Abstract
The oligodendrocyte transcription factor Olig2 plays a crucial role in the neurogenesis of both spinal cord and brain. In the cerebellum, deletion of both Olig2 and Olig1 results in impaired genesis of Purkinje cells (PCs) and Pax2+ interneurons. Here, we perform an independent study to show that Olig2 protein is transiently expressed in the cerebellar ventricular zone (VZ) during a period when PCs are specified. Further analyses demonstrate that Olig2 is expressed in both cerebellar VZ progenitors and early-born neurons. In addition, unlike in the ganglionic eminence of the embryonic forebrain where Olig2 is mostly expressed in proliferating progenitors, Olig2+ cells in the cerebellar VZ are in the process of leaving the cell cycle and differentiating into postmitotic neurons. Functionally, deletion of Olig2 alone results in a preferential reduction of PCs in the cerebellum, which is likely mediated by decreased neuronal generation from their cerebellar VZ progenitors. Furthermore, our long-term lineage tracing experiments show that cerebellar Olig gene-expressing progenitors produce PCs but rarely Pax2+ interneurons in the developing cerebellum, which opposes the “temporal identity transition” model of the cerebellar VZ progenitors stating that majority of Pax2+ interneuron progenitors are transitioned from Olig2+ PC progenitors.
Similar content being viewed by others
Introduction
The basic helix-loop-helix (bHLH) transcription factors Olig genes, i.e. Olig1 and Olig2, are first identified in the spinal cord and named as such because of their pivotal function in oligodendrocyte development throughout the central nervous system (CNS)1,2. Subsequently, the two groups who discovered the genes reported simultaneously that Olig2 and Olig1 are also required for spinal motor neuron (MN) specification3,4. Similarly in the developing forebrain, Olig2-expressing progenitors in the medial ganglionic eminence (MGE) give rise to subpopulations of GABAergic interneurons where the homeobox transcription factor Dlx2 counteracts with Olig2 to specify neuronal versus oligodendroglial cell fate5. In addition, at least some cholinergic neurons in the basal forebrain are also derived from Olig2-expressing progenitors, and Olig2 deletion leads to a severe reduction of these neurons6.
In the cerebellum, Purkinje cells (PCs) are GABAergic projection neurons that, along with deep cerebellar nuclei (DCN) GABAergic projection neurons, are derived from the ventricular zone (VZ) of the early cerebellar primordium7. GABAergic inhibitory interneurons are also derived from the VZ, and yet in a more ventral region that can be defined by Pax2 expression8. Importantly, pancreas transcription factor 1a (Ptf1a) plays an indispensable role in the generation of all VZ-derived cerebellar GABAergic neurons including PCs and Pax2+ interneurons9,10. Although several other transcription factors, namely Mash111, Ngn112, Ngn213, NeuroD114, Gsx115 and Olig1, 214,15 have been described to express in the cerebellar VZ with distinct micro-domains16, the mechanisms involved in the specification and generation of VZ-derived GABAergic neurons including PCs are still partially understood. In a recent report, Seto et al. demonstrated that Olig2 counteracts with Gsx1 in the early cerebellar VZ to maintain PC progenitor identity, and that deletion of both Olig1 and Olig2 leads to reduction of PCs and increase of Pax2+ interneurons while deletion of Olig2 alone shows no obvious phenotypic defects15. However, in the present study, we show that deletion of Olig2 alone results in a significant reduction of PCs and no change of Pax2+ interneurons, indicating that Olig2 function is required for a complete specification of PCs. Mechanistically, we also show that Olig2 is expressed in the late-phase of the VZ progenitor cell cycle and controls the rate of neurogenesis from cerebellar VZ progenitors, but not their proliferation. Furthermore, our long-term lineage tracing analysis indicates that Olig2+ progenitors give rise to PCs and DCN neurons, but rarely Pax2+ interneurons, challenging the “temporal identity transition” model of the cerebellar VZ progenitors that was recently proposed15.
