Abstract
The circadian system is composed of a set of clock-genes including PERIOD, CLOCK, BMAL1 and CRY. Disrupting this system promotes cancer development and progression. The expression levels of miR-206, miR-219, miR-192, miR-194 and miR-132 regulating clock-genes and three functional polymorphisms rs11133373 C/G, rs1801260 T/C, rs11133391 T/C in CLOCK sequence were associated with the survival of 83 mCRC patients (50 males and 33 females). Longer overall survival (OS) was observed in women compared to men, 50 versus 31 months. This difference was associated with rs11133373 C/C genotype (p = 0.01), rs1801260 T/C+C/C genotype (p = 0.06) and rs11133391 T/T genotype (p = 0.06). Moreover women expressing high levels (H) of miR-192 (p = 0.03), miR-206 (p = 0.003), miR-194 (p = 0.02) and miR-219 (p = 0.002) had a longer OS compared to men. In women longer OS was reinforced by the simultaneous presence of two or more H-miR, 58 months versus 15 months (p = 0.0008); in this group of women an OS of 87 months was reached with the additional presence of rs11133391T/T genotype (p = 0.02). In this study we identified a subgroup of female patients who seems to have a better prognosis. Personalized medicine should prospectively take into account both genetic and gender differences.
Similar content being viewed by others
Introduction
The circadian system is a coordinated network of biological clocks that drive 24 h changes in metabolism and proliferation with the suprachiasmatic nucleus (SCN) of the hypothalamus synchronizing circadian oscillations in most peripheral tissues, including the gastrointestinal tract. A molecular clock, based on circadian variation in a set of clock-genes, has been validated at a cellular level where two transcriptional factors Clock and Bmal1 (CLOCK and BMAL1 gene) activate their targets, Per (PERIOD1, 2 and 3 gene) and Cry (CRY1 and 2 gene)1,2. The Clock plays a central role in regulating the circadian rhythm. It is a transcription factor of the basic helix-loop-helix (bHLH) which contains DNA binding histone acetyltransferase activity. The clock forms a heterodimer with Bmal1 that binds E-box enhancer elements upstream of PERIOD and CRY and activates transcription of these genes. Subsequently Cry and Per interfere with Clock/Bmal1 activity repressing their own transcription by interacting in a negative feedback loop. Experimental data show that the disruption of the circadian rhythm is responsible for several metabolic-related diseases where increasing evidence has raised the possibility that the circadian rhythm genes may play an important role in human cancer development and progression1.
Large population-based studies have documented a relatively high degree of polymorphisms in the human CLOCK and have shown how several of them affect the subject’s phenotype3,4,5,6,7. Previous genetic association studies have proven that there are three functional single nucleotide polymorphisms in the CLOCK sequence linked to cancer and phenotype, these include: rs11133373 C/G and rs11133391 T/C, linked to prostate and breast cancer and rs1801260 T/C, linked to several phenotypes, such as sleep disturbance, mood weight loss bipolar disorders, breast and CRC2,4,6,7,8,9. Karantanos et al.7, in a case-control study in 402 patients and 480 healthy controls showed that the 311T>C CLOCK gene polymorphism was associated with an increased risk of colorectal cancer development but with no difference in outcome. Alexander et al.6 confirmed an increased risk for adenoma in another case-control study when the PERIOD3 gene length polymorphism was evaluated.
Clock-genes are regulated by different miRNAs and among these miR-192, miR-194, miR-206, miR-132 and miR-219 are associated with different types of cancer such as breast, lung and gastrointestinal cancer10,11,12,13,14,15,16. MiRNAs are small non coding RNA molecules that regulate gene expression by binding partially to complementary target sites within the 3′-untranslated regions (3′-UTRs) of select mRNAs. MiRNAs act as potent negative regulators of protein translation either by directing the mRNA for degradation or inhibiting its translation into the encoded protein. This level of regulation can affect virtually all aspects of the circadian system, from the core timing mechanism and input pathways that synchronize clocks to the environment and output pathways that control overt rhythmicity11.
Recently it has been reported that differences in survival are related to gender and to specific genetic profiling17,18.
In this study, for the first time, three functional polymorphisms in the CLOCK sequence and five miRNAs regulating CLOCK, BMAL1 and PERIOD in advanced colorectal cancer patients and their effect on survival were analyzed.
