Abstract
Type III secretion systems (T3SSs) are essential devices in the virulence of many Gram-negative bacterial pathogens. They mediate injection of protein effectors of virulence from bacteria into eukaryotic host cells to manipulate them during infection. T3SSs involved in virulence (vT3SSs) are evolutionarily related to bacterial flagellar protein export apparatuses (fT3SSs), which are essential for flagellar assembly and cell motility. The structure of the external and transmembrane parts of both fT3SS and vT3SS is increasingly well-defined. However, the arrangement of their cytoplasmic and inner membrane export apparatuses is much less clear. Here we compare the architecture of the cytoplasmic regions of the vT3SSs of Shigella flexneri and the vT3SS and fT3SS of Salmonella enterica serovar Typhimurium at ~5 and ~4 nm resolution using electron cryotomography and subtomogram averaging. We show that the cytoplasmic regions of vT3SSs display conserved six-fold symmetric features including pods, linkers and an ATPase complex, while fT3SSs probably only display six-fold symmetry in their ATPase region. We also identify other morphological differences between vT3SSs and fT3SSs, such as relative disposition of their inner membrane-attached export platform, C-ring/pods and ATPase complex. Finally, using classification, we find that both types of apparatuses can loose elements of their cytoplasmic region, which may therefore be dynamic.
Similar content being viewed by others
Introduction
Type III secretion systems (T3SSs) are essential devices in the virulence of many Gram-negative bacterial pathogens. They mediate injection of protein effectors of virulence from bacteria into eukaryotic host cells to manipulate them during infection. T3SSs involved in virulence (vT3SSs) are evolutionarily related to bacterial flagella assembly apparatuses (fT3SSs), which allow cell motility. Both types of macromolecular machines are composed of three major parts: a hollow extracellular portion (needle or flagellum), a basal body -known as the “needle complex” (NC) base for vT3SSs or hook-basal body (HBB) for fT3SSs- spanning both bacterial membranes and containing a membrane-inserted protein export apparatus, and a cytoplasmic export complex. The entire apparatus serves as a conduit for secretion of virulence proteins or flagellar filament components.
The structure of the external and transmembrane parts of both fT3SS and vT3SS is increasingly well-defined1, with the identity and location of the majority of the outer and inner membrane components being well-established. However, the arrangement of their cytoplasmic and inner membrane export apparatuses (CIMEA) is much less clear. The cytoplasmic region of one of the five inner membrane export apparatus (IMEA) components, MxiA of Shigella flexneri vT3SS and FlhA of Salmonella enterica serovar Typhimurium fT3SS, is required to form a nonameric toroid immediately underneath the basal body2. Within the cytoplasmic export apparatus (CEA), the cytoplasmic F1-like export ATPase Spa47/FliI forms a homohexameric complex positioned ~5 nm underneath the MxiA/FlhA toroid and 10 to 15 nm down from the membrane-embedded portion of the IMEA3. In fT3SSs, FliM and FliN form a large and robust ring surrounding the cytoplasmic face of the basal body, known as the C-ring. Recent electron cryotomography (ECT) studies in Shigella flexneri identified densities for six cytoplasmic pods surrounding the base of its vT3SS with the sole FliM-FliN homolog Spa33 being required for their presence and that of the ATPase complex. MxiN, a homolog the dimeric flagellar ATPase inhibitor FliH was only required for the presence of the ATPase complex and was hence postulated to form six additional “linkers” seen to connect the ATPase complex to the pods4 (Fig. 1d). Whether such an arrangement is also found in other vT3SSs is unknown. Furthermore, despite the conservation within this protein family, whether FliH forms similar links to the C-ring in fT3SS5, where the rotational symmetry of the C-ring shows variable symmetries from 32 to 366, is not yet clear.
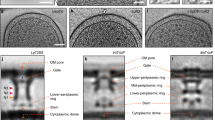
ECT of Shigella flexneri minicell and subtomogram averaging maps of its Needle Complex with various symmetries.
(a) Central slice of a tomographic reconstruction of a typical minicell showing two needle complexes embedded in the cell membranes. Scale bar indicates 100 nm. (b) Schematic diagram of a NC embedded in the cell membrane. Purple color indicates in vitro structures from isolated NC, blue color indicates known position of a component of the inner membrane export apparatus (export platform) and orange color indicates cytoplasmic export apparatus (CEA), as defined by a new study4: ATPase, pods and linkers. Abbreviations are as follows: inner membrane, IM; outer membrane, OM; peptidoglycan layer, PG; outer region, OR; inner regions, IR1 and IR2; IMR inner membrane region; OMR, outer membrane region. (c) Top, central sections of subtomogram averaging maps of NCs with C1, C2, C3 and C6 symmetry, respectively, and below each corresponding cross sections at height indicated by the dashed yellow line. The cross section of map with C1, C2, C3 and C6 symmetry shows six “blobs”, corresponding to the pod-like regions. Scale bars indicate 10 nm.