Results
Olig2 is co-expressed with neuronal and progenitor markers in the early cerebellum
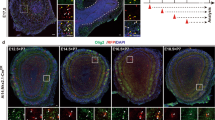
Olig2 is transiently expressed in the cerebellar VZ during E11.5∼E13.5, a time window of PC specification14. To further delineate neurogenic function of Olig2 in the early cerebellum, we performed a co-staining of Olig2 with a marker of early postmitotic neurons, Doublecortin (DCX), at E12.5 when Olig2 expression is strong (Fig. 1A). We found that DCX staining is prevalent in the cerebellar plate at this stage but absent from the rhombic lip (RL) and the VZ, the two major germinal zones of the developing cerebellum (Fig. 1B). The nuclear transitory zone (NTZ) Olig2+ cells mostly co-express DCX suggesting that they are postmitotic neurons (Fig. 1C,C1). In contrast, Olig2 expression in the VZ shows a largely non-overlapping pattern with DCX (Fig. 1C2). Occasionally, we were able to find DCX and Olig2 double-positive cells at the boundary between the DCX+ and Olig2+ zones (Fig. 1C2’, arrow), suggesting that DCX+ neurons are derived from VZ Olig2+ progenitors by downregulating Olig2 expression. Similar expression pattern has been found with Olig2 and an early postmitotic neuronal marker Lhx1/517,18 previously14. A 2-hour bromodeoxyuridine (5-bromo-2′-deoxyuridine, BrdU)-pulse labeling analysis showed that 25.2 ± 3.3% of the VZ Olig2+ cells are also BrdU+ (Fig. 1D and arrow in 1D’) confirming that they are dividing progenitors, whereas the NTZ Olig2+ cells are nearly all BrdU− (Fig. 1D). Therefore, the dynamic expression pattern of Olig2 during early cerebellar development suggests its potential role in the genesis and differentiation of several cerebellar neuronal types including PCs that are differentiated from the VZ progenitors at this developmental stage.
Co-immunostaining is performed to analyze the expression of Olig2 (A) and a neuronal marker, DCX (B) on sagittal sections of the E12.5 cerebellum. The overlay image (C) reveals differential DCX expression patterns of the Olig2+ cells in the VZ and NTZ. The enlarged images of the boxed regions in (C) are shown in (C1) and (C2), respectively. Double-positive cells are pointed by arrows in (C1) and an arrow in a higher power confocal image (C1’). Double-positive cells (indicated by an arrow in C2’, a higher power confocal image of C2 region) are very rarely seen in (C2) where Olig2 and DCX show mostly a non-overlapping staining pattern. (D) Co-immunostaining of Olig2 and BrdU in the VZ of the E12.5 cerebellum that has been treated by a BrdU pulse-labeling. (D’) The confocal image of the VZ in (D) showing a double-positive cell (arrow). (E) Co-expression analysis of Olig2 with markers of proliferation (Ki67), radial glia (BLBP) and PC differentiation (Lhx1/5) in the cerebellar VZ at E13.5. (F) A diagram depicting Olig2 expression range during the transition of VZ progenitors to early PCs. (G) Co-immunostaining with anti-Olig2 and anti-Pax2 on sagittal cerebellar sections of the E13.5 wild-type mouse. (G’) is the enlarged image of boxed regions in (G) showing a non-overlapping staining pattern. VZ, ventricular zone; RL, rhombic lip; NTZ, nuclear transitory zone. Scale bar: 80 μm in A,B,C and G; 25 μm in C1 and C2; 15 μm in C1’ and D’; 20 μm in C2’, E and G’; 40 μm in D.
Next, we examined Olig2 expression in the VZ in further detail to determine its potential function in PC generation. In the E13.5 cerebellar VZ, a small fraction (7.1 ± 3.6%) of Olig2+ cells co-express the proliferation marker Ki67 (Fig. 1E). In addition, we found that the Olig2+/Ki67+ cells are rarely epithelial progenitors that can be labeled by Ki67 and a radial glial marker brain lipid binding protein (BLBP) (Fig. 1E). On the other hand, 35.9 ± 3.6% of Olig2+ cells are positive for an early marker of postmitotic PCs, Lhx1/5, which shows a non-overlapping expression pattern with Ki67 (Fig. 1E). Therefore, these immunostaining data indicate that Olig2 is expressed in both non-epithelial VZ progenitors and newborn PCs (Fig. 1F), defining the specification process of PCs from the VZ progenitors. Similar expression patterns have also been found with some of these markers in the E12.5 cerebellum previously14,15.