Results
Patients data
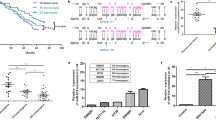
Patient data are shown in Table 1. Most of the patients (78%) received first line triplet chemotherapy (fluorouracil plus oxaliplatin and irinotecan) ± monoclonal antibody, 82% were KRAS wild-type and 55% of them were resected for liver metastases after induction chemotherapy. Median progression free survival (PFS) was 14 months and median overall survival (OS) was 35 months. Women were observed to have better median PFS and OS than men, 19 months versus 12 months (2 years-PFS 25.8% vs 12.3%, p = 0.03) and 50 months versus 31 months (3 years-OS 61.5% vs 35.9%, p = 0.03), respectively (Fig. 1 and Table 1). Differences in prognostic factors between the female and male groups were also observed for age and chronomodulated therapy (p = 0.04), as reported in Table 1.
Polymorphisms
Seventy six of the 83 the patients were genotyped for rs11133373 C/G, rs1801260 T/C and rs11133391 T/C polymorphisms. The genomic DNA of 7 cases was either consumed or degraded. No departures from Hardy-Weinberg equilibrium were observed and the frequencies did not show any differences when compared to the Italian population (TSI, Tuscany) (data not shown). No associations were observed in the entire population between polymorphisms and patient and tumour characteristics nor with clinical outcome, PFS and OS. Only performance status was linked to rs11133391 T/T genotype with a relative risk of 1.2 (95% CI 1–1.5; p = 0.04).
rs11133373 C/G
When analysed by gender, results showed a better survival rate in women compared to men with rs11133373 C/C, median PFS of 23 months vs 11 months (2 years-PFS 33.3% vs 6.3%, p = 0.006) and median OS of 57 months vs 18 months (3 years-OS 70.1% vs 25.0%, p = 0.01), respectively (Fig. 2A). In women the C/C genotype was also associated with a longer PFS and OS compared to C/G+G/G genotype (23 months vs 16 months and 57 months vs 44 months, respectively) while the opposite effect happened in men (11 months vs 14 months and 18 months vs 32 months, respectively) (supplementary Table S1).
rs1801260 T/C
Results showed a better survival rate in women compared to men with rs1801260 T/C+C/C genotype, median PFS of 20 months vs 11 months (2 years-PFS 17.6% vs 7.1%, p = 0.03) and median OS of 50 months vs 17 months (3 years-OS 50.7 vs 16.5%, p = 0.06), respectively (Fig. 2B).
rs1133391 T/C
The rs11133391 T/T genotype was associated with longer median PFS of 23 months vs 10 months (2 years-PFS 33.3% vs 12.4%, p = 0.04) and median OS of 87 months vs 25 months (3 years-OS 70.1 vs 25.4% p = 0.06), respectively (Fig. 2C). We also found that the T/T genotype was associated with a longer OS compared to T/C+C/C genotype (87 months vs 44 months) (see supplementary Table S1). We did not find any differences between performance status and T/T genotype in patients divided by gender.
miRNA expression levels
The expression levels of miR-192, miR-206, miR-194, miR-219 and miR-132 were successfully performed in 81 cases (49 males and 32 females). The expression levels of each miRNA were separated into High (H) and Low (L) categories and the association analysis between H and L of each miRNA and survival of patients were analyzed. No significant associations were found between miRNAs and patient data with the exception of hepatic resection: we found 12 patients with hepatic resection vs 29 without hepatic resection in the L-miR-194 group and 24 patients with hepatic resection vs 16 without hepatic resection in the H-miR-194 group reaching a relative risk of 1.77 (95% CI 1.15–2.71; p = 0.009).
miRNAs expression levels and gender effect
When patients were divided by sex, women expressing H-miR-192, H-miR-206, H-miR-194 and H-miR-219 had a longer survival compared to men.
miR-192
Women with H-miR-192 showed a median PFS of 16 months vs 12 months in men (2 years-PFS 28.6% vs 5.4%, p = 0.09) and a median OS of 51 months vs 31 months (3 years-OS 61.5% vs 28.7%, p = 0.03), respectively (Fig. 3A).