Two other CEA components are known, but have not been localised within the cytoplasmic structures so far described via ECT. These are FliJ/Spa13, a homolog of the F1-ATPase γ-subunit, which binds the fT3SS ATPase homohexamer’s central cavity in vitro7, and MxiK, which interacts with the ATPase8 but lacks a clear fT3SS homolog. All components are essential for high efficiency type III secretion. The organisation of the other components within the structures described by ECT is increasingly clear, especially in the better-studied fT3SS. The smaller FliN is similar to the C-terminus of the larger FliM, each being composed of one sequence-related SPOA domain, known as SPOA2 and SPOA1, respectively9,10,11. Most vT3SS systems encode a single protein in place of FliM and FliN. This protein is as large as FliM and resembles FliM at its N-terminus but is more like FliN at its C-terminal domain where it carries both a SPOA1 and a SPOA2 domain. However, in Yersinia, Salmonella and Shigella vT3SSs, smaller proteins that are similarly sized to FliN and contain just the SPOA2 domain are generated from the same genes through unconserved internal translational initiation start sites, and they are essential for function12,13,14. Recently, it was shown that the smaller protein homodimerises and forms a heterotrimeric complex with the C-terminal SPOA2 domain of the larger one11,14. Thus, this complex contains four SPAO domains. The ratio of FliM to FliN in the flagellar C-ring in situ was estimated as 1:315, and this was confirmed recently in vitro14. Because FliM contains only a single SPAO domain, the number of SPAO domains in the FliM-FliN complex is also four. This implies that the fT3SS C-ring is structurally equivalent with the vT3SSs counterpart.
However, what remains unclear is whether and how these structurally similar complexes form rings in fT3SSs and pods in vT3SSs. The full length Yersinia vT3SS Spa33 homolog was found in 22 + /−7 to 8 copies per basal body. This suggests there is one copy of the SPAO domain heteroteramer per pod. However, the full composition of the pod and its mode of binding to the vT3SS inner membrane-spanning MxiG ring with 24-fold rotational symmetry remain unclear. Furthermore the Shigella Spa33 full length (Spa33-FL) and its C-terminal fragment (Spa33-C) associate as Spa33-FL/C2 that further oligomerises into elongated arrays in vitro, which can be manually positioned in the EM density corresponding to the cytoplasmic edge of the Salmonella Typhimurium fT3SS C-ring with the 34-fold rotational symmetry6,14. This lead to questioning of whether the six pods seen might just represent an incomplete view of a dynamic vT3SSs C-ring14. Indeed, full-length YscQ from Yersinia, which is a homolog of Spa33, was shown to be dynamic, with a turnover half-time of about 2 min, which was reduced by an active export ATPase16. This finding, the considerable physical distance separating the export ATPase from the export platform and the uncertainty over the route that exported proteins take across the system, also suggest several states of the export apparatus might be discernable.
Here we investigate using ECT the structures of the cytoplasmic regions of the vT3SSs of Shigella flexneri and Salmonella enterica serovar Typhimurium, where the latter is also known as SPI1, and that of the fT3SS Salmonella Typhimurium and compare them to determine the common and different structural features. We show that the cytoplasmic regions of vT3SSs display conserved six-fold symmetric features including pods, linkers and an ATPase complex, whereas fT3SSs probably display the six-fold symmetry only in their ATPase region. In addition, we identify morphological differences between vT3SSs and fT3SSs, such as relative disposition of their inner membrane-attached export platform, C-ring/pods and ATPase complex. Finally, using classification, we find that both types of apparatuses can loose elements of their cytoplasmic region, which may therefore be dynamic.
Results and Discussion
The cytoplasmic region of the Shigella flexneri vT3SS displays C6 symmetry
We generated minicells by over expression of Salmonella FtsZ in Shigella flexneri. We confirmed the presence of the needle complexes in the minicells (Fig. 1a), as found in previous reports for Yersinia, Salmonella and Shigella spp.4,17,18. Recently, Hu et al. reported six “pod” densities, corresponding to the C-ring structure in HBBs, in the cytoplasmic region of the Shigella flexneri NC visualized in situ. This included also six linker densities joining the ATPase and the pods (Fig. 1b). This had never been visualized in NCs, even from other species. We picked 265 subtomogram particles, manually aligned them by eye, and used Dynamo19 for subtomogram averaging. We performed CTF-correction using TOMOCTF. When we applied C50 symmetry, we, like others17,18, could not detect densities of the pods and linkers. However, when we applied C6 symmetry in our procedure we could detect six pod densities and six linkers (Fig. 1c), very similar in dimensions (width of cytoplasmic region 32 nm and height 20 nm between top of MxiAc and bottom of Spa47), and shape, to those identified by Hu et al.4 in the cytoplasmic region of the Shigella vT3SS. Furthermore, when we generated three-dimensional (3D) averages without imposing symmetry (C1; Fig. 1c) or by imposing two and three-fold rotational symmetry (C2 and C3; Fig. 1c), we also detected six pod densities in the cytoplasmic region. When we applied C1, C2 and C3 symmetry, we used the volume resulting from a 3D-reconstruction applying C101 symmetry as a reference volume to avoid reference bias. The fact that the six pods are visible no matter what symmetry and starting model strongly supports their existence in situ. How the six pods are associated at regular intervals with the cytoplasmic face of MxiG inner membrane ring, which displays C24 symmetry20, remains to be determined. There must be a specific structural arrangement for Spa33 heterotrimers binding to a group of four MxiG subunits to build up an architecture with such a symmetry mismatch.