Besides PCs, GABAergic inhibitory interneurons are also generated from cerebellar VZ and can be identified by their expression of Pax28. We found that Pax2 and Olig2 exhibit a nearly non-overlapping expression pattern at E13.5 (Fig. 1G,G’) similarly to E12.515 suggesting that Olig2 plays little role in the specification of GABAergic inhibitory interneurons.
The NTZ Olig2+ cells represent a distinct subpopulation of DCN neurons in the developing cerebellum
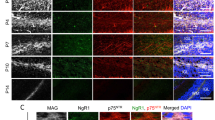
The expression pattern of Olig2 in the NTZ of the developing cerebellum suggests that these positive cells are derived from the RL. In addition, our result indicates that these cells are DCX+ newborn neurons (Fig. 1). To further characterize the NTZ Olig2+ cells, we performed co-immunostaining analyses with anti-Olig2 and markers of RL derivatives. A previous report has indicated that Pax6, Tbr2 and Tbr1 are expressed in differentiating gltutamatergic DCN neurons in a sequential manner19. At E12.5 when Olig2 is highly expressed in the NTZ, we found that some Pax6+ cells are sparsely distributed along the dorsal pial surface, and that Pax6+ and Olig2+ cells do not overlap (data not shown). As development proceeds, Pax6+ cells are more prominent along the pial surface as shown at E14.5 (Fig. S1A). However, the NTZ Olig2 and Pax6 still show a non-overlapping pattern (Fig. S1A, insert). Between the Olig2+ and Pax6+ domains, there apprear to be a group of cells that are negative for both markers, but positive for Tbr1 (Fig. S1B). Although the Olig2+ and Tbr1+ domains are closer in position with double positive cells being occasionally seen (Fig. S1B, insert, arrowhead), they are also largely non-overlapping. In addition, the NTZ Olig2+ cells are nearly all postmitotic as indicated in a 2-hour BrdU-pulse labeling experiment at E14.5 (Fig. S1C). Notably, a significant number of BrdU+ cells can still be found in the VZ, and yet Olig2 expression has already disappeared in this region at this developmental stage (Fig. S1C). At E18.5, while Tbr1+ neurons concentrate to the medial part of the DCN, Olig2 expression is more towards the lateral region (Fig. S1D). Two morphologically distinct Olig2+ cells are found with smaller cells being located in the periphery of the Olig2+ domain and bigger cells being in the core (Fig. S1D, inserts). While the small Olig2+ cells are often NG2+ suggestive of oligodendrocyte progenitor cells (OPCs), the big Olig2+ cells are NeuN+ (data not shown). In sum, these expression patterns suggest that Olig2 expression labels a distinct group of DCN neurons that locate more lateral to the Tbr1+ neurons in the DCN, which is consistent with a previous observation14.
Olig2-null mice show reduced PC generation
To explore directly the function of Olig2 during cerebellar neurogenesis, we examined mice carrying an Olig2-null mutation. Since Olig2-null mice die soon after birth3, neurogenesis has to be analyzed during embryonic stages in this animal. First, immunostaining with an antibody against Calbindin, a specific PC marker, showed a siginificant reduction in PC number on mid-sagittal sections of Olig2-null cerebella at E15.5 compared with controls (Fig. 2A). Normally, PCs that are generated from ventral cerebellar VZ migrate dorsally and spread tangentially underneath the cerebellar pial surface around this developmental stage20. We observed numerous Calbindin+ cells within the cerebellar parenchyma in the control mouse (presumably the migrating PCs), but very few in the Olig2-null mouse (Fig. 2A). This observation may suggest that PC migration is disrupted in the absence of Olig2. However, this could also be explained by reduced number of PCs and thus early termination of the migration process in the Olig2-null cerebella. Second, to confirm that the PC reduction on mid-sagittal cerebellar sections is not due to a redistribution of PC in the Olig2-null cerebellum, we examined acute cultures that were dissociated from whole E17.5 cerebellar tissues and demonstrated a significant decrease in the number of Calbindin+ cells per field in Olig2-null cultures (Fig. 2B), but not in Olig1-null cultures (Fig. 2C), while the percentage of Pax6+ granule neuron precursors among total cells showed no change in either mutant cultures compared with controls (Fig. 2D,E). These results indicate a specific effect of Olig2 ablation on PC generation. Lastly, gene expression comparison at E18.5 indicated that the mRNA levels of Calbindin