Abstract
The generation of human neural tissue-like 3D structures holds great promise for disease modeling, drug discovery and regenerative medicine strategies. Promoting the establishment of complex cell-cell interactions, 3D culture systems enable the development of human cell-based models with increased physiological relevance, over monolayer cultures. Here, we demonstrate the establishment of neuronal and astrocytic metabolic signatures and shuttles in a human 3D neural cell model, namely the glutamine-glutamate-GABA shuttle. This was indicated by labeling of neuronal GABA following incubation with the glia-specific substrate [2-13C]acetate, which decreased by methionine sulfoximine-induced inhibition of the glial enzyme glutamine synthetase. Cell metabolic specialization was further demonstrated by higher pyruvate carboxylase-derived labeling in glutamine than in glutamate, indicating its activity in astrocytes and not in neurons. Exposure to the neurotoxin acrylamide resulted in intracellular accumulation of glutamate and decreased GABA synthesis. These results suggest an acrylamide-induced impairment of neuronal synaptic vesicle trafficking and imbalanced glutamine-glutamate-GABA cycle, due to loss of cell-cell contacts at synaptic sites. This work demonstrates, for the first time to our knowledge, that neural differentiation of human cells in a 3D setting recapitulates neuronal-astrocytic metabolic interactions, highlighting the relevance of these models for toxicology and better understanding the crosstalk between human neural cells.
Similar content being viewed by others
Introduction
There has been an increasing demand for a paradigm shift from animal cells towards robust human cell models of high physiological relevance1. This is due to the fact that over the last years, the drug discovery and development pipelines have registered low success rates both during preclinical and clinical phases, with only 23 small molecules and 2 biologics being approved by the Food and Drug Administration (FDA) in 20132,3. These is even more dramatic for neurological disorders, such as Alzheimer’s disease for which there is an estimated overall success rate of only 0.5%4. Although several factors have been described to contribute to this, the current lack of adequate and predictive preclinical models plays an important role. Most of the available preclinical models are based on animals or immortalized cell lines, which do not accurately recapitulate key events of human pathologies5,6,7. In this context, the generation of human neural 3D cell models holds great promise for drug discovery, toxicology and disease modeling.
Multiple human neural cell sources have become available, including immortalized cell lines, embryonic or adult stem cells and induced pluripotent stem cells (iPSC)8. The human Ntera2/clone D1 (NT2) cell line has been extensively characterized, demonstrating its ability to generate neuronal and astrocytic cells9,10,11. NT2 neural differentiation has been shown to recapitulate developmental processes observed during in vivo neurogenesis12, generating neuronal cells able to establish functional synapses and elicit action potentials following depolarization13,14. Given these properties, the NT2 cell line has been reported to be a promising human cell source for toxicological applications15,16,17,18. Traditional cell culture systems based on cell monolayers do not reflect the in vivo architecture complexity, mechanical and biochemical cues, failing to mimic important features of the target tissue/organ. Conversely, 3D culture systems allow to recapitulate the in vivo cell-cell and cell-extracellular matrix interactions (ECM), while presenting an intermediate degree of complexity between traditional 2D cultures and the organ19. Different strategies can be explored for 3D cell culture, including embedding cells in artificial matrices/scaffolds20 or as cell aggregates21. Neural differentiation of human stem cells in these 3D systems results in a heterogeneous culture, composed of the different neural cell lineages present in the brain: neurons, astrocytes and oligodendrocytes15,22,23. Complex cell-cell interactions and networks can be established within these cell aggregates, or neurospheres, making it possible to mimic key brain features, such as synaptic activity or electrophysiological properties. Recently, Choi et al. have been able to recapitulate amyloid-β and tau pathological events of Alzheimer’s disease in a human 3D cell model, which was not possible to model in rodents nor in 2D cultures24. Human neural 3D cell models can thus be valuable tools to study human neural metabolism25, as this field has traditionally relied on rodent models, using both in vivo studies and primary cultures, which can diverge considerably from the human phenotype and make it impossible to develop appropriate treatment approaches.
The brain’s homeostasis depends on multiple metabolic cycles between the different cell populations that represent major hallmarks of neural metabolism. For instance, as neurons are continually loosing tricarboxylic acid (TCA) cycle intermediates, mainly for the synthesis of neurotransmitters (α-ketoglutarate is converted into glutamate and subsequently GABA), compensatory mechanisms have to act in order to replenish the carbon skeleton for maintaining TCA cycle activity26. Some metabolic neural features have been showed to be present in 2D cultures of stem cell-derived neural cells, as glycogen synthesis in astrocytes or the ability to modulate glucose consumption in response to glutamate addition27. The presence of astrocytes in co-cultures with neurons has been shown also to contribute to a closer recapitulation of in vivo features, such as the dopaminergic neurodegeneration process following administration of neurotoxicants28,29. In vitro co-culture systems have also been proposed to be relevant tools for studying neurodevelopmental processes, including neuritogenesis and synaptogenesis30,31. Still, other important neural metabolic hallmarks, including the glutamine-glutamate-GABA cycle between neurons and astrocytes32 have not been previously reported for these stem-cell derived co-cultures cultures. This specific neuron-astrocyte shuttle is one of the major metabolic specialization and compartmentation in brain tissue, where astrocytes are able to take up glutamate or GABA released by neurons into the synaptic cleft and convert glutamate to glutamine or GABA to Succinyl CoA, entering the TCA cycle via the GABA shunt. Astrocyte derived-glutamine is then transported back to neurons, acting as precursor for neurotransmitter synthesis. These cycles are critical in order to control the excitotoxic effects of glutamate as well as to feed neurons with a neurotransmitter precursor.
The use of 13C-labeled substrates and nuclear magnetic resonance (NMR) spectroscopy are powerful tools for the study of neural metabolic networks, having provided important insights into the biochemical mechanisms and compartmentation of neural metabolism33,34,35. Different 13C-labeled substrates have been used to study neuronal-glial metabolic interactions, taking advantage of cell specific metabolic features. An example of this metabolic specialization is the fact that glucose is taken up mostly by neurons36, while astrocytes have been shown to metabolize acetate37,38.
In this work, we combined the use of a human 3D in vitro neural model with 13C-NMR spectroscopy to study the metabolic features of human neuronal-astrocytic networks. Furthermore, to study neuronal-glial metabolic interactions in detail the cells were challenged with compounds targeting specifically astrocytic or neuronal populations, using methionine sulfoximine (MSO) and acrylamide, respectively. This study demonstrates that some of the main metabolic features and specializations found in brain tissue are recapitulated in human neuron-astrocyte 3D in vitro networks.