Structural variation within the cytoplasmic region of the Shigella flexneri vT3SS
Since a vT3SS cytoplasmic region protein Spa33 homolog displays dynamic behavior in Yersinia as monitored by super-resolution fluorescence microscopy16, we investigated morphological variation within the NC cytoplasmic region. For this we used classification by multireference alignment within the cytoplasmic region only, having masked out the rest of the structure, using the subtomogram average obtained during 3D-averaging with C6 symmetry (Fig. 2a). The shape of the inner membrane profile, the appearance of the MxiAC toroid and the height of the ATPase complex differ between classes (Fig. 2b). We also noticed that the densities of ATPase in class 1 and that of the pods and ATPase in class 3 were weaker than those in class 1. The degree and size of inter-class variation suggests that the cytoplasmic region of vT3SSs undergoes large-scale conformational changes.
Classification of the Needle Complex subtomogram averages from Shigella flexneri and superimposition of in vitro map on in situ map.
(a–c) Central/cross section maps of original subtomogram averaging map of the NC (a) and three classes (b), middle and bottom each corresponding cross sections at height indicated by the dashed yellow line and the dashed pink line, respectively. The number of particles in each class is indicated in the top right corner. (c) Refined map excluding class 3, which shows weak densities in cytoplasmic region. (d,e) The 3D rendering maps show pod-like densities, linker densities between membrane associated export apparatus (export apparatus) and pods, export apparatus and cytoplasmic export platform (export platform). (f,g) Superposition of in vitro NC map obtained from single particle image analysis (g, yellow) on in situ map from subtomogram averaging (f, grey). In situ and in vitro NC map show OR, Neck, IR1 and IR2. Scale bar indicates 10 nm.
Comparison between in situ and in vitro NC structure
As the cytoplasmic region did not appear clearly in class 3, we refined the subtomogram averaging excluding class 3 (Fig. 2b). The cytoplasmic region became clearer than before in the new 3D map (Fig. 2c–e). We used this map for comparison with the density map of the NC obtained from single particle image analysis of cryoEM data (Fig. 2g; Kajimura, Cheung, Blocker and Namba, unpublished). The periplasmic regions of both maps are morphologically in good agreement (Fig. 2f). Liu’s group proposed that the MxiG cytoplasmic region protrudes into the pod region4. We see similar features in both tomograms and single particle image reconstructions. As previously reported17,18, the NC volume obtained by single particle reconstruction is ~5 nm shorter in its outer membrane region than the in situ NC volume obtained from ECT data. The map from Liu’s group shows more detail in the outer membrane region (OMR), which extends into the OR (Fig. 1b) of NC associated elements, and this part may be formed by the pilotin MxiM21,22.
Co-instability of Shigella vT3SS CEA components
Analysis of differences between ECT maps from WT and deletion mutants as a means to assign the location of the missing components4 relies on the assumption that in the absence of one, the others are stable, and then correctly localized. We tested this first assumption for all CEA proteins, mainly using FLAG-tagged constructs as we failed to generate antibodies against most of these proteins that detect them at native levels in whole bacteria. Evidently, co-stabilisation does not equate to incorporation into the same multi-protein complex(es). However, we previously reported that His6-Spa47, when expressed in Δspa47 at its native level, co-purifies in complex with Spa33, MxiN and MxiK23.
Firstly, we checked the activity of Shigella vT3SSs from deletion mutants, using Congo red (CR) as a selective, artificial inducer of secretion in this species (Fig. 3a 24). In the ΔmxiK, ΔmxiN, Δspa33, Δspa13 and Δspa47 mutants, no secretion of early effector proteins (IpaA, IpaB, IpaC, IpaD and IpgD) was observed, as previously reported, due to lack of secretion of any components of the apparatus beyond the inner membrane8,25,26. We also confirmed restoration of inducible secretion activity of vT3SSs from these deletion mutant strains complemented with the corresponding constructs encoding wild-type (mxiN) or FLAG-tagged proteins (Fig. 3a). This indicates all mutants are non-polar on downstream mxi genes and the FLAG-tagged constructs are functional.
Analysis of the stability or expression of components of the Shigella T3SS cytoplasmic export apparatus in mutants of this portion of the apparatus.
(a) Complementation of cytoplasmic export apparatus knock mutants by in-trans expression of wild-type or FLAG-tagged proteins. Induced secretion of Ipa proteins in the presence (+) or absence (−) of Congo red (CR) was analysed by silver staining. The wild-type (WT) and the deletion mutants were used as controls, and bacterial numbers were normalized by optical density. The positions of the major Ipa proteins detected by silver staining are indicated on the side. The data shown here are representative of 2 independent experiments giving similar results. (b–f) Expression of native MxiN (b) and FLAG-tagged MxiK (c), Spa33 (d), Spa13 (e) and Spa47 (f) in different mutant backgrounds. Pellets of overnight grown cultures with equivalent bacterial numbers were analysed by immunoblotting with either a polyclonal rabbit antiserum against MxiN (b) or a monoclonal antibody against the FLAG tag (c–f), as indicated at the side. The data shown here are representative of 2 independent experiments giving similar results. The blots in b-f were cropped to show only the area where signal was detected. (g) Table semi-quantitatively summarising results of co-stability experiments in (b–f).
