded rats than EGCG (100 mg/kg) at the point of 6 and 8 h.
In vivo activity of DOTD.
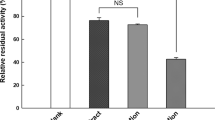
(a) Effects of DOTD on TG levels in the plasma of rats loaded with olive oil; (b) Effects of DOTD on lipase activity in the mucosa and the contents of small intestine of rats loaded with olive oil. Values are expressed as mean ± S.D. (n = 7 in each group). *P < 0.05, **P < 0.01 vs. control group and †P < 0.05, ††P < 0.01 vs. EGCG group.
Inhibitory effects of DOTD on lipid absorption in intestine of olive oil-loaded rats
The effects of DOTD on TG level in olive oil-loaded rats might be related with the lipase inhibition. As shown in Fig. 5b, the activities of lipase in both intestine and mucosa contents were obvious higher in intestine section 1 than in section 2 and 3. When DOTD (100 mg/kg) was administrated to rats, the lipase activities were significantly inhibited. These results indicated DOTD with potent lipase inhibitory effect in vivo.
Discussion
In the present study, the developed structure-activity relationships on CoMFA, CoMSIA and docking models emphasize different preference of substituent group in all five substituent position (Fig. 4a). Among these indications, the favor groups in R1 and R2 position gave us major guidelines in our rational improvement of compounds since most field contours are located around this part. Hydrophilic, hydrogen-bond acceptor groups are favorable at R1 position. According to the docking analysis, R1 substituent form key interaction with lipase by H-bonding. The pyrogallol here with its multiple hydroxy is more beneficial. The bulky, electron withdrawing groups at R2 position are most vital in affecting activity. As can be summarized from both activity data and QSAR contour maps, without bulky, electron withdrawing group like galloyl moieties in R2 position, the activity is diminished or even vanished. These conclusions also verify the assumption of Nakai et al.10.
Along with the information of substituent preference in R3 and R5 position, we found that the EGCG dimers polymerized in a head-to-head style are of more prominent activity for they precisely match our QSAR results above which indicated bulky, minor and hydrophilic or electron withdrawing groups in the specific substitute position. As illustrated in Fig. 6, head-to-head polymerization violates no SAR instructions however dimers polymerized in head-to-tail way disobey some inevitably. As to the bulky group preference in R4 position, we assumed that galloyl moieties can benefit activity. However, we haven’t obtained a 5-O-gallate dimer yet, this hypothesis needs further verification.
The present research indicated that the beneficial effects of EGCG on obesity and associated pathologies are significantly related to its direct and acute effects on lipid metabolism and energy balance. Evidences we obtained demonstrated that EGCG played a predominant role on dietary lipid metabolism both through stimulating their oxidation and reducing their absorption. In this study, the chemical constituents of tea flower, including petal, stamen and pollen were analyzed. The inhibitory activities of TFE against pancreatic lipase by 37 polyphenols from tea flower were examined in vitro and obtained satisfactory results. We also established the docking and 3D-QSAR models of lipase inhibition by CoMSIA and CoMFA methods. Statistical significance was obtained in CoMFA (q2 = 0.622, r2 = 0.956, F = 261.463, SEE = 0.069) and CoMISA (q2 = 0.631, r2 = 0.932, F = 75.408, SEE = 0.122) model. DOTD was obtained from EGCG oxidative polymerization catalyzed by crude polyphenol oxidase and showed a potent lipase inhibitory activity. The content of DOTD in tea flower and tea leaves was comparatively lower, which was difficult to be extracted. Akizawa et al. extracted DOTD from oolong and named it oolongtheanin 3′-O-gallate10. However, it could be obtained through enzymatic oxidation under a mild condition. This method could be further optimized to allow mass production.
Materials and Methods
Chemicals and Regents
C, EGC, EC, ECG, EGCG, CG, GC, GCG and all other compounds used in Table 3 were obtained from Wako Ind. (Osaka, Japan). Olive oil, 4-methylumbelliferyl (4-MU) oleate and type VI-S pancreatic lipase were bought from Sigma-Aldrich (St. Louis, MO, USA).
Animals
Six-week-old SD male rats were obtained from Guangdong Medical Laboratory Animal Center (Guangzhou, China). All the rats were kept in cages and fed with standard chow and water in animal room with controlled illumination (12 h light/dark cycle), humidity (50 ± 5%) and temperature (23 ± 1 °C). All the animal treatments were performed under the “Guideline for the Care and Use of Laboratory Animals” (No. 85–23, revised 1985), published by National Institutes of Health of U.S. All the animal experimental procedures were approved by the Laboratory Animal Ethics Committee of Jinan University (20120910001).