Materials and Methods
Cell culture and 3D neural differentiation
Ntera-2 (NT2) cells were routinely cultivated in standard tissue culture flasks (Sarstedt) and maintained in DMEM (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone) and 1% (v/v) penicillin-streptomycin (P/S; Life Technologies), as previously described39. NT2 neural differentiation was performed as previously described15. Briefly, 125 mL silanized spinner flasks (Wheaton) equipped with ball impeller were inoculated with a single cell suspension at 6.7 × 105 cell/mL in 75 mL of DMEM with 10% FBS and 1% P/S. On the day after, 50 mL of fresh medium were added. At day 3, neural differentiation was induced adding 20 μM retinoic acid (RA; Sigma-Aldrich), by performing a 50% medium exchange. RA treatments were performed every 2–3 days, during 3 weeks. After this period, cells were posteriorly maintained in DMEM with 5% (v/v) FBS, 1% (v/v) P/S and absence of RA during 2 weeks, for neuronal maturation. Spinner flasks stirring speed was gradually increased from 40 to 100 rpm throughout culture time. Cells were maintained in a humidified atmosphere of 5% CO2 in air at 37 °C.
Incubation with 13C labeled substrates
After neural differentiation process (day 38), two 50% media exchanges were performed in the following two days of culture with a low glucose DMEM medium (5.5 mM glucose), 5% FBS and 1% P/S. At day 3 the medium was completely removed and replaced by 100 mL of: (i) DMEM with 5.5 mM [1-13C]glucose, 3 mM acetate and 5% FBS or (ii) DMEM with 5.5 mM glucose, 3 mM [2-13C]acetate and 5% FBS. After 12 hours incubation, neurospheres were harvested, centrifuged at 300 × g for 2 minutes and washed 2 times in ice-cold PBS followed by centrifugation. The obtained cell pellets were the immersed in liquid nitrogen and 2 mL of 70% (v/v) ethanol was added. Complete cell lysis was achieved by ultrasound sonication (Sonifier 250D, Branson). The cell extracts were centrifuged two times at 20,000 × g for 15 minutes. Cell pellets were stored at −80 °C for total protein quantification.
Fate of [1-13C]glucose and [2-13C]acetate
A simplified representation of the main neuronal and astrocytic metabolic networks, as well as the labeling patterns resulting from the metabolism of [1-13C]glucose and [2-13C]acetate, is shown in Fig. 1. Through glycolysis, [1-13C]glucose can be metabolized into [3-13C]pyruvate, which can then be reduced to [3-13C]lactate or transaminated to [3-13C]alanine. The labeled pyruvate molecules can enter the tricaboxylic acid (TCA) cycle in the mitochondria via pyruvate dehydrogenase (PDH) with formation of [2-13C]acetyl-CoA, which is converted to [2-13C]citrate. With the progression of the TCA cycle, α-[4-13C]ketoglutarate is formed and can exit the cycle for the synthesis of [4-13C]glutamate. This can act as neurotransmitter, in case of glutamatergic neurons, or be used to fuel the synthesis of [4-13C]glutamine in astrocytes or [2-13C]GABA in GABAergic neurons. In case α-[4-13C]ketoglutarate remains in the TCA cycle, the label will be scrambled by the formation of succinate that is a symmetric molecule, resulting in equal amounts of [2-13C]- and [3-13C]succinate that gives rise to fumarate, malate and oxaloacetate labeled in the same positions. [2-13C]- and [3-13C]oxaloacetate can also exit the cycle by being converted into aspartate, which will have the same carbon positions labeled. If the cycle continues with the labeled oxaloacetate, this will be condensed with an acetyl-CoA molecule, which can be unlabeled or labeled. In presence of unlabeled acetyl-CoA, [3-13C]- or [2-13C]glutamate/glutamine and [3-13C]- or [4-13C]GABA will be generated from the second turn of TCA cycle. In case the of labeled acetyl-CoA, the generated molecules will also present labeling in the positions described previously for the first turn, resulting in [3,4-13C]- or [2,4-13C]glutamate/glutamine and [2,3-13C]- or [2,4-13C]GABA.
(A) Schematic representation outlining the main neuronal-astrocytic metabolic networks and glutamine-glutamate/GABA cycles. (B) 13C-labeling patterns of metabolites derived from [1-13C]glucose and [2-13C]acetate. 13C-label can enter the TCA clycle via pyruvate dehydrogenase (PDH; orange circle) or via pyruvate carboxylase (PC; blue circle).
Alternatively, [3-13C]pyruvate can enter the TCA cycle through the anaplerotic pathway, which in neural cells is achieved via pyruvate carboxylase (PC), leading to the formation of [3-13C]oxaloacetate. This can exit the cycle by transamination generating [3-13C]aspartate or proceed in the cycle forming α-[2-13C]ketoglutarate, which can act as precursor for the synthesis of [2-13C]glutamate/glutamine and [4-13C]GABA. In case the label remains in the cycle for a second turn, [1-13C]glutamate/glutamine will be formed and the decarboxylation of glutamate into GABA will result in the loss of the labeled carbon. It should be noted that backflux of labeled oxaloacetate after carboxylation of pyruvate will lead to scrambling of the label from the C3 position in oxaloacetate to the C2 position and make it indistinguishable from oxaloacetate obtained from cycling. Thus, pyruvate carboxylation assessment using [1-13C]glucose can be underestimated, since backflux is not taken into account40.
Incubation with [2-13C]acetate results in its conversion into [2-13C]acetyl-CoA, which will then enter the TCA cycle and lead to a labeling pattern similar to what was described above for glucose.
Incubation with toxic compounds
For the experiments with toxic compounds, two 50% media exchanges with low glucose medium were also performed in two consecutive days after neural differentiation (day 38). For acrylamide incubation experiments, at day 3 after differentiation, the medium was completely removed and replaced by DMEM with 5.5 mM glucose, 3 mM acetate, 2.5 mM acrylamide and 5% FBS. After 36 hours of incubation, the medium was completely exchanged with a medium of similar composition but with [1-13C]glucose and maintained for further 6 hours. For methionine sulphoximine (MSO) experiments, at day 3 after differentiation, the medium was completely removed and maintained for 12 hours with DMEM containing 5.5 mM glucose, 3 mM [2-13C]acetate, 20 μM MSO and 5% FBS. Following incubation with labeled substrates, cells were harvested as described above. Control experiments were performed in parallel under the same experimental conditions except for the absence of the toxic compounds.