Next, we checked the stability of each of the native or FLAG-tagged components in the WT strain and deletion mutants of the others five components (Fig. 3b–g). MxiN, for which we have a quality antibody, was detected in all mutants, except in Δspa33, where it was completely absent (Fig. 3b). This indicates MxiN requires Spa33 for stability and a Δspa33 mutant is phenotypically identical to a Δspa33 ΔmxiN double mutant. In mutants of all the other FLAG-tagged components expressed, the FLAG-tagged proteins were more abundant than in the WT strain (Fig. 3: panel c, lanes 2 and 3; panel d, lanes 2 and 5; panel e, lanes 2 and 6; panel f, lanes 2 and 7). This suggests there is competition for CEA complex formation between the WT proteins and their tagged equivalents in the WT background and that excess, uncomplexed, tagged molecules are unstable. Whilst can assume from Fig. 3a that the FLAG-tagged proteins are equally functional to the WT ones and hence compete equally well with them, we have no way of estimating what each of their relative expression ratios are. But, we can assume, since the expression of all FLAG tagged proteins is from the lac promoter in pUC19, that each individual tagged protein is expressed at the same level in each strain. Therefore the fact that FLAG-MxiK is detected inΔmxiK but not in WT (Fig. 3c, lanes 2 and 3), suggests that the stability of FLAG-MxiK is enhanced in a background which does not express WT MxiK. FLAG-MxiK is also stabilized, to a lesser extent, in Δspa47 but not in the other deletion mutants (Fig. 3c, lane 7). Taken together with the first observation, this suggests WT MxiK is partially destabilised in Δspa47 but not in the other mutants. Together these data suggest that native MxiK requires Spa47 for stability. This is supported by the fact that FLAG-Spa47 is more stable in Δspa47 and ΔmxiK than in WT or any of the other mutants (Fig. 3f, lanes 3 and 7). This also suggests, conversely, that Spa47 requires MxiK for stability.
Spa33_FLAG is less stable in WT and Δspa13 than in all the other mutants (Fig. 3d, lanes 2 and 6). This conversely suggests native Spa33 requires MxiK, MxiN and Spa47 for stability. Using the same reasoning, Spa13 seems to require primarily Spa47 and Spa33 for stability, but also perhaps MxiK and MxiN (Fig. 3e). To summarize (Fig. 3g), in the Δspa33 mutant examined by Hu et al.4, in which the entire NC cytoplasmic region was lacking, it is likely that MxiN and Spa13 levels were substantially reduced, and that, as a consequence, any expressed Spa47 was mislocalized. In the ΔmxiN mutant Hu et al. examined, MxiK, Spa33 and Spa47 would have been present but Spa13 was likely unstable and the ATPase complex hence mislocalized. This explains why not only the linker regions, probably formed of MxiN, but also the ATPase portion is lacking in their ECT map. A Spa13 monomer is too small and/or dynamic to be localised by ECT at currently achievable resolutions and deletion of mxiK has pleiotrophic effects, most notably on Spa47 stability, which itself might affect the localization of other CEA components. Therefore, we chose not to attempt to localise these components using difference maps here. However, our past co-precipitation data23, and now co-stability data, suggest they are closely associated with Spa47.
Subtomogram averaging in the Salmonella vT3SS with C6 symmetry
To determine whether the six-fold symmetry of the CEA was found also in other vT3SSs, we re-picked 347 NC subtomogram particles from the Salmonella minicell ECT data, with which Kawamoto et al.17 have already published a structure obtained by subtomogram averaging with C100 (cylindrical) symmetry and without CTF correction. We then performed CTF correction using TOMOCTF and subtomogram averaging using Dynamo as for the Shigella vT3SS, with C6, C1, C2 and C3 symmetry imposed.
We confirmed the six-fold rotational symmetry of the Salmonella vT3SS cytoplasmic region, which we had not observed so far, even in the maps with C2 and C3 symmetry imposed (Fig. 4a). It is likely that the structure of the C-ring region was smeared in the previous study by the C100 symmetry applied17 to smooth the cylindrical features of the basal body. Next, we performed 3D classification by multireference alignment using Dynamo. Several of the classes showed variation in their inner membrane and InvAC regions (Fig. 4b). We removed class 1 and 4 (Fig. 4b), which seemed to lack the pods, for refinement. The result showed six linkers and pods (Fig. 4c,d). In addition, class 3 and the refined volume showed extra density under the ATPase, not seen in the Shigella vT3SS structure. The difference is shown in Fig. 4e by direct comparison of these two structures by superposition. The extra density may represent hexameric chaperone/effector complexes docked to the ATPase complex prior to substrate export, as proposed for SopB/SigE in Salmonella by Roblin et al.27.
ECT subtomogram averaging map of Salmonella Typhimurium.