and another PC marker RORα21 are significantly reduced in Olig2-null cerebella, while Pax2 and Pax6 mRNA levels remain unchanged (Fig. 2F). Furthermore, the levels of the Bergmann glial marker BLBP and the DCN neuron markers Tbr1 and Brn219 remained unchanged in the Olig2-null cerebella indicating that Olig2 ablation has little effect on these two cell types (Fig. 2F). Gsx1 has been shown to promote Pax2+ interneuron generation15 and did not display a change in expression (Fig. 2F). The effect of Olig1 ablation on cell type marker expression was also analyzed at E17.5, and, except for Olig1 itself, no significantly changed genes were observed between Olig1-null and control cerebellar samples (Fig. 2G). To further confirm the unchanged level of Pax2 expression in Olig2-null cerebella (Fig. 2F), we also performed immunostaining on E18.5 mid-sagittal cerebellar sections and found no significant difference in the number of Pax2+ cells per section between Olig2-null and control cerebella (Fig. 3A). Pax2+ cells were also quantified on E18.5 cerebellar sections of the Olig1-null mice and showed no significant difference compared with controls (Fig. 3B). Taken together, Olig2, but not Olig1, ablation specifically reduces PC generation and has no detectable effects on other neuronal types and astroglia in the embryonic cerebellum.
(A) Immunostaining with an anti-Calbindin antibody on E15.5 cerebellar mid-sagittal sections of Olig2+/+ and Olig2−/− mice shows notably reduction of PCs in the cerebellum (CB). Quantification on confocal images shows a significant reduction in the number of Calbindin+ cells per cerebellar section in Olig2−/− mice compared with controls (n = 3 for both genotypes). DAPI is used to label nuclei. MB, midbrain. The number of Calbindin+ cells per field in the acute cultures of dissociated E17.5 Olig2−/− (B) and E18.5 Olig1−/− (C) cerebella are compared with their controls (n = 3 for both mutant and control genotypes). In these same acute cultures, the percentage of Pax6+ cells among total cells (DAPI+) are also compared for E17.5 Olig2−/− (D) and E18.5 Olig1−/− (E) cerebella. Gene expression levels in dissected cerebellar tissues are compared by qRT-PCR between Olig2+/− and Olig2−/− mice (n = 4 for both genotypes) at E18.5 (F), or between Olig1+/+,− and Olig1−/− mice (n = 3 for both genotypes) at E17.5 (G). Scale bar: 200 μm in A and B; 100 μm in D. *P < 0.05; **P < 0.01; n.s., not significant by Student’s t-test.
(A) Immunnostaining analysis is performed on mid-sagittal cerebellar sections of the E18.5 Olig2-null mice. The number of Pax2+ cells per cerebellar section is quantified and compared. No significant change in the number of Pax2+ interneuron progenitors is detected between Olig2-null and control mice (n = 3 for both genotypes). (B) Similar immunostaining was also performed and quantified on cerebellar sections of E18.5 Olig1-null mice. DAPI is used to label nuclei. Scale bar: 200 μm. n.s., not significant by Student’s t-test.
Olig2 deletion impairs PC generation from their cerebellar VZ progenitors without affecting overall cell proliferation
To check if Olig2-ablation affects progenitor proliferation in the cerebellar VZ, we performed a 2-hour BrdU-pulse labeling at E12.5 and compared BrdU-labeled cells. We found no significant difference in the percentage of BrdU+ cells among total cells between Olig2-null and wild-type animals (Fig. 4A,B). The co-staining of BrdU and the postmitotic neuronal marker Lhx1/5 in the dorsal portion of cerebellar VZ at this stage allowed us to identify newborn PCs during the 2-hour BrdU labeling period. In wild-type mice, we observed a small number of BrdU and Lhx1/5 double-positive cells that are usually localized along the boundary between BrdU and Lhx1/5-positive zones in the cerebellar VZ (Fig. 4C, arrows). However, we could hardly detect any such double-positive cells in Olig2-null mice, despite that numerous Lhx1/5+ PCs are present (Fig. 4C,D). In addition, we detected very few caspase-3+ cells in the E12.5 cerebella of both types of mice (data not shown) suggesting that apoptosis is not involved in the PC defect of Olig2-null mice. Thus, Olig2 may play little role in progenitor proliferation per se, but is required for normal rate of PC generation from their progenitors in the cerebellar VZ. In a given time period of PC generation, fewer PCs may be specified from VZ progenitors and therefore moderate loss of PCs in the Olig2-null cerebellum (Fig. 2).