miR-206
Women with H-miR-206 showed a median PFS of 20 months vs 10 months in men (2 years-PFS 33.3% vs 11.6%, p = 0.006) and median OS of 56 months vs 20 months (3 years-OS 75.6% vs 22.6%, p = 0.003), respectively (Fig. 3B). Women with H-miR-206 had a longer survival compared to those with L-miR-206 with an 11 month difference for PFS and 25 months for OS. In contrast men with H-miR-206 had a shorter survival compared to those with L-miR-206 with a 4 month difference for PFS and 12 months for OS (supplementary Table S2).
miR-194
Women with H-miR-194 showed a median PFS of 19 months vs 12 months in men (2 years-PFS 25.0% vs 0.0%, p = 0.01) and median OS of 56 months vs 32 months (3 years-OS 67.0% vs 32.4%, p = 0.02), respectively (Fig. 3C).
miR-219
Women with H-miR-219 showed a median PFS of 20 months vs 11 months (2 years-PFS 30.8% vs 6.5%, p = 0.02) and median OS of 58 months vs 25 months (3 years-OS 75.0% vs 26.4%, p = 0.002), respectively (Fig. 3D). Women with H-miR-219 had a longer survival compared to those with L-miR-219 with a 4 month difference for PFS and 8 months for OS. On the contrary men with H-miR-219 had a shorter survival compared to those with L-miR-219 with an 8 month difference for PFS and 6 months for OS (supplementary Table S2).
miR-132
Women with L-miR-132 had a longer PFS and OS compared to those expressing H-miR-132 with a difference of 5 months and 13 months, respectively (supplementary Table S2).
Subsequently, an additional analysis was performed in women expressing two or more H-miR simultaneously versus none and we found that the first group had a median OS of 58 months versus 15 months (3 years-OS 68.9% vs 25.0%, p = 0.0008). This effect was further increased by the presence of rs11133391T/T genotype (2 years-OS 73.3% vs 50.7%, p = 0.02) with a median OS of 87 months vs 44 months, as shown in Fig. 4 (supplementary Table S3).
Discussion
This study highlights that functional rs11133373 C/G, rs1801260 T/C, rs11133391 T/C CLOCK polymorphisms as well as expression levels of miR-192, miR-206, miR-194 and miR-219 related to clock-genes, are differently associated with survival in women and men with mCRC, suggesting that the circadian system plays a role in this gender-effect result. Moreover this study indicates that these genetic variants could be used as sex-specific prognostic biomarkers because an opposite expression effect in women and in men related to survival was found.
Treatment of advanced colorectal cancer has profoundly evolved over the last twenty years from mono chemotherapy with 5-fluorouracil modulated by folinic acid to double or triple combination chemotherapy plus a monoclonal antibody. Moreover the selection of RAS mutation has identified a subset of patients who cannot benefit from anti-EGFR antibodies such as cetuximab or panitumumab. These changes, together with the use of resection of liver metastases, was the reason for the survival shift from 12 months to more than 30 months19,20 as in this series with a median PFS of 14 months and median OS of 35 months. With reference to drugs delivery in cancer patients according to circadian rhythms, three trials compared conventional versus time-dependent administration of 5-fluorouracil-folinic acid and oxaliplatin. In the last of these trials improved survival was found in men treated with chronomodulated infusion and in women treated by conventional flat infusion21. When analyzed together, a meta-analysis confirmed a significant three-month median survival improvement in men versus women with chronoinfusion and the contrary in female patients receiving flat infusion suggesting a gender effect in the modulation of time-delivered treatment schedule22.
On the contrary, opposite results were found in these 83 advanced colorectal cancer patients. Most of them were administered a three drug combination ± cetuximab, female patients were significantly younger than men and were mostly treated by chronomodulated infusion. In this population female patients had significantly better median PFS (19 versus 12 months) and OS (50 versus 31 months) than men. Thus a possible explanation for relying on the different signatures in clock-genes regulation according to gender could be plausible.