(a) central sections from the subtomogram averaging map of the NC in Salmonella with C1, C2, C3 and C6 symmetries, respectively and below each corresponding cross sections at height indicated by the dashed yellow line. (b) Central and cross section of four classes and middle/bottom each corresponding cross sections at height indicated by the dashed yellow line and the dashed pink line, respectively. The number of particles for each class is indicated in the top right corner. (c) Refined map excluding class 1 and 4, which show only weak densities in cytoplasmic region. (d) 3D surface rendering from in situ map of Salmonella showing NC and cytoplasmic regions. Below, the map inclined 60° to show the export apparatus, including extra density and linkers. Top right, a central section of the map. (e) Comparison of in situ maps of Shigella flexneri (grey) and Salmonella Typhimurium T3SSs (yellow). Below, the maps inclined 60° to show the export apparatus and linkers. Top right, overlay of central sections of these maps. Scale bar indicates 10 nm.
Subtomogram averaging of the Salmonella fT3SS with C6 symmetry
FliH is a homolog of the MxiN linker protein, forms dimers and binds one FliI ATPase molecule28 to build a FliH12FliI6FliJ ring below the FlhAC toroid29. This indicates that two FliH molecules make one linker and suggests they extend to the C-ring region, like in vT3SSs. Therefore, we also performed subtomogram averaging with C6 symmetry on ECT data from the Salmonella fT3SS17 to test for the appearance of six linker densities between the ATPase and C-ring. But, no linker densities could be observed in the cytoplasmic region below the HBB (Supplementary Figure S3a, right). Interestingly, classification of these particles also reveals a class (class 3) lacking clear features for the C-ring and ATPase complex and one (class 1), lacking a clear ATPase complex.
Conclusions from comparison of the cytoplasmic region of different T3SSs
As a result of subtomogram averaging with C6 symmetry, we respectively confirmed and revealed six pods, corresponding to the fT3SS C-ring, and six linker densities in the cytoplasmic regions of the vT3SSs of Shigella and Salmonella species. However, we could not observe linker densities in the flagellar HBB. The fT3SS C-ring displays 32 to 36-fold symmetry although there are constantly only six linkers given the hexameric nature of the ATPase. Given that these linkers are readily seen in the vT3SS maps, which are at similar resolution to the HBB one, the fT3SS linkers may be more flexible and hence able to interact dynamically with several nearby C-ring subunits, leading them to become smeared out by the imposition of C6 symmetry.
Parts of the T3SS export apparatuses have common features and function. However, the structure of those parts can display different morphologies (Fig. 5). In particular, the position of FlhAC/MxiAC/InvAC toroid, also termed the export platform, differs by ~5 nm between fT3SS and vT3SS. The position of the ATPase also differs between them, being ~6 nm further below the export platform in fT3SSs. Furthermore, the fT3SS C-ring is ~12 nm wider and ~4 nm shorter than the vT3SS pods. We presume the fT3SS C-ring is positioned closer to the cytoplasmic membrane in order for its top portion to interact with the MotA-MotB stator complex unit located therein for motor torque generation30. This function is not found in vT3SS.
Comparison between fT3SS and vT3SSs.
Centrally sectioned maps of Salmonella HBB, NC and Shigella NC. Gray dotted lines indicate the position of, from top, outer and inner membranes, export platform, export apparatus. Red numbers indicate distances between export platform and apparatus, width of the pod/C-ring region and size of the ATPase region. At the bottom, numbers show percent of particles of each kind entirely lacking features in their cytoplasmic region.
The ATPase is supported into its position by the FliH/MxiN family of linkers. Therefore, Abrusci et al. proposed that the position of the ATPase is controlled by the length of the FliH/MxiN family proteins, as well as by the height and width of the C-ring/pods2. However, despite quite different relative positions of the ATPase complex in their species of origin, FliH and MxiN are both ~230 residues long. This suggests that additional factors control the morphological differences we observe. For instance, FliJ, the γ-subunit equivalent in Salmonella flagella, is 147 residues long when its Shigella vT3SS homolog Spa13 is only 112 residues long. This length difference would lead ~53 Å reduction in length in vT3SS homologs, assuming 1.5 Å per residue of α-helix, reduced maximally to ~26 Å, given that these proteins partially fold upon themselves to form intramolecular α-helical coiled coils7. As FliJ inserts into the central cavity of the FliI ATPase hexamer and binds FlhA7,31, it may connect the CEA to the FlhA export platform, perhaps explaining the ~30 Å height difference in position we see between fT3SSs and vT3SSs ATPase complexes.
Between the vT3SSs we analysed, the main difference is the extra density under the ATPase region of the Salmonella vT3SS, which may be formed by hexameric chaperone-substrate complexes27. Indeed, the vT3SS of Salmonella shows greater secretion activity under the growth conditions we used32 than that of Shigella, which requires induction. However, from our data, we cannot determine which map best reflects the state of active export. For this, we need to analyse in parallel mutants locked in specific secretion modes33, something we have tried but failed to obtain quality data for so far.