(A) Proliferation of cerebellar VZ progenitors at E12.5 is analyzed by their ability to incorporate BrdU in a 2-hour pulse-labeling treatment and compared between Olig2+/+ and Olig2−/− mice. (B) The percentage of BrdU+ cells among total cells (DAPI+) in the cerebellar VZ (as indicated) is calculated and compared between the two genotypes (n = 3 for both genotypes). (C) PC generation from the proliferating cerebellar VZ progenitors at E12.5 is also analyzed by co-staining of the postmitotic neuronal marker Lhx1/5 and BrdU in Olig2+/+ and Olig2−/− mice. (D) Percentage of Lhx1/5+ cells among BrdU+ cells in the dorsal portion of the cerebellar VZ is quantified and compared between the two genotypes (n = 3 for both genotypes). VZ, ventricular zone. Scale bar: 20 μm in A and C. *P<0.05 by Student’s t-test.
Unlike in the embryonic forebrain, Olig2-expressing progenitors in the cerebellar VZ are in the late phase of the cell cycle
To further examine the proliferative characteristic of the Olig2-expressing progenitors in the cerebellar VZ, we performed a 30-min BrdU-pulse labeling at E12.5 to label exclusively the cells in the S-phase of the cell cycle. Consistent with our observation by immunostaining that Olig2 is expressed in progenitors that are leaving the cell cycle (Fig. 1), we found that Olig2-expressing cells are rarely labeled by BrdU in the 30-min pulse (Fig. 5A,A’). On the contrary, nearly 60% of the Olig2+ cells are labeled by BrdU in the ganglionic eminence (GE) of the forebrain of the same animal (Fig. 5B,B’), suggesting that most Olig2+ cells in the forebrain are progenitors in the S-phase, actively dividing. Furthermore, a more detailed cell cycle analysis of E12.5 cerebellar VZ progenitors by BrdU-pulse labeling shows that Olig2 is preferentially expressed in the late phase of the progenitor cell cycle (14h after BrdU pulse) (Fig. 5D). Therefore, these results confirm that Olig2-expressing progenitors in the cerebellar VZ are distinct from the ones in other CNS regions and may exhibit unique neurogenic properties during development.
Co-immunostainings were done on mid-sagittal sections of the cerebellum (A) and forebrain (B) of E12.5 embryos that have been pulse-labeled with BrdU for 30 min before sacrifice. (A’,B’) are enlarged images of boxed regions in (A,B), respectively. The arrow indicates a double-positive cell in (B’). (C) Comparison of BrdU labeling in the VZ Olig2+ cells between cerebellum and forebrain. Note that BrdU labels more than 50% of the Olig2+ cells (BrdU+/Olig2+) in the GE of the forebrain, but nearly none in the cerebellar VZ. (D) A series of BrdU-pulse labeling analyses of E12.5 cerebellar VZ progenitors. Percentages are shown to represent BrdU+ and BrdU- fractions among Olig2+ cells in the cerebellar VZ. VZ, ventricular zone; GE, ganglionic eminence.