Sexual dimorphism has been observed in most human diseases23,24,25. Both hormonal and genetic differences between men and women can lead to differences in gene expression patterns that can influence disease risk and course even in cancer. Although CRC is more frequent in men, prognosis seems to be better in women. Majek et al.26 showed in a German population of 164,996 patients an age-adjusted 5-year relative survival higher in women (64.5% vs 61.9%, p < 0.0001). This was then confirmed at the multivariate analysis in subjects under 65 years of age with an excess attributable risk of death lower than 14%. In this group, women have fewer adenomas but roughly equivalent to the number of colorectal cancers found in men. Simon et al.27 reported that women randomized to receive estrogen/progesterone in the Women’s Health Initiative had a lower rate of CRC but a greater number of positive lymph nodes than women in the placebo group. Female patients treated in the adjuvant setting benefit more than men after resection of colon cancer26,27,28. Gender related differences in survival were advocated for urothelial and kidney cancer29. Stage and gender were found to be prognostic factors for survival in lung cancer30. In a series of 11,774 melanoma cases a significant female advantage was observed for melanoma specific survival. They were at a lower risk of progression and maintained a significant survival advantage after first progression and lymph node metastases31.
Functional polymorphisms of EGFR are inversely related to gender specific OS in mCRC: the EGFR (CA)n repeat polymorphism was associated with OS in female patients, with females carrying fewer than (CA)20 repeats having better OS and was associated to increased EGFR expression, while in males the opposite was observed18. Also estrogen receptor ß (Erß) polymorphisms have been shown to be relevant and gender related in advanced colorectal cancer32. Press et al.18 showed that female patients with less than (CA)22 repeat alleles had short OS compared to female patients that had in homozygous more than (CA)22 repeat alleles, while the opposite difference in survival was found in men. Finally a link between EGFR signaling and the circadian system has been shown. Lauriola et al.33 showed that in animals glucocorticoids inhibit signaling downstream of EGFR through a circadian control mechanism: EGFR signals are suppressed by glucocorticoids during the active phase (night-time in rodents) while EGFR signals are enhanced during the resting phase. It was also demonstrated that KRAS overexpression can disrupt the circadian clock in metastatic cell lines34. We used the miRDB data base (mirdb.org/miRDB) for predicting the potential of miRNA-mRNA interaction also in the EGFR pathway. We found that miR-192, miR-206 and miR-194 have binding sites on EGFR mRNA; miR-206, miR-132, miR-194 and miR-219 on KRAS mRNA; miR-192 on ESR2 (Erß) mRNA; and miR-219 on EGF mRNA. Thus our hypothesis is that the biological framework of colorectal cancer in women seems to be characterized by Erß signaling, EGFR activation and circadian clock control, all interacting mechanisms, involved in tumour growth.
This study has several limitations. It is a retrospective study with a limited number of patients, so it can only be used to generate a hypothesis. Whereas the main strength of the study is that all the patients were treated by the same clinical group and follow-up was sufficiently long. Prospective, cooperative studies are needed to explore the mechanisms by which CLOCK polymorphisms and miRNA expression levels related to clock-genes exert their effect on EGFR or on other pathways. In the era of precision medicine the goal of treating men and women differently with advanced colorectal cancer is a promising challenge.
Methods
Patients
The cohort of patients was retrospective and consisted of 83 patients diagnosed with mCRC treated in one single center. Patients were required to have a histologically confirmed diagnosis of advanced CRC and available Formalin-Fixed, Paraffin-Embedded tissue (FFPE) samples of the primary tumor and peripheral blood samples. All experiments were performed in accordance with relevant guidelines and regulations. The study was approved by the Ethics Commitee of the Regina Elena National Cancer Institute in Rome and signed informed consents were collected.
The primary end-point of the study was the evaluation of survival outcome according to the SNPs and miRNAs expression levels. Additional analyses were addressed to compare miRNAs and SNPs according to clinical features.
The genotype analysis of rs11133373 C/G, rs1801260 T/C and rs11133391 T/C polymorphisms was performed on genome DNA (gDNA) extracted from peripheral blood cells in patients. The miRNA expression level analysis of miR-192, miR-206, miR-132, miR-194 and miR-219 was performed on miRNAs extracted from FFPE samples.
See under “supplementary Material and Methods” the Table S4 for nucleic acids extraction (gDNA and miRNA) and PCR conditions, for the miRNAs expression level analysis see Figure S1(A–E).