Data set
All polyphenols and their bioactivities were showed in Table 4. The demarcation of training and test sets was based on the following criteria as previous report11: (I) Molecule diversity was statistically significant. (II) The chosen compounds should be of precise and concise information of activity ranges and structure features to eliminate bias or redundancy. (III) The compounds with most and least activities should be arranged in the training set. With the considerations above, all thirty-seven molecules were assigned randomly with the compounds 6, 11, 14, 15, 21, 31 and 37 chosen as the test set. Table 4 displays the structures of all compounds in the training and test sets. For the convenience in analyzing, the activities of the polyphenols were converted to pIC50.
Molecular docking
Molecular docking is now widely used in studying ligand-protein interaction and new drug discovery12,13,14,15,16. Many different algorithms were applied in different docking software to explore the affinity of ligands and receptor. In this case, Surflex-Dock in SYBYL 8.1 equipped with an empirical scoring function and a patented search engine was used to dock small-molecules into a protein-active pockets13 and applied to investigate the interactions between the compounds and lipase (PDB code: 1LPB)17. The idealized hypothetical ligand which makes all potential interactions with the binding site was generated as the protomol with the threshold of 0.5 and bloat of 1 Å and other parameters are default. Surflex-Dock uses CH4, N-H and C=O fragments as probes to detect into the cavity and gather the information of steric effects and hydrogen-bond sites in the receptor pocket18,19. Before docking, we repaired the side chain, deleted all the water molecules in the crystal structure, then all hydrogen atoms and Gasteiger charges were added20,21. A staged minimization was conducted to optimize the receptor. Using the conditions optimized above, compound 8 was docked into the binding pocket. The total scores are expressed as the logarithm of dissociation constant (lgKd) to characterize binding affinities. Ten conformers with highest scores from docking results were ranked in a spreadsheet file and we took the configuration with highest total score into further investigation in protein-ligand interaction. The CCP4mg program was applied to depict the interactions between compound 8 and lipase graphically22,23.
Molecular modeling and alignment
All 37 polyphenols’ structures shown in Fig. 7 and Table 3 were prepared in sketch module of SYBYL 8.1 (Tripos, Inc., St. Louis, MO, USA). After the construction of molecules, structural energy minimization was conducted by conjugate gradient methods under the conditions of a gradient convergence of 0.05 kcal/(Å mol) in Gasteiger-Hückel charge. The maximum steps of minimization were set to 20000. Alignment is the most essential procedure in building a reliable model. The predictive ability and the statistic quality of 3D-QSAR models are closely related to the alignment24. Normally, three different aligning methods such as pharmacophore, common substructure and docking overlaps are used to obtain convinced alignment25,26. The compound 8 retrieved from the docking result with the highest active value and reasonable energy (16.205 kcals/mol) was used as the template molecule with benzopyrone ring as the common substructure for its core position and rigid structure.
CoMFA and CoMSIA setup
CoMSIA and CoMFA are widely used computational methods in 3D-QSAR modeling which could help us to understand the structures of polyphenolic compounds and guide rational structure modification to obtain novel compounds with more potent activity. With the 3D grid spacing of 2 Å in x, y and z axis, the Coulomb potentials and the Lennard-Jones were applied to calculate the energies of steric and electrostatic field27. The field values of CoMFA model were detected by a probe atom of a sp3 hybridized carbon with the value cut off set at 30 kcal/mol28,29. For the CoMSIA method, two additional different fields of hydrophobic, hydrogen-bond acceptor were also calculated as an extension of steric and electrostatic fields30. In CoMSIA method, similarity indices were introduced to make all grid points calculable. The similarity indices were calculated by equation (1):

where q is the grid point and k indicates the physicochemical properties of electrostatic and steric descriptors; Wprobe,k represents the probe atom and the attenuation factor is a default value of 0.3; i means summation index calculating on all atoms of the molecule j and Wik represents the actual value of physicochemical property k of the atom i31.
Partial least squares (PLS) analysis
The PLS analysis was applied to linearly correlate CoMSIA and CoMFA fields to pIC50 values. For this analysis32, “leave-one-out” was applied in calculating the cross-validation coefficient (q2) to investigate the robust of the training set. The q2 is a good indicator of the model fitting and in general and the q2 value above 0.5 means an acceptable model. Whereafter, the correlation coefficient (r2) was calculated by a non-cross-validation procedure and the optimized model of 3D-QSAR was generated by using the optimum number of components determined in “leave-one-out”. SEE was calculated by equation (2) to test the model.