Olig gene-expressing progenitors give rise to PCs and DCN neurons, but rarely Pax2+ interneurons, in the cerebellum
To obtain a full spectrum of neuronal types that may involve Olig2 function in the developing cerebellum, we performed a long-term lineage tracing analysis using both Olig1-Cre and Olig2-Cre reporter mouse lines. Since Olig1 and Olig2 expressions are coordinately regulated in the developing CNS22 and are co-expressed in the cerebellum15, we expected to see similar labeling patterns in these reporter lines. The Olig1-Cre faithfully labels oligodendrocyte lineage cells throughout the CNS in our previous studies23,24. Indeed, in the P18 cerebellum of the Olig1-Cre;Z/EG reporter mouse, we observed many GFP+ cells in the white matter (WM) where most oligodendrocytes reside (Fig. 6A). Besides, we also observed strong GFP+ cells with big cell bodies and complex dendritic arbors in the molecular layer (ML) (Fig. 6A). Further immunohistochemical analysis confirmed their identity as PCs that are both Calbindin+ and Parvalbumin+ (Fig. 6B,C), but negative for Bergmann glial markers, GFAP (Fig. 6D) and BLBP (Fig. 6E). The rarely seen small GFP+ cells in the ML are Parvalbumin− (Fig. 6C, arrow and insert), but Olig2+ (Fig. 6F, arrow), indicative of cells of oligodendrocyte lineage, but not small interneurons. Nearly all GFP+ cells in the WM co-express Olig2 (Fig. 6G) but none co-express Pax2, a pan marker of GABAergic inhibitory interneurons (Fig. 6H). Next, we examined if Olig1-Cre could label any neuronal cell types in the DCN. For that purpose, we engaged the Olig1-Cre;LacZ reporter line where cellular labeling revealed by anti-β-gal staining is mostly nuclear and easy to identify. In the DCN of this reporter line, a significant number of big β-gal+ cells are NeuN+ (Fig. 6I, arrow), some of which are also GABA+ (Fig. 6I, insert, arrow) suggestive of GABAergic DCN projection neurons. One thing to note is that we do observe variable labeling efficiency between the two reporter lines. For example, the labeling efficiency of PC is higher in the Olig1-Cre;LacZ (65.5 ± 3.8%) than in the Olig1-Cre;Z/EG reporter line (31.6 ± 7.6%). We reason that this variation may be due to the distinct genetic structure of the reporter loci in each reporter mouse line. Nevertheless, these results indicate that most, if not all, PCs are derived from Olig gene-expressing progenitors in the cerebellum. In parallel, we also performed lineage tracing analysis with the Olig2-Cre;tdTomato reporter line, in which labeled cerebellar cell types can be identified by their expression of tdTomato (Fig. 6J). Again, PCs are identified as a labeled cell type by their expression of Calbindin and Parvalbumin (Fig. 6K,L) but not GFAP (Fig. 6M). Interestingly, some small tdTomato+ cells in the ML of this reporter line shows co-expression of Parvalbumin (arrows, Fig. 6L) suggestive of small interneuron identity. In the WM, Olig2-Cre-labeled cell types are nearly all of oligodendrocyte lineage being positive for Olig2 and CC1 (Fig. 6N,O), and negative for Pax2 (Fig. 6P). Together, our lineage tracing analysis demonstrates that Olig gene-expressing progenitors give rise to PCs and DCN neurons, but rarely Pax2+ interneurons, in the cerebellum.
Olig1-Cre lineage tracing analysis is performed in the cerebellum of either Z/EG (A–H) reporter line at P18 or ROSA26;LacZ (I) reporter line at P16. Labeled cells with different morphology in the cerebellum of Olig1-Cre;Z/EG mice are revealed by their GFP expression (A). The identity of GFP+ cells are examined by co-staining with cell-type specific markers Calbindin (B), Parvalbumin (C), GFAP (D), BLBP (E), Olig2 (F,G) and Pax2 (H). Some GFP+ cells are indicated as either negative (arrows in C and H) or positive (arrows in F,G) for marker staining. The high magnification insert in (C) confirms the cellular identity of the indicated GFP+ cell by showing the presence of DAPI+ nucleus. Many labeled cells in the cerebellar DCN of Olig1-Cre;LacZ reporter mice are observed by anti-β-gal staining, some of which are NeuN+ (arrow, I) and GABA+ (arrow, insert in I). Olig2-Cre lineage tracing analysis is also performed in the cerebellum of ROSA26;tdTomato (J–P) reporter line at P22. Labeled cells with different morphology in the cerebellum of Olig2-Cre;tdTomato mice are revealed by their tdTomato expression (J). The identity of tdTomato+ cells are examined by co-staining with cell-type specific markers Calbindin (K), Parvalbumin (L), GFAP (M), Olig2 (N), CC1 (O) and Pax2 (P). Note that two tdTomato+ cells are co-labeled with Parvalbumin suggestive of small interneurons (arrows in L). DAPI is used to label nuclei. ML, molecular layer; IGL, internal granule layer; WM, white matter; DCN, deep cerebellar nuclei. Scale bar: 100 μm in (A,J); 20 μm in (B–H,K–P); 25 μm in I.