NMR sample preparation and analysis
Lyophilized samples were prepared as previously decribed41, by dissolving in 120 mL D2O containing 0.25% ethylene glycol and 0.002% TSP. Samples were then transferred to SampleJet tubes (3.0 ± 103.5 mm). All samples were analyzed on a 14.1T Ultra shielded Plus Avance III magnet (Bruker BioSpin GmbH, Rheinstetten, Germany) operating at 600 MHz (for 1H) using QCI CryoProbe (Bruker BioSpin GmbH) and equipped with SampleJet auto sampler (Bruker BioSpin GmbH). 1H-NMR spectra were accumulated with a pulse angle of 90°, 2.7 s acquisition time, and 10 s relaxation delay. The number of scans was 128. Proton decoupled 13C-NMR spectra were obtained using an acquisition time of 1.7 s, 0.5 s relaxation delay, and a 30° flip angle. Scans were accumulated at 30 kHz spectral width with 98 K data points. The number of scans was typically 2000. Relevant peaks in the spectra were identified and integrated using MNova software (Mestrelab Research, Santiago de Compostela, Spain). Concentrations of metabolites were calculated from the integrals of the peaks using TSP (1H-NMR spectra) or ethylene glycol (13C-NMR spectra) as internal quantification standards. Concentrations from the 1H-NMR spectra were corrected for the numbers of protons that constituted the peak. Correction factors for incomplete relaxation and nuclear Overhauser effects in the 13C spectra were obtained by acquiring scans with completely relaxed nuclei (relaxation delay of 20 s) and only a brief proton decoupling during the radiofrequency pulse shortly before the signal was recorded to avoid heteronuclear splitting of signals, but no proton decoupling for the remainder of the acquisition to avoid nuclear Overhauser effects. The singlets in the 13C-nuclear magnetic resonance spectroscopy (13C-NMRS) data were corrected for naturally abundant 13C by subtracting 1.1% of the total cellular contents obtained from 1H spectra or HPLC data. All amounts were corrected for total protein weight. Percent excess enrichment was calculated after subtracting natural abundance where appropriate, and is in the following referred to as percent enrichment.
HPLC analysis
Amino acids were quantified by high performance liquid chromatography (HPLC) using a pre-column derivatization method based on the Waters AccQ. Tag Amino Acid Analysis (Waters, USA) as previously described42. Briefly, sample proteins were precipitated by adding an equal volume of acetonitrile and removed by centrifugation at 13,000 × g for 15 minutes. The obtained supernatants were used for primary and secondary aminoacid derivatization by mixing with 6-aminoquinolyl N-hydroxysuccinimidyl-carbamate, which allows their separation in a reversed phase column (Waters, USA) and the detection of fluorescence at 395 nm. As internal standard, α-aminobutyric acid was added to ensure consistency between runs. Mobile phases were prepared following the manufacturer’s instructions, filtered and degassed in an ultrasound bath before usage.
Total protein quantification
Total cell biomass was evaluated by quantification of total protein using the bicinchoninic acid (BCA) protein assay kit (Pierce), after dissolving the cell pellet in Tris buffer (50 mM Tris, 5 mM EDTA, 150 mM NaCl, pH 7.4).
Immunofluorescence microscopy
Neurospheres were fixed in 4% (w/v) paraformaldehyde (PFA) + 4% (w/v) sucrose in PBS for 20 min and processed for immunostaining as previously described43. Primary and secondary antibodies were used as follows: mouse anti-βIII-tubulin (1:200; Millipore Darmstadt, Germany, MAB1637); rabbit anti-GFAP (1:200; Millipore, AB5804); AlexaFluor® 488 goat anti-mouse IgG (1:500; Life Technologies, A11001) and AlexaFluor® 594 goat anti-rabbit IgG (1:500; Life Technologies, A11012). Cell nuclei were counterstained with TO-PRO-3 (Life Technologies). Samples were visualized using point-scan confocal microscopy (SP5, Leica, Wetzlar, Germany) or light-sheet microscopy44,45. Merge between channels and maximum z-projections, as well as linear brightness and contrast adjustments of the images, were performed using the open source FIJI software46.
Real time RT-PCR
Total RNA was extracted with High Pure RNA Isolation Kit (Roche), according to the manufacturer’s instructions. RNA was quantified in a NanoDrop 2000c (Thermo Scientific) and used for cDNA synthesis. Reverse transcription was performed with High Fidelity cDNA Synthesis Kit (Roche), using Anchored-oligo(dT)18 Primer (Roche) or with the Super Script III First Strand synthesis system (Invitrogen), using random hexamers (Invitrogen). qPCRs were performed in triplicates using LightCycler 480 SYBR Green I Master Kit (Roche) with the following primers: βIII-tubulin (TUBB3; fwd 5′-GGGCCTTTGGACATCTCTTC-3′ and rev 5′-CCTCCGTGTAGTGACCCTTG-3′), glial fibrillary acidic protein (GFAP; fwd 5′-AGAGAGGTCAAGCCCAGGAG-3′ and rev 5′-GGTCACCCACAACCCCTACT-3′) and ribosomal protein L22 (RPL22; fwd 5′-CACGAAGGAGGAGTGACTGG-3′ and rev 5′-TGTGGCACACCACTGACATT-3′). The reactions were performed with LightCycler 480 Instrument II 96-well block (Roche). Quantification cycle values (Cq’s) and melting curves were determined using LightCycler 480 Software version 1.5 (Roche). All data were analyzed using the 2−ΔΔCt method for relative gene expression analysis47. Changes in gene expression were normalized using the housekeeping gene RPL22 as internal control.
Synaptic activity assessment
Neurospheres plated on poly-D-lysine (PDL)-coated multi-well plates were washed with PBS prior to a 5 minutes incubation with 100 mM KCl Buffer (5 mM HEPES-NaOH, pH 7.4; 10 mM glucose; 2.5 mM calcium chloride; 1 mM magnesium chloride; 37 mM sodium chloride; 100 mM potassium chloride). Afterwards, neurospheres were incubated for 15 minutes with 10 mM FM1–43 dye (Life Technologies) dissolved in normal saline buffer (5 mM HEPES-NaOH, pH 7.4; 10 mM glucose; 2.5 mM calcium chloride; 1 mM magnesium chloride; 37 mM sodium chloride; 5 mM potassium chloride). Neurospheres were then washed with ADVASEP-7 (Sigma) dissolved in 5 mM KCl buffer for 1 min, followed by three washes of 1 min with 5 mM KCl buffer. Exocytosis was stimulated with 100 mM KCl buffer and samples were imaged live in a fluorescence microscope (DMI6000; Leica) to monitor synaptic vesicle release. Fluorescence intensity was measured using the open source FIJI software46. FM1-43 unloading analyses were performed after normalization by dividing the fluorescence intensity of each frame by the initial fluorescence intensity. FM1-43 fluorescence decrease rate was determined by a linear regression analysis of the normalized fluorescence intensity time profile for the 15 minutes following KCl stimuli.