Statistical Method
rs11133373 C/G, rs1801260 T/C and rs11133391 T/C were checked for any deviations from the Hardy-Weinberg equilibrium. The frequencies of each SNP in the cohort of patients were compared with the general Italian population (TSI, http://www.ensembl.org/info/genome/variation/index.html). According to the frequency of the minor allele and the low frequency of the homozygous variant genotype, we decided to collapse the heterozygous and minor allele homozygous in all the analyses performed. The expression levels of miR-192, miR-206, miR-132, miR-194 and miR-219, were expressed as values obtained from the ΔCt equation of each miRNA to test with respect to RNU-6-2 used as reference gene (ΔCt = Cttarget − Ctreference). Median ΔCt values were employed for splitting miRNA expression levels into two groups, High expression level (H) and Low expression level (L).
The Chi-Square or Fisher Exact tests were used to estimate all associations between categorical variables. Quantitative variables were compared using the non-parametric Mann–Whitney test. The Odds Ratio (OR) and the 95% confidence intervals (95% CI) were estimated for each variable using the logistic univariate regression model.
Overall survival (OS) and Progression-free Survival (PFS) were calculated by the Kaplan-Meier product-limit method. PFS was calculated as the time from the start date of the first line therapy until date of progression or date of last follow-up evaluation. OS survival was calculated as the time from the start date of the first line therapy until to the date of death or last contact. Log-rank test was used to assess the differences between subgroups.
All reported P-values were based on two-sided tests and a P-value of less than or equal to 0.05 was considered statistical significant.
SPSS software was used for all statistical evaluations (SPSS version 21.0, SPSS Inc., Chicago, Illinois, USA).
Additional Information
How to cite this article: Garufi, C. et al. Gender effects of single nucleotide polymorphisms and miRNAs targeting clock-genes in metastatic colorectal cancer patients (mCRC). Sci. Rep. 6, 34006; doi: 10.1038/srep34006 (2016).
References
Yu, E. A. & Weaver, D. R. Disrupting the circadian clock: gene-specific effects on aging, cancer and other phenotypes. Aging (Albany. NY). 3, 479–493 (2011).
Innominato, P. F., Lévi, F. A. & Bjarnason, G. A. Chronotherapy and the molecular clock: Clinical implications in oncology. Adv. Drug Deliv. Rev. 62, 979–1001 (2010).
Hawkins, G. A., Meyers, D. A., Bleecker, E. R. & Pack, A. I. Identification of coding polymorphisms in human circadian rhythm genes Per1, Per2, Per3, Clock, Arntl, Cry1, Cry2 And Timeless in a multi-ethnic screening panel. DNA sequence. 19, 44–49 (2009).
Zhou, F. et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer 118, 937–946 (2012).
Ciarleglio, C. M. et al. Genetic differences in human circadian clock genes among worldwide populations. J. Biol. Rhythms 23, 330–340 (2008).
Alexander, M. et al. Case-control study of the PERIOD3 clock gene length polymorphism and colorectal adenoma formation. Oncol Rep. 33, 935–941 (2015).
Karantanos, T. et al. Association of the clock genes polymorphisms with colorectal cancer susceptibility. J Surg Oncol. 108, 563–567 (2013).
Zhu, Y. et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 69, 9315–9322 (2009).
Hoffman, A. E. et al. CLOCK in breast tumorigenesis: genetic, epigenetic and transcriptional profiling analyses. Cancer Res. 70, 1459–1468 (2010).
Savvidis, C. & Koutsilieris, M. Circadian rhythm disruption in cancer biology. Mol. Med. 18, 1249–1260 (2012).
Lim, C. & Allada, R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 16, 1544–1550 (2013).
Nagel, R., Clijsters, L. & Agami, R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 276, 5447–5455 (2009).
Zhou, W., Li, Y., Wang, X., Wu, L. & Wang, Y. MiR-206-mediated dynamic mechanism of the mammalian circadian clock. BMC Syst. Biol. 5, 141 (2011).
Vickers, M. M. et al. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin. Exp. Metastasis 29, 123–132 (2012).
Alvarez-Saavedra, M. et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet 20, 731–751 (2011).
Cheng, H.-Y. M. et al. microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829 (2007).