Finally, upon classification, we revealed that 26~36% particles of fT3SS and vT3SSs lack clear CEA features. Therefore, not all T3SSs have C-ring/pod and ATPase regions, and these are found at similar ratios for each T3SS studied. Interestingly, this is also observed in Borrelia flagellum, where a similar fraction appeared to lack part of the C-ring34. One possibility is that these particles have lost their C-ring/pods and ATPase structure because they are damaged by the preparation of minicells or in a non-active state for other artefactual reasons. However, vT3SSs are known to have dynamic components in their cytoplasmic domains16. Therefore, this finding could also support the notion that the entire CEA is dynamic in most T3SSs. This notion is strengthened by recent study by ECT and sub-tomogram averaging to examine the structure of the Chlamydia trachomatis T3SS in the presence and absence of host membrane contact. Comparison of the averaged structures indicated that a ~4nm compaction of the basal body occurs when the needle tip contacts the host cell membrane. This compaction was coupled to a stabilization of the cytoplasmic export apparatus35. We did not see an overall compaction in classes containing the ATPase and associated complexes. But, given that 2/3 less particles were collected for the host cell contact condition, the enhancement of the signal seen from the cytoplasmic parts of the T3SS during host cell contact may correspond to an increased number of T3SS possessing these parts when they are likely to be performing effector transport.
Methods
Bacterial strains and cell culture
All bacterial strains and plasmids used in this study are listed in Supplementary Table S1. S. flexneri strains were maintained and selected on CR agar plates36 and grown at 37 °C in trypticase soy broth (TCSB; Becton Dickinson) supplemented with antibiotics when necessary (100 μg of ampicillin ml−1, 50 μg of kanamycin ml−1, 20 μg of chloramphenicol ml−1; Sigma).
Molecular cloning
All primers used in this study are listed in Supplementary Table S2 and all constructs were verified by DNA sequencing (Eurofins). Constructs encoding N-terminally FLAG-tagged MxiK, Spa13, Spa33 and Spa47 were obtained via PCR using primers listed in Supplementary Table S2 and ligated into pUC19 via XbaI/BamHI. The C-terminally FLAG-tagged Spa33-encoding construct was also made as an N-terminally FLAG-tagged Spa33-encoding construct but failed to complement Δspa33.
Analysis of Ipas secretion and protein expression
The secretion of Ipa proteins after Congo red induction was carried our as previously described37. The total level of protein expression was revealed via western blot using either monoclonal anti-FLAG (Sigma) or polyclonal anti-MxiN23. Goat anti-mouse DyLight 800 (Fisher Scientific) or goat anti-rabbit Alexa 680 (Invitrogen) conjugates were used as secondary antibodies. The membranes were then visualized using an Odyssey infrared imaging system (LI-COR Biosciences).
Preparation of minicells
We transformed WT Shigella flexneri M90T serotype 5a with the pBAD24::ftsZ plasmid17. 1 ml of overnight culture was added to 100 ml of TCSB, including the appropriate antibiotic. The culture was incubated at 35 °C, shaking at 160 rpm until an OD600 of ~0.5 was reached. Then, 0.2% v/v arabinose was added. The culture was further incubated at 35 °C shaking at 160 rpm until an OD600 of ~1.0 was reached. The culture was centrifuged at 3,000 g for 7 min to remove the large cells. To collect the minicells, the supernatant was centrifuged at 20,000 g for 5 min and the pellet resuspended in 30 μl of TCSB. To supply a gold fiducial maker, 150 μl of 10 nm nanogold (Sigma) were mixed with 50 μl of 5% w/v BSA, centrifuged at 14000 rpm for 15 min, and the supernatant removed. The sample solution was mixed with gold pellet.
ECT data collection and tomogram reconstruction
We performed ECT data collection as previously described17, with some modifications. Quantifoil molybdenum, 200 mesh, R0.6/1.0 micromachined holey carbon grids (Quantifoil Micro Tools) were glow discharged. 2.7 μl of sample solution supplemented with 10 nm diameter of nanogold was applied to the grid, blotted with filter paper, and plunged into liquid ethane using a Vitrobot (Mk2, FEI), setting blot time to 5 seconds, humidity 100% at 4 °C. Images were collected at liquid-nitrogen temperature using a Titan Krios FEG transmission electron microscope (FEI) operated at 300 kV on a Falcon II direct electron detector (FEI). The pixel size was 0.57 nm. Single-axis tilt series were collected covering an angular range from −65° to +65° with a nonlinear Saxton tilt scheme at 5–10 μm underfocus using the Xplore 3D software package (FEI). A total dose of 200 e−/A2 or less was used for each tilt series. Images were binned two fold and 3D reconstructions were calculated using the IMOD software package38. We applied CTF correction to those tilt series as calculated by TOMOCTF39. The Salmonella minicell data used here were collected in previous work17.
Subtomogram averaging
Individual NCs and HBBs were manually picked up using the IMOD software package with 100 × 100 × 100 pixel box size for Shigella NCs, 128 × 128 × 128 pixel box size for Salmonella NCs, 128 × 128 × 128 pixel box size for Salmonella HBBs. By this procedure, the orientations of particles were visually aligned, so that the needle/hook portion pointed vertically upwards (z-axis) and the base faced downwards, and were recorded in text files to use them as initial parameters in the Dynamo software package (19), which performed subtomgram averaging. These parameters were also used in this package to correct for the missing wedge. The initial volume for subtomogram average was produced manually in Dynamo using a sum of aligned NCs or HBBs with C101 symmetry.