where, n represents the compounds quantity; c represents the counts of components; PRESS means the sum of squared deviations between the observed and predicted values of test compounds.
External validation
The coefficient q2 is often identified as a benchmark in verifying the model, however it’s not always adequate. A model with high q2 and r2 values doesn’t meant to be absolutely accurate. Even though a model predict the test set precisely enough, the model might perform poorly in predicting when it was applied to another new set of molecules33. Therefore, a new parameter of external test coefficient (r2pred) was introduced in assessing the model’s predictive ability34. Predictive values r2pred were calculated by the equation (3):

where, SD represents the sum of squared deviations between the activities of test set and training set.
Preparation of oxidized polymerization from tea polyphenols
The leaves of green tea (C. sinensis) were collected in April from tea plants which grow in the mountain in Longmen County, Guangdong Province, China. Fresh tea leaves (300 g) of C. sinensis were homogenized in a 1000 mL acetone under cooling. The insoluble components were removal by filtration with gauze and the remaining extract were freeze-dried and stored in −80 °C freezer. Five grams of above extract was suspended in cysteine phosphate buffer (50 mM, pH 6.0) by stirring for 2 h under 5 °C. The solution was centrifuged (12, 000 rpm, 30 min) and the supernatant was treated with 1.0 M ammonium sulfate. The solution was purified with Sephacryl S-300 and Phenyl Sepharose CL-4B. The targeted fraction was dissolved in citric acid–potassium phosphate buffer (0.01 M: 0.02 M, 600 mL, pH 5.6) and applied to the polyphenol oxidase reaction. EGCG (600 mg) was added to the solution of polyphenol oxidase and incubated for 3 h at 32 °C. Then, 90% of CH3CN containing 1% TFA (600 mL) was added to stop this reaction. The reaction product was then diluted 5 times with distilled water and eluted on an HP-20 column. After washing with distilled water, 90% CH3CN containing 0.1% TFA was used to elute catechins. Three oxidized polyphenols were isolated and purified by prep-HPLC. The structures of the oxidized compounds were identified by MS and NMR analyses as DOTD (18 mg), prodelphinidin B2 (5 mg) and prodelphinidin A2-3′-O-gallate (4 mg). The experiments above were repeated to obtain enough compounds for further studies. MS and NMR data of DOTD were supplied in the Supporting Information.
Inhibition of pancreatic lipase in vitro
As a substrate, 4-MU oleate was used to test the activity of pancreatic lipase. A mixture of 4-MU oleate solution (50 μL) and different concentrations of test sample (25 μL) were added in the 96-well microtiter plate. Then, lipase solution (50 U/mL, 25 μL) was added and incubated (30 min, 25 °C). To stop the reaction, sodium citrate solution (0.1 M, 0.1 mL, pH 4.2) was added. The level of the product 4-methylumbelliferone was measured by a fluorescence reader (Tecan, Switzerland, Ex/Em = 355 nm/460 nm). The lipase inhibition rate (%) of each test was calculated by equation (4):

where, Ftest and Ftest blank respectively represent the fluorescence values of test samples with and without the substrate 4-MU oleate. Fcontrol and Fcontrol blank respectively represent the fluorescence values of control with and without the substrate 4-MU oleate.
IC50 of the test samples was calculated on the basis of least-squares regression line of plots of different concentrations (log) against the lipase inhibition rate (%).
Plasma lipid tolerance test
The influence of DOTD on lipid absorption was determined by plasma lipid tolerance test as we previously described35. Before this experiment, SD rats were starved for 18 h. DOTD (50, 100 mg/kg) was orally administrated to to rats. Thirty min later, olive oil solution was administered orally at a dosage of 5 mL/kg. Rats in control group received the same volume of distilled water. The chronological changes of blood TG levels in rats were analyzed in rats at time point of 0, 2, 4, 6 and 8 h (n = 7 in each group). Blood samples were distilled from an arterial cannula implanted in the abdominal aorta through the femoral artery of rats under anesthesia. The blood was contained in a tube with sodium heparin (2%) and centrifuged (5,000 rpm, 5 min). The supernatants were used to determine plasma TG level by a commercial kit obtained from Nanjing Jiancheng Bioengineering Ltd. (Nanjing, China).