Statistical Analysis
All values are presented as means ± standard error of the mean (s.e.m.). All analyticals were performed as triplicates from samples of two independent experiments. Student’s t-tests were used to compare means. Before choosing the adequate type of t-test, Levene’s test for equal variances was performed. P < 0.05 was chosen as the level of significance. All comparisons were made using two-tailed statistical tests.
Results
Neuronal and glial metabolic signatures and trafficking
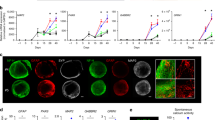
NT2 aggregates differentiation towards the neural lineage can be achieved upon RA induction and subsequent maturation (Fig. 2A), resulting in highly viable (Fig. 2B) differentiated neurospheres composed mainly of βIII-tubulin-positive neurons and GFAP-positive astrocytes (Fig. 2C), as previously reported by our group15. Phenotypic characterization demonstrated that the neuronal population was comprised by glutamatergic (vGluT1-positive) and GABAergic (GAD65/67-positive) neurons (Fig. 2D,E). In this study, differentiated NT2 neurospheres were incubated with [1-13C]glucose or [2-13C]acetate, and the fate of the 13C label was observed by 13C-NMR spectroscopy for the identification of major neural metabolic signatures.
(A) Schematic experimental workflow for NT2 3D differentiation. Cells were inoculated in a stirred suspension culture system and aggregated for 3 days. Neural differentiation was induced by exposure to retinoic acid (RA) for 3 weeks, followed by 2 weeks without RA. Differentiated neurospheres were further cultured in presence of 13C-labeled substrates for 6-12 hours. (B) Immunofluorescence light-sheet microscopy of neurospheres stained for βIII-tubulin. Scale bar, 50 μm. (C–E) Immunofluorescence light-sheet microscopy of neurospheres stained for βIII-tubulin and GFAP (C), βIII-tubulin and vGluT1 (D), βIII-tubulin and GAD65/67 (E). Scale bars, 20 μm.
Typical 13C-NMR spectra after incubation with each 13C labeled substrate are presented in Fig. 3. Incubation with [1-13C]glucose resulted in detectable levels of labeled glutamine, glutamate, lactate, alanine, aspartate and GABA (Fig. 4A and Table 1). After 12 hours of incubation, glutamate and glutamine were mainly labeled at C4, which results from one turn in the TCA cycle (Fig. 1). Additionally, the presence of labeling in C2 and C3 positions indicates label cycling in the TCA cycle. The amounts of [2-13C]glutamine were higher relative to [2-13C]glutamate, while the labeling in C3 was similar for both. This reflects the different contributions of the anaplerotic (via PC) and oxidative (via PDH) pathways for the synthesis of these two metabolites. These contributions can be expressed as the ratio PC/PDH, which can be estimated by dividing the difference between C2 and C3 by C4 ((C2-C3)/C4)48. This was found to be 2.3-fold higher for glutamine in comparison with glutamate (Fig. 4B), revealing an increased contribution of PC over PDH for glutamine synthesis. GABA labeling was only observed in the C2 position, which derives from the decarboxylation of the most abundant glutamate isotopomer (C4). Aspartate was labeled in the C2 and C3 positions with no significant differences between the two. Labeling of alanine and lactate in the C3 position was also observed, which derive from [3-13C]pyruvate molecules generated from glycolysis.
(A) Intracellular amounts of 13C labeled metabolites in cultures incubated with [1-13C]glucose (blue bars) or [2-13C]acetate (orange bars). (B) Metabolic ratios for PC/PDH described by the (C2-C3)/C4 ratio of labeled glutamate (Glu) and glutamine (Gln) in cultures incubated with [1-13C]glucose (blue bars) or [2-13C]acetate (orange bars). Data are mean ± s.e.m. P values are given for PC/PDH ratio (analysis of glutamate versus glutamine): ***P < 0.001. PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase.
Incubation with [2-13C]acetate during 12 hours resulted also in significant labeling of glutamate, glutamine, aspartate and GABA (Fig. 4A and Table 1). The percentage of 13C enrichment was similar or slightly lower than that from [1-13C]glucose in all metabolites, except in glutamine, where a higher 13C incorporation from [2-13C]acetate is in accordance with acetate being a major carbon source for glutamine synthesis in astrocytes. As observed with [1-13C]glucose, labeling in C4 was the most common in glutamate and glutamine. Moreover, significant differences were not observed for the PC/PDH ratio for glutamine and glutamate, as in this case there is not contribution from the PC pathway for the labeling pattern, since [2-13C]acetate is converted into [2-13C]acetyl-CoA that enters directly the TCA cycle (Fig. 4B). Finally, labeling of GABA in the C2 position was observed (Figs 3 and 4A), suggesting the trafficking of labeled glutamine from astrocytes to neurons that acts as precursor for GABA synthesis.
Inhibition of astrocytic glutamine synthesis
To isolate the contribution of astrocytic-derived glutamine as precursor for neuronal neurotransmitter synthesis, following the glutamine-glutamate/GABA shuttles between neurons and astrocytes, differentiated neurospheres were incubated with MSO, a specific glutamine synthase (GS) inhibitor.
The presence of MSO in the culture medium resulted in an almost complete absence of 13C incorporation in glutamine coming from [2-13C]acetate (Fig. 5A) and a 2.8-fold decrease in the intracellular glutamine pool (Fig. 5B). In accordance with the role of astrocytic glutamine as a key precursor for GABA synthesis in neurons, decreased [2-13C]GABA labeling and a 50% reduction in intracellular GABA content was also observed (Fig. 5A,B). As for glutamate, its total intracellular pool was not affected in MSO-treated cultures. Nevertheless, a modest increase in 13C labeled glutamate was observed, mostly visible in the amount of [3-13C]glutamate, suggesting a slower degradation or accumulation, as its condensation with ammonia to generate glutamine is impaired by MSO. The catabolism of branched chain amino acids (BCAA), comprising valine, isoleucine and leucine, was decreased in cultures exposed to MSO, as shown by the significantly higher intracellular pools detected (Fig. 5B).
(A) Intracellular amounts of 13C labeled metabolites in cultures incubated with [2-13C]acetate alone (blue bars) or [2-13C]acetate plus MSO (red bars). (B) Total concentration of intracellular metabolites in control (blue bars) and MSO-treated cultures (red bars). Data are mean ± s.e.m. Asterisks indicate significant difference: *P < 0.05; **P < 0.01; ***P < 0.001; near significant P values (<0.1) are given in numbers. Gln, glutamine; Val, valine; Ile, isoleucine; Leu, Leucine.