Discussion
In this study, we aim to determine the neurogenic role of Olig2 gene in the early developing cerebellum. We independently show that Olig2 is dynamically expressed in both the VZ and NTZ of the cerebellar primordium at early embryonic stages labeling different neuronal lineages during cerebellar neurogenesis. In particular, we show that deletion of Olig2 alone, but not Olig1 alone, specifically impairs PC generation of the cerebellum without substantially affecting other neuronal types. Furthermore, our long-term lineage tracing analysis indicates that Olig gene-expressing progenitors give rise to both DCN neurons and PCs, but rarely Pax2+ interneurons (Fig. 6), arguing against the “temporal identity transition” model of the cerebellar VZ progenitors that were recently proposed15.
Potential mechanisms of Olig2 function in PC specification
The expression pattern relative to that of Pax2 has constrained Olig2 to the “pc2” domain of the cerebellar VZ where it could interact with other transcription factors16. Ptf1a-null mice exhibit a substantial loss of PCs and Pax2+ interneurons9, whereas Mash1-null mice do not show major defects on PC development25. Although Ngn1 deletion has not been studied in the developing cerebellum in terms of its functional contribution to PC genesis26, Ngn2 deletion results in a reduction of total number of PC as shown in a recent report13. In that report, Florio and colleagues conducted a detailed analysis of cell-cycle kinetics on cerebellar VZ progenitors and demonstrated that Ngn2 is mainly expressed in progenitors in the G1 phase getting ready to exit the cell cycle, and rapidly down-regulated in differentiated neurons13. We also showed that Olig2 is expressed in the late phase of the progenitor cell cycle (Fig. 5D) and also maintained even after cell cycle exit, since we observed 35.9% of Olig2+ cells overlapped with Lhx1/5+ postmitotic neurons in the E13.5 cerebellar VZ (Fig. 1E). Prolonged expression may allow Olig2 interact with different transcription factors including Ngn2 to function in PC generation.
Previous reports indicated that Olig2 and Ngn2 interact to regulate MN specification27,28,29. Olig2 function is believed to expand and prime pMN progenitors to MN fate whereas Ngn2 is to promote cell cycle exit and neuronal differentiation. Consistently, while Ngn2-null mice exhibit only impaired MN differentiation30, Olig2 deletion abolishes MN phenotype3,4. The proliferative function of Olig2 is also demonstrated in the forebrain where a Olig1/2 triplication mutant exhibits increased Ki67+ cells in the embryonic MGE, which can be rescued by a Olig1/2+/− genotype31. Mechanistically, Olig2 and Ngn2 may interact at different levels. First, Olig2 protein can turn on Ngn2 gene transcription3,4,29; second, Olig2 and Ngn2 can compete for binding to E-box DNA enhancer elements27. Therefore, these two factors may interact directly to coordinate neurogenesis. In a different scenario, Olig2 has been shown to suppress the transcription of cell cycle dependent kinase inhibitor 1a (CDKN1A) in neural stem cells, which in turn facilitates cell proliferation32. In addition, CDKs phosphorylates Ngn2 protein on multiple sites and inhibits its binding to E-box DNA fragments and thus transcriptional activity33. Therefore, these studies provide an alternative mechanistic pathway by which Olig2 and Ngn2 interact indirectly through cell cycle regulators to balance cell proliferation and neurogenesis. Olig2 and Ngn2 are both expressed and colocalized in the “pc2” domain of the cerebellar VZ14, allowing these similar mechanisms to occur during PC specification. In fact, Olig2 deletion results in a slower rate of PC generation from the cerebellar VZ progenitors (Fig. 4) suggesting a defect of cell cycle kinetics, and Ngn2-null cerebellar VZ progenitors stall in the G1 phase of the cell cycle and thus produce fewer PCs13. This suggests that Olig2 and Ngn2 may functionally converge to cell cycle regulation of the cerebellar VZ progenitors. An interesting observation worthy of noting is that Olig2 deletion induces only impaired PC generation in the cerebellum (this study) but a complete absence of MNs in the spinal cord3,4. This underlines differential contribution of Olig2 in the generation of PCs vs. MNs, and may be reflected by the distinct proliferative characteristic of Olig2-expressing progenitors in the cerebellar VZ (Fig. 5).