Lipase activity in intestinal mucosa
Before this experiment, rats were starved for 18 h. Rats were orally loaded with olive oil solution at a dosage of 5 mL/kg. DOTD (100 mg/kg) was administered to rats at 0 or 30 min before olive oil load, or at 30 or 60 min after olive oil load. All the rats were anesthetized 2 h after olive oil administration. The small intestines were taken out and cut into 3 sections. The intestinal digesta and mucosa were obtained and suspended as 5% PBS solution in tubes and centrifuged (10,000 rpm, 10 min)36. The supernatants were collected to determine the lipase activities as described above. The protein level was determined by Coomassie brilliant blue protein quantitative kit obtained from Nanjing Jiancheng Bioengineering Institute (Jiangsu, China).
Statistical analysis
Docking and QSAR statistical results were summarizing together in Sybyl MDE module37,38. Data are expressed as means ± S.D. and analyzed by one-way ANOVA followed by Tukey’s post hoc test (SPSS Inc, USA). Differences at P < 0.05 are considered as significance.
Additional Information
How to cite this article: Li, Y.-F. et al. Prediction and evaluation of the lipase inhibitory activities of tea polyphenols with 3D-QSAR models. Sci. Rep. 6, 34387; doi: 10.1038/srep34387 (2016).
References
Wu, C. & Wei, G. Tea as a functional food for oral health. Food constituents and oral health: current status and future prospect Cambridge: Woodheading Publishing 396-417 (2009).
Preedy, V. R. Tea in health and disease prevention (Academic Press, 2012).
Wu, C. H. et al. Relationship among habitual tea consumption, percent body fat and body fat distribution. Obes Res 11, 1088–1095 (2003).
Shimada, K. et al. Oolong tea increases plasma adiponectin levels and low-density lipoprotein particle size in patients with coronary artery disease. Diabetes Res Clin Pract 65, 227–234 (2004).
He, R. R. et al. Beneficial Effects of Oolong Tea Consumption on Diet-induced Overweight and Obese Subjects. Chin J Integr Med 15, 34–41 (2009).
Yuda, N. et al. Polyphenols Extracted from Black Tea (Camellia sinensis) Residue by Hot-Compressed Water and Their Inhibitory Effect on Pancreatic Lipase in vitro. J Food Sci 77, H254–H261 (2012).
Crespy, V. & Williamson, G. A review of the health effects of green tea catechins in in vivo animal models. J Nutr 134, 3431S–3440S (2004).
Lambert, J. D., Hong, J., Yang, G. Y., Liao, J. & Yang, C. S. Inhibition of carcinogenesis by polyphenols: evidence from laboratory investigations. Am J Clin Nutr 81, 284S–291S (2005).
Kurihara, H. et al. Evaluation of the hypolipemic property of Camellia sinensis var. ptilophylla on postprandial hypertriglyceridemia. J Agric Food Chem 54, 4977–4981 (2006).
Nakai, M. et al. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J Agric Food Chem 53, 4593–4598 (2005).
Zheng, J. et al. Insight into the Interactions between Novel Isoquinolin-1,3-Dione Derivatives and Cyclin-Dependent Kinase 4 Combining QSAR and Molecular Docking. Plos One 9 (2014).
Wang, J., Kollman, P. A. & Kuntz, I. D. Flexible ligand docking: a multistep strategy approach. Proteins 36, 1–19 (1999).
Jain, A. N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46, 499–511 (2003).
Jain, A. N. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility and knowledge-based search. J Comput Aided Mol Des 21, 281–306 (2007).
Spitzer, G. M., Wellenzohn, B., Laggner, C., Langer, T. & Liedl, K. R. DNA minor groove pharmacophores describing sequence specific properties. J Chem Inf Model 47, 1580–1589 (2007).
Ambure, P. S., Gangwal, R. P. & Sangamwar, A. T. 3D-QSAR and molecular docking analysis of biphenyl amide derivatives as p38 alpha mitogen-activated protein kinase inhibitors. Mol Divers 16, 377–388 (2012).
Egloff, M. P. et al. The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate. Biochemistry 34, 2751–2762 (1995).
Ruppert, J., Welch, W. & Jain, A. N. Automatic identification and representation of protein binding sites for molecular docking. Protein Sci 6, 524–533 (1997).
Holt, P. A., Chaires, J. B. & Trent, J. O. Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J Chem Inf Model 48, 1602–1615 (2008).
Muthas, D., Sabnis, Y. A., Lundborg, M. & Karlén, A. Is it possible to increase hit rates in structure-based virtual screening by pharmacophore filtering? An investigation of the advantages and pitfalls of post-filtering. J Mol Graph Model 26, 1237–1251 (2008).
van Westen, G. J. P. & Overington, J. P. A ligand’s-eye view of protein similarity. Nat Methods 10, 116–117 (2013).
Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K. & Noble, M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr 58, 1955–1957 (2002).