Impairment of neuronal metabolism
The NT2 neural cells were further challenged by incubating the neurospheres with [1-13C]glucose in the presence of acrylamide, a toxic compound that specifically targets the neuronal population. The specificity of acrylamide toxicity towards neurons was confirmed by gene expression analysis of neuronal (TUBB3 and SYP) and glial (GFAP) specific transcripts. After incubation with acrylamide, TUBB3 and SYP expression levels were significantly lower in comparison with control, with a 4- and 5.3-fold decrease respectively, while no modulation on GFAP expression was observed (Fig. 6B).
(A) Intracellular amounts of 13C labeled metabolites in cultures incubated with [1-13C]glucose alone (blue bars) and [1-13C]glucose plus acrylamide (red bars). (B) Gene expression analysis of differentiated neurospheres in control (blue bars) and acrylamide-treated cultures (red bars). Gene expression fold changes (normalized to expression levels previous to incubation) of neuronal markers (TUBB3, SYP) and glial marker (GFAP). (C) Synaptic activity assessment by FM1-43 unloading of differentiated neurospheres in control (blue trace) and acrylamide-treated cultures (red trace). Data represents a single exponential fit and standard deviation of the measured fluorescence values. (D) FM1-43 fluorescence decrease rate determined for control (blue bar) and acrylamide-treated cultures (red bar). Data are mean ± s.e.m. Asterisks indicate significant difference: *P < 0.05; **P < 0.01; ***P < 0.001; near significant P values (<0.1) are given in numbers.
13C-NMRS analysis revealed significant changes in the amounts of labeled glutamate and GABA after incubation with acrylamide (Fig. 6A). While the amounts of labeled glutamate in the C2, C3 and C4 positions were significantly higher in comparison with the control, [2-13C]GABA amounts significantly decreased in the presence of acrylamide. Additionally, a significant increase of [2-13C]glutamine was observed, suggesting an increased contribution of the anaplerotic pathway for glutamine synthesis upon acrylamide exposure. As for the total intracellular metabolite pools, no significant differences were detected between acrylamide exposed neurospheres and control (data not shown).
The effects of acrylamide on neuronal functionality were also assessed. For this, synaptic activity of differentiated neurospheres was evaluated using the fluorescent probe FM1-43, which provides a direct measurement of synaptic vesicle release following a depolarizing KCl stimulus49,50. The analysis of FM1-43 unloading kinetics demonstrated that the same stimuli induced a lower dimming of fluorescence intensity in acrylamide-treated neurospheres (Fig. 6C), which was reflected by a significantly lower fluorescence decrease rate in comparison with control neurospheres (Fig. 6D).
Discussion
We have previously demonstrated that 3D cultures of human mature neurons and functional astrocytes can be derived from NT2 cells15. The use of this 3D differentiation protocol was also shown to attain higher cell yields in half of the culture time, as compared with 2D cultures16,43. Importantly, NT2 differentiated neurospheres present a cellular organization that better mimics the in vivo tissue structural features, with an interspersed distribution of neurons and astrocytes. In 2D cultures, these neuron-glia interfaces are much less pronounced, resulting in a compartmentalized cellular organization where neurons bundle in small aggregates on top of a monolayer of glial cells51,52. Recent reports support the increased inter-cellular communications established in 3D neural models via secreted mediators, as the extracellular deposition of amyloid-β that was only observed in 3D cultures of human differentiated neurons expressing familial Alzheimer’s disease (FAD) mutations24; neurodevelopment regional specification53 and the effects of viral pathogen ZIKV exposure during neurodevelopment54. Therefore, metabolic data from 3D neural cell models can provide valuable insights, by offering the possibility to better understand the biochemical changes induced by compounds of interest in a controlled setting25. In this context, the metabolic specialization and establishment of functional metabolic shuttles between the different cell compartments are major hallmarks of brain functionality. Thus, the main goal of this work was to investigate whether human neural cells generated in 3D cultures were able to acquire the main metabolic features and interactions found in the human brain.
To analyze the metabolic profile of the differentiated NT2 neurospheres, we followed the fate of two different 13C-labelled substrates: [1-13C]glucose and [2-13C]acetate. While glucose acts as carbon source mainly for neurons, acetate is only taken up and metabolized by astrocytes37,38. Using both substrates it is possible to assess the presence of the two different cellular compartments by following the incorporation of the label by 13C NMR spectroscopy. This approach had previously been explored by our group for deciphering the main neural metabolic pathways in mono- and co-cultures of primary rodent neural cells in 3D culture systems55,56,57.
Incubation with [1-13C]glucose revealed the presence of cell-specific metabolic features, in agreement with phenotypic data that showed heterogeneous differentiated neurospheres composed of neurons and astrocytes15. The observed amounts of C2 13C labeled glutamate and glutamine indicate differential contributions of anaplerotic and oxidative pathways to pyruvate incorporation into the TCA cycle for the synthesis of these two metabolites. This is expressed as the PC/PDH ratio48, which was found to be 2.3-fold higher for glutamine synthesis, in comparison with glutamate. These observations are in agreement with previous reports in 2D primary cultures of murine cells and rat brains, showing the exclusive activity of PC in murine astrocytes58,59, the cellular compartment responsible for glutamine synthesis60. When analyzing the 13C-labeling in C4, similar amounts of [4-13C]glutamine and [4-13C]glutamate were observed, suggesting that astrocytes are present in higher numbers than neurons in these 3D cultures. Moreover, the higher contribution of the oxidative pathway for glutamate synthesis, relative to glutamine, and the presence of labeled GABA strongly suggest the presence of mature neuronal populations comprising glutamatergic and GABAergic lineages.
Incubation with [2-13C]acetate resulted in labeling patterns reflecting glial specific metabolism, in agreement with previous reports describing that acetate is mostly metabolized by astrocytes, in 2D murine primary cultures37,38. The overall percent enrichment in 13C labeling with [2-13C]acetate was similar to [1-13C]glucose incubation, demonstrating the importance of oxidative pathways for astrocytic metabolism. Our data demonstrate the presence of C2 labeled GABA, for which astrocyte-derived [4-13C]glutamine acts as precursor37. This indicates the establishment of such an important neuron-glia interaction as the glutamine-glutamate-GABA shuttle32. These findings, together with the observation of PC activity in astrocytes, reveal the presence of typical neural metabolic features important for neural tissue function. To our knowledge, this is the first report demonstrating these features in human stem-cell derived neural cells.