We also prepared a tight mask containing the needle/hook structures, inner, outer membrane, basal body and cytoplasmic region in real space (Supplementary Figure S1a). The tight mask was applied to each particle during each iteration. In the first four iterations, the searching parameters were roughly with 2 times the binning volume (C6: in plane rotation 30 degree, axial rotation 60 degree, 2 degree increments, C1-C3: in plane rotation 30 degree, axial rotation 360 degree, 5 degree increments) and then the searching parameters were refined in a further 8 iterations without binning (in plane rotation 10 degrees, axial rotation 10 degrees, 1-degree increments and maximum allowed searching 2 voxels). During processing, we applied C1, C2, C3 or C6 symmetry. The resolution was estimated by Fourrier Shell Correlation (FSC) using the gold-standard procedure in Dynamo, in which the two independent data sets produced by dividing the particles contributing to the final average into two halves were used in subtomgram averaging independently (Supplementary Figure S2). The 0.143 threshold criterion was used. The resulting 3D maps were rendered using UCSF Chimera.
Classification of subtomogram averages
Classification by multireference alignment in Dynamo was performed by first generating 3–4 initial references by randomly adding 1–2 sigma Gaussian noise using EMAN40 to an average yielded from the final result of subtomgram averaging with C6 symmetry. During the procedure, particles were masked in the cytoplasmic region (Supplementary Figure S1b), and fixed to prevent rotation and translation.
Single particle cryoEM map of Shigella NC
NCs were affinity-purified from strains with mxiG−/N-His6-mxiG, using a protocol described previously from Cheung et al. Here, Ni-NTA agarose beads incubation was shortened to 2 hours. A 3.0 μl sample was applied onto a Quantifoil holey carbon molybdenum grid (R 0.6/1.0), which was layered with continuous thin carbon to ~10 nm thickness in house, at 4 °C and was plunge-frozen into liquid ethane using a fully automated vitrification device (Vitrobot, FEI). The specimen was observed at temperatures of 80 K using a JEOL JEM3200FSC electron microscope, an Ω-type energy filter, and a field-emission electron gun operated at an accelerating voltage of 200 kV. Micrographs were recorded under low-dose conditions on a CCD camera (TemCam-F415MP, TVIPS) at a magnification of 91,000 and an electron dose of ~40 electrons/Å2. Defocus and astigmatism in the images were determined using CTFFIND3. Approximately 8,000 images of NCs were boxed into ~50,000 particle images with side-views of 320 × 320 pixels using a boxer program from EMAN40. Those images were then normalized. Image analysis was performed largely with SPIDER41 and RELION42 packages. After 2D-classification in RELION to select good particles and side-views, we applied C12 symmetry during the three-dimensional reconstruction by using SPIDER package since we previously reported that this is the largest common denominator between OMR and IMR symmetries20.
Additional Information
Accession Codes: The three sub-tomogram averages of T3SSs reported in this study have been deposited in the EM databank with accession numbers EMD-8096 (Shigella vT3SS), EMD-8122 (Salmonella vT3SS), EMD-8121 (Salmonella fT3SS).
How to cite this article: Makino, F. et al. The Architecture of the Cytoplasmic Region of Type III Secretion Systems. Sci. Rep. 6, 33341; doi: 10.1038/srep33341 (2016).
References
Burkinshaw, B. J. & Strynadka, N. C. Assembly and structure of the T3SS. Biochim Biophys Acta 1843, 1649–1663, doi: 10.1016/j.bbamcr.2014.01.035 (2014).
Abrusci, P. et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20, 99–104, doi: 10.1038/nsmb.2452 (2013).
Chen, S. et al. Structural diversity of bacterial flagellar motors. Embo J 30, 2972–2981, doi: 10.1038/emboj.2011.186 (2011).
Hu, B. et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci USA 112, 1047–1052, doi: 10.1073/pnas.1411610112 (2015).
Zhao, X., Norris, S. J. & Liu, J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53, 4323–4333, doi: 10.1021/bi500059y (2014).
Thomas, D. R., Francis, N. R., Xu, C. & DeRosier, D. J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188, 7039–7048 (2006).
Ibuki, T. et al. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol 18, 277–282, doi: nsmb.1977 10.1038/nsmb.1977 (2011).
Jouihri, N. et al. MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol Microbiol 49, 755–767 (2003).
Brown, P. N., Mathews, M. A., Joss, L. A., Hill, C. P. & Blair, D. F. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol 187, 2890–2902, doi: 10.1128/JB.187.8.2890-2902.2005 (2005).
Park, S. Y., Lowder, B., Bilwes, A. M., Blair, D. F. & Crane, B. R. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci USA 103, 11886–11891, doi: 10.1073/pnas.0602811103 (2006).
Notti, R. Q., Bhattacharya, S., Lilic, M. & Stebbins, C. E. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun 6, 7125, doi: 10.1038/ncomms8125 (2015).
Yu, X. J., Liu, M., Matthews, S. & Holden, D. W. Tandem translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J Biol Chem 286, 36098–36107, doi: 10.1074/jbc.M111.278663 (2011).
Bzymek, K. P., Hamaoka, B. Y. & Ghosh, P. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 51, 1669–1677, doi: 10.1021/bi201792p (2012).
McDowell, M. A. et al. Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol 99, 749–766, doi: 10.1111/mmi.13267 (2016).
Zhao, R., Pathak, N., Jaffe, H., Reese, T. S. & Khan, S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol 261, 195–208, doi: 10.1006/jmbi.1996.0452 (1996).