Gordon, M. A. et al. Labonte M, Wilson P, Sherrod A, Ladner RD, Lenz HJ. Gender-specific genomic profiling in metastatic colorectal cancer patients treated with 5-fluorouracil and oxaliplatin. Pharmacogenomics. 12, 27–39 (2011).
Press, O. A. et al. Gender-related survival differences associated with EGFR polymorphisms in metastatic colon cancer. Cancer Res. 68, 3037–3042 (2008).
Garufi, C. et al. Cetuximab plus chronomodulated irinotecan, 5-fluorouracil, leucovorin and oxaliplatin as neoadjuvant chemotherapy in colorectal liver metastases: POCHER trial. Br. J. Cancer 103, 1542–1547 (2010).
Cremolini, C. et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet. Oncol. 16, 1306–1315 (2015).
Giacchetti, S. et al. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: the European Organisation for Research and Treatment of Can. J. Clin. Oncol. 24, 3562–3569 (2006).
Giacchetti, S. et al. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis. Ann. Oncol. 23, 3110–3116 (2012).
Cook, M. B., McGlynn, K. A., Devesa, S. S., Freedman, N. D. & Anderson, W. F. Sex disparities in cancer mortality and survival. Cancer Epidemiol. Biomarkers Prev. 20, 1629–1637 (2011).
Wang, Y., Freemantle, N., Nazareth, I. & Hunt, K. Gender differences in survival and the use of primary care prior to diagnosis of three cancers: an analysis of routinely collected UK general practice data. Plos One 9, e101562 (2014).
Barford, A., Dorling, D., Davey Smith, G. & Shaw, M. Life expectancy: women now on top everywhere. BMJ 332, 808 (2006).
Majek, O. et al. Sex differences in colorectal cancer survival: population-based analysis of 164,996 colorectal cancer patients in Germany. Plos One 8, e68077 (2013).
Simon, M. S. et al. Estrogen plus progestin and colorectal cancer incidence and mortality. J. Clin. Oncol. 30, 3983–3990 (2012).
Smith, R. E., Colangelo, L., Wieand, H. S., Begovic, M. & Wolmark, N. Randomized trial of adjuvant therapy in colon carcinoma: 10-year results of NSABP protocol C-01. J. Natl. Cancer Inst. 96, 1128–1132 (2004).
Lucca, I., Klatte, T., Fajkovic, H., de Martino, M. & Shariat, S. F. Gender differences in incidence and outcomes of urothelial and kidney cancer. Nat. Rev. Urol. 12, 653 (2015).
Yoshida, Y. et al. Gender Differences in Long-Term Survival after Surgery for Non-Small Cell Lung Cancer. Thorac. Cardiovasc. Surg. Sep 14 Epub ahead of print (2015)
Joosse, A. et al. Gender differences in melanoma survival: female patients have a decreased risk of metastasis. J. Invest. Dermatol. 131, 719–726 (2011).
Press, O. A. et al. Gender-related survival differences associated with polymorphic variants of estrogen receptor-β (ERβ) in patients with metastatic colon cancer. Pharmacogenomics J. 11, 375–382 (2011).
Lauriola, M. et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 5, 5073 (2014).
Relógio, A. et al. Ras-mediated deregulation of the circadian clock in cancer. Plos Genet 10, e1004338 (2014).
Acknowledgements
This work was supported by Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.) Project 11770. We would thank Maria Assunta Fonsi for administrative assistance in the study, Tania Merlino and Maria Guttinger for the review of the English version.
Author information
Authors and Affiliations
Contributions
C.G., A.T., E.G. and A.R. conceived and performed the study design, the manuscript preparation and data interpretation; E.G., T.R. and A.R. performed the research experiments; I.S. performed statistical analysis, data interpretation and manuscript preparation; C.G., E.G., A.T., E.M., Marcella M., T.R., M.Z., G.M.E., F.C., Mauro M. and A.R. treated the patients, collected samples and patient’ data and commented the manuscript; All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Garufi, C., Giacomini, E., Torsello, A. et al. Gender effects of single nucleotide polymorphisms and miRNAs targeting clock-genes in metastatic colorectal cancer patients (mCRC). Sci Rep 6, 34006 (2016). https://doi.org/10.1038/srep34006
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep34006
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.