The presence of neuronal-astrocytic metabolic trafficking was further investigated by specifically challenging either astrocytic or neuronal populations, through MSO or acrylamide exposure, respectively. MSO has been widely described to act as an irreversible inhibitor of GS activity61, and has been linked to debilitating neurological disorders when ingested by animals or humans62. The presence of 20 μM MSO in differentiated neurospheres led to an almost complete inhibition of glutamine synthesis, as observed by the extensive reduction in the amount of 13C labeled glutamine. Also, the intracellular pools of BCAAs were found to be higher in MSO exposed cultures, suggesting a decreased catabolism of these amino acids. BCAAs have been described to have an important role in nitrogen turnover, contributing with their amino group to de novo glutamate biosynthesis, mainly through the activity of branched-chain aminotransferase (BCAT)63,64,65. Still, in the adult human brain and contrary to rodent neural cells, this enzyme is absent in astrocytes and is only expressed as a cytosolic isoform in neurons, mostly glutamatergic and GABAergic, and as a mitochondrial isoform in endothelial cells of the vasculature66. Our results suggest that MSO exposure can modulate BCAA metabolism in human neurons, where MSO can inhibit BCAT activity, although less potently than GS. This modulatory effect on BCAT had been proposed in studies on rodent brains67, but hadn’t been previously described for 2D neural cultures or in human neural cells. More importantly, MSO-treatment resulted in a significant decrease (50%) of GABA intracellular pool, likely due to shortage of glutamine from astrocytes. These results further support the establishment of the glutamine-glutamate-GABA cycle in the human cell model employed, and are in line with previous reports in 2D primary cultures of murine cells, arguing that GABAergic neurons are highly dependent on astrocyte-derived glutamine for neurotransmitter synthesis37.
Neurospheres were exposed to acrylamide to further challenge the neuronal-astrocytic metabolic interactions, in this case by targeting specifically the neuronal population. Acrylamide, a water-soluble alkene, causes cumulative neurotoxicity and neurodegeneration in adult humans and animals68. The mechanisms of acrylamide toxicity have been proposed to involve interference with kinesin-related motor proteins in neurofilaments, impairing axonal transport and ultimately leading to axonal degeneration and cell death69. More recently, the ability of acrylamide to form covalent adducts with highly nucleophilic cysteine thiolate groups located in active sites of presynaptic proteins has been suggested as the molecular basis for acrylamide-induced neuronal impairment68. Gene expression analysis on differentiated neurospheres exposed to 2.5 mM acrylamide confirmed the cell specificity of this compound, leading to significant negative modulation on expression levels of neuronal markers, while glial markers did not show any significant modulation. 13C-NMR spectroscopy data revealed significant alterations in the intracellular pools of 13C labeled glutamate and GABA, demonstrating the detrimental effects of acrylamide exposure on the neuronal populations. The amounts of labeled glutamate increased upon acrylamide exposure whereas labeling in GABA decreased. Although downregulation of GABA synthesis was likely to occur as outcome of the acrylamide-induced neurodegenerative processes, the increased glutamate synthesis was unexpected. One possible mechanism to explain glutamate accumulation is the impairment of synaptic activity in acrylamide-treated neurospheres, indicated by lower FM1-43 unloading kinetics. Acrylamide-induced neurotoxicity has been reported to affect synaptic vesicle trafficking and cycling, with observations of pre- and postsynaptic machinery structural70 and functional abnormalities, such as reduced number of presynaptic vesicles in the active zone, decreased neurotransmitter release and reduced neurotransmitter response15,68,69,70,71. These alterations are likely to impact the delicate neuronal-astrocytic contacts at synaptic sites, leading to impairment of metabolic interactions between these cell types. An imbalance in these interactions may also contribute to lower GABA synthesis upon acrylamide exposure, due to the high dependence of this neuronal sub-type on astrocyte-derived glutamine for GABA synthesis37. For astrocytes, a decrease in neurotransmitter release by neurons would represent a significant loss of carbon skeletons influx. Consequently, a continuous loss of TCA cycle intermediates to feed biosynthesis processes would occur in astrocytes, as for instance glutamine synthesis. In brain, the main enzyme responsible to replenish carbon skeletons loss due to exit of TCA cycle intermediates is PC72, and cells modulate its activity to supply the needs of de novo synthesis of these intermediates, as oxaloacetate59,73. This would explain our observation of an increase in [2-13C]glutamine (corrected for cycling, see Fig. 1), which derives from PC activity, in acrylamide-exposed neurospheres.
To our knowledge, this work demonstrates for the first time that in vitro differentiation of human neurons and astrocytes in a 3D setting allows the development of important and typical neural metabolic features found in the brain, such as the glutamine-glutamate-GABA cycle. Moreover, the combined data on acrylamide and MSO treatments showed that this human cell model and approach can be employed for toxicological studies, enabling the discrimination of the toxic effects on neuronal and astrocytic populations. The human 3D neural in vitro model employed can therefore contribute to a better understanding of human neural metabolism, namely neuron-astrocyte metabolic coupling, in pathological phenotypes that cannot be recapitulated in 2D cultures.
Additional Information
How to cite this article: Simão, D. et al. Functional metabolic interactions of human neuron-astrocyte 3D in vitro networks. Sci. Rep. 6, 33285; doi: 10.1038/srep33285 (2016).
References
Giri, S. & Bader, A. A low-cost, high-quality new drug discovery process using patient-derived induced pluripotent stem cells. Drug Discov. Today 20, 37–49 (2014).
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Mullard, A. 2013 FDA drug approvals. Nat. Rev. Drug Discov. 13, 85–89 (2014).
Calcoen, D., Elias, L. & Yu, X. What does it take to produce a breakthrough drug? Nat. Rev. Drug Discov. 14, 161–162 (2015).
Jucker, M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 16, 1210–1214 (2010).
Begley, C. G. & Ellis, L. M. Drug development: Raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Mak, I. W., Evaniew, N. & Ghert, M. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 6, 114–118 (2014).
Schüle, B., Pera, R. a R. & Langston, J. W. Can cellular models revolutionize drug discovery in Parkinson’s disease? Biochim. Biophys. Acta 1792, 1043–1051 (2009).
Andrews, P. W. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev. Biol. 103, 285–293 (1984).
Pleasure, S. J., Page, C. & Lee, V. M. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J. Neurosci. 12, 1802–1815 (1992).
Goodfellow, C. E., Graham, S. E., Dragunow, M. & Glass, M. Characterization of NTera2/D1 cells as a model system for the investigation of cannabinoid function in human neurons and astrocytes. J. Neurosci. Res. 89, 1685–1697 (2011).
Przyborski, S. A., Morton, I. E., Wood, A. & Andrews, P. W. Developmental regulation of neurogenesis in the pluripotent human embryonal carcinoma cell line NTERA-2. Eur. J. Neurosci. 12, 3521–3528 (2000).