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr 60, 2288–2294 (2004).
Cho, S. J. & Tropsha, A. Cross-validated R2-guided region selection for comparative molecular field analysis: a simple method to achieve consistent results. J Med Chem 38, 1060–1066 (1995).
Afantitis, A. et al. A novel simple QSAR model for the prediction of anti-HIV activity using multiple linear regression analysis. Mol Divers 10, 405–414 (2006).
Zheng, J. et al. Exploring QSARs for 5-Lipoxygenase (5-LO) Inhibitory Activity of 2-Substituted 5-Hydroxyindole-3-Carboxylates by CoMFA and CoMSIA. Chem Biol Drug Des 78, 314–321 (2011).
Murumkar, P. R., Giridhar, R. & Yadav, M. R. 3D-quantitative structure-activity relationship studies on benzothiadiazepine hydroxamates as inhibitors of tumor necrosis factor-alpha converting enzyme. Chem Biol Drug Des 71, 363–373 (2008).
Lan, P., Huang, Z. J., Sun, J. R. & Chen, W. M. 3D-QSAR and Molecular Docking Studies on Fused Pyrazoles as p38 alpha Mitogen-Activated Protein Kinase Inhibitors. Int J Mol Sci 11, 3357–3374 (2010).
Reddy, R. N., Mutyala, R. R., Aparoy, P., Reddanna, P. & Reddy, M. R. An analysis of hydrophobic interactions of thymidylate synthase with methotrexate: Free energy calculations involving mutant and native structures bound to methotrexate. J Mol Model 16, 203–209 (2010).
Klebe, G. & Abraham, U. Comparative molecular similarity index analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries. J Comput Aided Mol Des 13, 1–10 (1999).
Sivan, S. K. & Manga, V. Molecular docking and 3D-QSAR studies on triazolinone and pyridazinone, non-nucleoside inhibitor of HIV-1 reverse transcriptase. J Mol Model 16, 1169–1178 (2010).
Zhang, N. & Zhong, R. Docking and 3D-QSAR studies of 7-hydroxycoumarin derivatives as CK2 inhibitors. Eur J Med Chem 45, 292–297 (2010).
Roy, K. & Paul, S. Docking and 3D-QSAR studies of acetohydroxy acid synthase inhibitor sulfonylurea derivatives. J Mol Model 16, 951–964 (2010).
Golbraikh, A. & Tropsha, A. Beware of q(2)! J Mol Graph Model 20, 269–276 (2002).
He, R.-R., et al. Effects of histamine on lipid metabolic disorder in mice loaded with restraint stress. J Pharmacol Sci 111, 117–123 (2009).
Rao, R. H. & Mansbach, C. M. 2nd . Alkaline lipase in rat intestinal mucosa: physiological parameters. Arch Biochem Biophys 304, 483–489 (1993).
Durdagi, S., Papadopoulos, M. G., Papahatjis, D. P. & Mavromoustakos, T. Combined 3D QSAR and molecular docking studies to reveal novel cannabinoid ligands with optimum binding activity. Bioorg Med Chem Lett 17, 6754–6763 (2007).
Durdagi, S., Mavromoustakos, T. & Papadopoulos, M. G. 3D QSAR CoMFA/CoMSIA, molecular docking and molecular dynamics studies of fullerene-based HIV-1 PR inhibitors. Bioorg Med Chem Lett 18, 6283–6289 (2008).
Acknowledgements
This study is supported by the research grant from the Major Project for the Innovation of Industry and Research of Guangzhou City (201508020128), National Natural Science Foundation of China (81473115 & 81402981 & 81560661), Guangdong Provincial Natural Science Foundation of China (S20120011316), Science and Technology Program of Guangzhou (2013J4501037), Medical Scientific Research Foundation of Guangdong Province (B2014228) and Project of Guangdong Administration of Traditional Chinese Medicine (20141064).
Author information
Authors and Affiliations
Contributions
Y.-F.L., Y.-Q.C., J.D., W.-X.L., J.-S.G. and X.W. performed the experiments. Y.-Q.C. and R.-R.H. and H.K. drafted the manuscript. The analysis and interpretation of data were supplied by J.J. and P.-H.S. and W.-X.L. provided the drug and chemical composition data. The conception and design of the study was supplied by H.G., P.-H.S. and R.-R.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Li, YF., Chang, YQ., Deng, J. et al. Prediction and evaluation of the lipase inhibitory activities of tea polyphenols with 3D-QSAR models. Sci Rep 6, 34387 (2016). https://doi.org/10.1038/srep34387
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep34387
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.