Diepold, A., Kudryashev, M., Delalez, N. J., Berry, R. M. & Armitage, J. P. Composition, formation, and regulation of the cytosolic c-ring, a dynamic component of the type III secretion injectisome. PLoS Biol 13, e1002039, doi: 10.1371/journal.pbio.1002039 (2015).
Kawamoto, A. et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Scientific reports 3, 3369, doi: 10.1038/srep03369 (2013).
Kudryashev, M. et al. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife 2, e00792, doi: 10.7554/eLife.00792 (2013).
Castano-Diez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol 178, 139–151, doi: 10.1016/j.jsb.2011.12.017 (2012).
Hodgkinson, J. L. et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol 16, 477–485 (2009).
Schuch, R. & Maurelli, A. T. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J Bacteriol 183, 6991–6998 (2001).
Okon, M. et al. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16, 1544–1554, doi: 10.1016/j.str.2008.08.006 (2008).
Johnson, S. & Blocker, A. Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol Lett 286, 274–278 (2008).
Bahrani, F. K., Sansonetti, P. J. & Parsot, C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun 65, 4005–4010 (1997).
Cherradi, Y., Hachani, A. & Allaoui, A. Spa13 of Shigella flexneri has a dual role: chaperone escort and export gate-activator switch of the type III secretion system. Microbiology, doi: 10.1099/mic.0.071712-0 (2013).
Schuch, R. & Maurelli, A. T. Spa33, a cell surface-associated subunit of the Mxi-Spa type III secretory pathway of Shigella flexneri, regulates Ipa protein traffic. Infect Immun 69, 2180–2189 (2001).
Roblin, P., Dewitte, F., Villeret, V., Biondi, E. G. & Bompard, C. A Salmonella type three secretion effector/chaperone complex adopts a hexameric ring-like structure. J Bacteriol 197, 688–698, doi: 10.1128/JB.02294-14 (2015).
Minamino, T. & MacNab, R. M. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol Microbiol 37, 1494–1503, doi: mmi2106 (2000).
Bai, F. et al. Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Scientific reports 4, 6528, doi: 10.1038/srep06528 (2014).
Minamino, T. & Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23, 267–274, doi: 10.1016/j.tim.2014.12.011 (2015).
Ibuki, T. et al. Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J Bacteriol 195, 466–473, doi: 10.1128/JB.01711-12 (2013).
Komoriya, K. et al. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol 34, 767–779 (1999).
Veenendaal, A. K. et al. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 63, 1719–1730, doi: MMI5620 10.1111/j.1365-2958.2007.05620.x (2007).
Kudryashev, M., Cyrklaff, M., Wallich, R., Baumeister, W. & Frischknecht, F. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169, 54–61, doi: 10.1016/j.jsb.2009.08.008 (2010).
Nans, A., Kudryashev, M., Saibil, H. R. & Hayward, R. D. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat Commun 6, 10114, doi: 10.1038/ncomms10114 (2015).
Meitert, T. et al. Correlation between Congo red binding as virulence marker in Shigella species and Sereny test. Roum Arch Microbiol Immunol 50, 45–52 (1991).
Martinez-Argudo, I. & Blocker, A. J. The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol Microbiol 78, 1365–1378, doi: 10.1111/j.1365-2958.2010.07413.x (2010).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76, doi: 10.1006/jsbi.1996.0013 (1996).
Fernandez, J. J., Li, S. & Crowther, R. A. CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–596, doi: 10.1016/j.ultramic.2006.02.004 (2006).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128, 82–97, doi: 10.1006/jsbi.1999.4174 S1047-8477(99)94174-6 (1999).
Shaikh, T. R. et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3, 1941–1974, doi: 10.1038/nprot.2008.156 (2008).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, doi: 10.1016/j.jsb.2012.09.006 (2012).
Acknowledgements
We thank Dr. Takayuki Kato (Osaka) for support with electron microscope maintenance, use and image processing and Dr. Morgan Beeby (Imperial College London) for discussions. This work was funded by UK MRC and Wellcome Trust project grants (MR-J002097-1 and WT088231, respectively) to AJB and KN and by JSPS KAKENHI Grant number 25000013 to KN.
Author information
Authors and Affiliations
Contributions
A.J.B. and F.M. and conceived and designed the experiments; P.P., M.P. and I.M. generated the minicell strains and optimised minicell generation; F.M. and A.K. optimised the minicell isolation, collected and processed the ECT data; N.K. provided the single particle cryoEM map of the Shigella NC; H.O. and D.S. constructed the FLAG-tagged strains and performed the co-stability analysis; F.M., K.N. and A.J.B. analysed the data; F.M. and A.J.B. wrote the paper, with comments from D.S. and K.N. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Makino, F., Shen, D., Kajimura, N. et al. The Architecture of the Cytoplasmic Region of Type III Secretion Systems. Sci Rep 6, 33341 (2016). https://doi.org/10.1038/srep33341
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep33341
This article is cited by
-
Type three secretion system in Salmonella Typhimurium: the key to infection
Genes & Genomics (2020)
-
A dynamic and adaptive network of cytosolic interactions governs protein export by the T3SS injectisome
Nature Communications (2017)
-
Assembly, structure, function and regulation of type III secretion systems
Nature Reviews Microbiology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.