Hartley, R. S., Margulis, M., Fishman, P. S., Lee, V. M. Y. & Tang, C. M. Functional synapses are formed between human NTera2 (NT2N, hNT) neurons grown on astrocytes. J. Comp. Neurol. 407, 1–10 (1999).
Coyle, D. E., Li, J. & Baccei, M. Regional differentiation of retinoic acid-induced human pluripotent embryonic carcinoma stem cell neurons. PLoS One 6, e16174 (2011).
Terrasso, A. P. et al. Novel scalable 3D cell based model for in vitro neurotoxicity testing: Combining human differentiated neurospheres with gene expression and functional endpoints. J. Biotechnol. 205, 82–92 (2015).
Hill, E. J., Woehrling, E. K., Prince, M. & Coleman, M. D. Differentiating human NT2/D1 neurospheres as a versatile in vitro 3D model system for developmental neurotoxicity testing. Toxicology 249, 243–250 (2008).
Laurenza, I. et al. A human pluripotent carcinoma stem cell-based model for in vitro developmental neurotoxicity testing: Effects of methylmercury, lead and aluminum evaluated by gene expression studies. Int. J. Dev. Neurosci. 31, 679–961 (2013).
Stern, M., Gierse, A., Tan, S. & Bicker, G. Human Ntera2 cells as a predictive in vitro test system for developmental neurotoxicity. Arch. Toxicol. 88, 127–136 (2014).
Pampaloni, F., Reynaud, E. G. & Stelzer, E. H. K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845 (2007).
Potter, W., Kalil, R. E. & Kao, W. J. Biomimetic material systems for neural progenitor cell-based therapy. Front Biosci 13, 806–821 (2008).
Moors, M. et al. Human neurospheres as three-dimensional cellular systems for developmental neurotoxicity testing. Env. Heal. Perspect 117, 1131–1138 (2009).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Simão, D. et al. Modeling human neural functionality in vitro: three-dimensional culture for dopaminergic differentiation. Tissue Eng. Part A 21, 654–668 (2015).
Choi, S. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
Ramirez, T. et al. Metabolomics in toxicology and preclinical research. ALTEX 30, 209–225 (2013).
Hertz, L., Dringen, R., Schousboe, A. & Robinson, S. R. Astrocytes: Glutamate producers for neurons. J. Neurosci. Res. 57, 417–428 (1999).
Tarczyluk, M. A. et al. Functional astrocyte-neuron lactate shuttle in a human stem cell-derived neuronal network. J. Cereb. Blood Flow Metab. 33, 1386–1393 (2013).
Efremova, L. et al. Prevention of the degeneration of human dopaminergic neurons in an astrocyte co-culture system allowing endogenous drug metabolism. Br. J. Pharmacol. 172, 4119–4132 (2015).
McNaught, K. S. & Jenner, P. Altered glial function causes neuronal death and increases neuronal susceptibility to 1-methyl-4-phenylpyridinium- and 6-hydroxydopamine-induced toxicity in astrocytic/ventral mesencephalic co-cultures. J. Neurochem. 73, 2469–2476 (1999).
Shi, P. et al. Synapse microarray identification of small molecules that enhance synaptogenesis. Nat. Commun. 2, 1–10 (2011).
Giordano, G. & Costa, L. G. Morphological assessment of neurite outgrowth in hippocampal neuron-astrocyte co-cultures. Curr. Protoc. Toxicol. Chapter 11, Unit 11.16 (2012).
Bak, L. K., Schousboe, A. & Waagepetersen, H. S. The glutamate/GABA-glutamine cycle: Aspects of transport, neurotransmitter homeostasis and ammonia transfer. J. Neurochem. 98, 641–653 (2006).
Rodrigues, T. B., Valette, J. & Bouzier-Sore, A.-K. 13C NMR spectroscopy applications to brain energy metabolism. Front. Neuroenergetics 5, 9 (2013).
Rothman, D. L., de Feyter, H. M., de Graaf, R. a., Mason, G. F. & Behar, K. L. 13C MRS studies of neuroenergetics and neurotransmitter cycling in humans. NMR Biomed. 24, 943–957 (2011).
Sonnewald, U. & Kondziella, D. Neuronal glial interaction in different neurological diseases studied by ex vivo 13C NMR spectroscopy. NMR Biomed. 16, 424–429 (2003).
Lundgaard, I. et al. Direct neuronal glucose uptake heralds activity-dependent increases in cerebral metabolism. Nat. Commun. 6, 6807 (2015).
Sonnewald, U. et al. Direct demonstration by [13C]NMR spectroscopy that glutamine from astrocytes is a precursor for GABA synthesis in neurons. Neurochem. Int. 22, 19–29 (1993).
Rae, C., Fekete, A. D., Kashem, M. a., Nasrallah, F. a. & Bröer, S. Metabolism, compartmentation, transport and production of acetate in the cortical brain tissue slice. Neurochem. Res. 37, 2541–2553 (2012).
Brito, C. et al. Increased levels of fucosyltransferase IX and carbohydrate Lewis(x) adhesion determinant in human NT2N neurons. J. Neurosci. Res. 85, 1260–1270 (2007).
Brekke, E. et al. Direct measurement of backflux between oxaloacetate and fumarate following pyruvate carboxylation. Glia 60, 147–158 (2012).
Brekke, E. M. F. et al. The pentose phosphate pathway and pyruvate carboxylation after neonatal hypoxic-ischemic brain injury. J. Cereb. blood flow Metab. 34, 724–734 (2014).
Carinhas, N. et al. Improving baculovirus production at high cell density through manipulation of energy metabolism. Metab. Eng. 12, 39–52 (2010).
Serra, M., Brito, C., Costa, E. M., Sousa, M. F. Q. & Alves, P. M. Integrating human stem cell expansion and neuronal differentiation in bioreactors. BMC Biotechnol. 9, 1–14 (2009).
Gualda, E. J. et al. OpenSpinMicroscopy: an open-source integrated microscopy platform. Nat Methods 10, 599–600 (2013).
Gualda, E. J., Simão, D., Pinto, C., Alves, P. M. & Brito, C. Imaging of human differentiated 3D neural aggregates using light sheet fluorescence microscopy. Front. Cell. Neurosci. 8, 1–10 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Hassel, B., Sonnewald, U. & Fonnum, F. Glial-neuronal interactions as studied by cerebral metabolism of [2-13C]acetate and [1-13C]glucose: an ex vivo 13C NMR spectroscopic study. J. Neurochem. 64, 2773–2782 (1995).
Gaffield, M. a & Betz, W. J. Imaging synaptic vesicle exocytosis and endocytosis with FM dyes. Nat. Protoc. 1, 2916–2921 (2006).
Hoopmann, P., Rizzoli, S. O. & Betz, W. J. Imaging synaptic vesicle recycling by staining and destaining vesicles with FM dyes. Cold Spring Harb. Protoc. 2012, 77–83 (2012).
Woehrling, E. K. et al. A Predictive In Vitro Model of the Impact of Drugs with Anticholinergic Properties on Human Neuronal and Astrocytic Systems. PLoS One 10, e0118786 (2015).
Woehrling, E. K., Hill, E. J. & Coleman, M. D. Evaluation of the importance of astrocytes when screening for acute toxicity in neuronal cell systems. Neurotox. Res. 17, 103–113 (2010).
Lancaster, M. A. & Knoblich, J. A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Garcez, P. P. et al. Zika virus impairs growth in human neurospheres and brain organoids. Science (80-.). 352, 816–818 (2016).
Sá Santos, S., Leite, S. B., Sonnewald, U., Carrondo, M. J. T. & Alves, P. M. Stirred vessel cultures of rat brain cells aggregates: characterization of major metabolic pathways and cell population dynamics. J. Neurosci. Res. 85, 3386–3397 (2007).
Sá Santos, S., Sonnewald, U., Carrondo, M. J. T. & Alves, P. M. The role of glia in neuronal recovery following anoxia: In vitro evidence of neuronal adaptation. Neurochem. Int. 58, 665–675 (2011).
Amaral, A. I. et al. Metabolic alterations induced by ischemia in primary cultures of astrocytes: Merging 13C NMR spectroscopy and metabolic flux analysis. J. Neurochem. 113, 735–748 (2010).
Yu, a C., Drejer, J., Hertz, L. & Schousboe, a. Pyruvate carboxylase activity in primary cultures of astrocytes and neurons. J. Neurochem. 41, 1484–1487 (1983).
Shank, R. P., Bennett, G. S., Freytag, S. O. & Campbell, G. L. Pyruvate carboxylase: an astrocyte-specific enzyme implicated in the replenishment of amino acid neurotransmitter pools. Brain Res. 329, 364–367 (1985).
Norenberg, M. D. & Martinez-Hernandez, A. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res. 161, 303–310 (1979).
Ronzio, R. a., Rowe, W. B. & Meister, a. Studies on the mechanism of inhibition of glutamine synthetase by methionine sulfoximine. Biochemistry 8, 1066–1075 (1969).
Shaw, C. A. & Bains, J. S. Did consumption of flour bleached by the agene process contribute to the incidence of neurological disease? Med. Hypotheses 51, 477–481 (1998).
Kanamori, K., Ross, B. D. & Kondrat, R. W. Rate of glutamate synthesis from leucine in rat brain measured in vivo by 15N NMR. J. Neurochem. 70, 1304–1315 (1998).
Lieth, E. et al. Nitrogen shuttling between neurons and glial cells during glutamate synthesis. J. Neurochem. 76, 1712–1723 (2001).
Hutson, S. M., Lieth, E. & LaNoue, K. F. Function of leucine in excitatory neurotransmitter metabolism in the central nervous system. J. Nutr. 131, 846S–850S (2001).
Hull, J. et al. Distribution of the branched chain aminotransferase proteins in the human brain and their role in glutamate regulation. J. Neurochem. 123, 997–1009 (2012).
Jessy, J. & Murthy, C. R. Branched chain amino acid transaminases in brain in methionine sulphoximine (MSI) toxicity. Biochem. Int. 16, 245–251 (1988).
LoPachin, R. M. & Gavin, T. Molecular mechanism of acrylamide neurotoxicity: Lessons learned from organic chemistry. Environ. Health Perspect. 120, 1650–1657 (2012).
LoPachin, R. M. The changing view of acrylamide neurotoxicity. Neurotoxicology 25, 617–630 (2004).
DeGrandchamp, R. L., Reuhl, K. R. & Lowndes, H. E. Synaptic terminal degeneration and remodeling at the rat neuromuscular junction resulting from a single exposure to acrylamide. Toxicol. Appl. Pharmacol. 105, 422–433 (1990).
Sisnaiske, J. et al. Acrylamide alters neurotransmitter induced calcium responses in murine ESC-derived and primary neurons. Neurotoxicology 43, 117–126 (2014).
Patel, M. S. The relative significance of CO2 fixing enzymes in the metabolism of rat brain. J. Neurochem. 22, 717–724 (1974).
Sibson, N. R. et al. In vivo 13C NMR measurement of neurotransmitter glutamate cycling, anaplerosis and TCA cycle flux in rat brain during [2-13C]glucose infusion. J. Neurochem. 76, 975–989 (2001).
Acknowledgements
iNOVA4Health-UID/Multi/04462/2013, a program financially supported by Fundação para a Ciência e Tecnologia/Ministério da Educação e Ciência, through national funds and co-funded by FEDER under the PT2020 Partnership Agreement is acknowledged. This work was also supported by PTDC/EBB-BIO/112786/2009 and PTDC/EBB-BIO/119243/2010, FCT, Portugal. The technical assistance from Hugo Pereira on Light-sheet imaging is gratefully acknowledged. D. Simão and A.P. Terrasso were recipients of PhD fellowships from FCT, Portugal (SFRH/BD/78308/2011 and SFRH/BD/52473/2014, FCT).
Author information
Authors and Affiliations
Contributions
D.S.: designed and performed the experiments, performed data analysis and interpretation, prepared the manuscript; A.P. Terrasso: implemented and performed the 3D cultures; A.P. Teixeira: performed data interpretation and critically revised the manuscript; C.B.: contributed to study design and critically revised the manuscript; U.S.: designed the experiments, performed data interpretation and critically revised the manuscript; P.M.A.: supervised the work and critically revised the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Simão, D., Terrasso, A., Teixeira, A. et al. Functional metabolic interactions of human neuron-astrocyte 3D in vitro networks. Sci Rep 6, 33285 (2016). https://doi.org/10.1038/srep33285
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep33285
This article is cited by
-
Advances in 3D neuronal microphysiological systems: towards a functional nervous system on a chip
In Vitro Cellular & Developmental Biology - Animal (2021)
-
MAM-2201, One of the Most Potent—Naphthoyl Indole Derivative—Synthetic Cannabinoids, Exerts Toxic Effects on Human Cell-Based Models of Neurons and Astrocytes
Neurotoxicity Research (2021)
-
Polyphenols journey through blood-brain barrier towards neuronal protection
Scientific Reports (2017)
-
NMR Metabolomics for Stem Cell type discrimination
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.