Function of Olig genes in the generation of interneurons and PCs in the cerebellum
Seto et al. demonstrated clearly that Olig1/2 double knockout (KO) drastically reduces (but not abolishes) PCs and induces an increase of Pax2+ interneurons generation in the developing cerebellum15. Furthermore, the authors did not observe any obvious phenotype in the Olig2 single KO. However, our results indicate that the Olig2 single KO mildly, but significantly, reduced PC generation with no detectable effect on cerebellar interneurons (Fig. 2). This raises an interesting possibility that the effect of Olig genes on PCs and interneurons generation is dose-dependent, i.e. PC generation is more sensitive to Olig gene function and prone to a decrease in Olig2 single KO, whereas interneuron generation is less sensitive and requires deletion of both Olig1 and Olig2 to show an increase in number. We showed that Olig genes (at least Olig2) function in the late phase of the progenitor cell cycle (Fig. 5) and maintain a proper generation of PCs from these progenitors without affecting their overall proliferation (Fig. 4). However, the inhibitory effect of Olig genes in interneurons generation is unclear, but unlikely through Gsx1, since no change of Gsx1 expression was observed in either Olig gene KO (Fig. 2) or overexpression (Fig. 7 in Seto et al.15). Further studies are required to delineate the underlying mechanisms.
Olig2, a mainly neurogenic factor in the cerebellar VZ?
Given the strong expression of Olig2 in the VZ of the early developing cerebellum, we asked if the Olig2+ VZ progenitors generate oligodendrocytes after they give rise to GABAergic neuronal cell types. However, we were unable to detect PDGFRα+ or NG2+ OPCs by immunostaining between E11.5 and E14.5 in the cerebellum (data not shown). Furthermore, the cerebellar OPCs gradually appear starting from E14.5 when Olig2 expression already disappears in the VZ (Fig. S1). This expression correlation suggests that oligodendrocytes may not be generated in the cerebellar VZ locally. In fact, cerebellar oligodendrocytes are thought to originate from extracerebellar sources25,34,35. By electroporating a GFP construct into the cerebellar VZ of the mouse embryos, Grimaldi et al. demonstrated that oligodendrocytes are not derived from cerebellar VZ progenitors at E14.5. In the same study, E12.5 solid cerebellar tissues were transplanted into the P2 GFP mouse cerebellum and examined 12 days after. The authors found that the majority of Olig2+ OPCs in the graft are also GFP+, indicating that they are not generated locally, but from the donor25. Extracerebellar origin of oligodendrocytes is also suggested in a quail/chick chimeric study35. Therefore, combining these previous reports and our results, we propose that, unlike in the spinal cord where it specifies both MNs and oligodendrocytes, Olig2 expressed in the VZ may be a mainly neurogenic factor during early cerebellar development, and that the classic Olig2-mediated neuron-oligodendrocyte linkage may be uncoupled in the developing cerebellum.
Long-term lineage tracing analysis does not support the “transition” model of cerebellar VZ progenitors
Based on a short-term lineage tracing and gene functional analyses, a “transition” model has been proposed to describe the relationship between cerebellar VZ progenitors of PCs and Pax2+ interneurons15. This model claims that during development, Olig2+ PC progenitors (PCPs) transition into Gsx1+ interneuron progenitors (PIPs) to generate Pax2+ interneurons in the cerebellar VZ, and that this timely transition is important to generate proper number of two cerebellar neuronal subtypes and is regulated positively by Gsx1 and negatively by Olig2 (Fig. 7 in Seto et al.15). According to this model, one would predict that lineally most, if not all, Pax2+ interneurons are derived from Olig gene-expressing PCPs. Unfortunately, our long-term genetic lineage tracing analysis does not support this model by showing that Pax2+ interneurons are rarely labeled in an either Olig1-Cre or Olig2-Cre reporter line indicating that Pax2+ interneurons may not be lineally related to Olig2+ PCPs. Furthermore, we, as well as Seto et al. themselves14, provide evidence that Olig2 is expressed in the late phase of cell cycle in progenitors that are ready to leave the cell cycle and differentiate into PCs. Therefore, we propose that Olig2+ cells are PC-committed progenitors and cannot transition to PIPs during cerebellar development. It is still possible that a transient expression of Olig2 during the presumptive PCP to PIP transition does not permit a threshold of cellular Cre accumulation to allow efficient Cre-mediated recombination and therefore negative reporter expression in the PIP of the Olig2-Cre reporter mice (Fig. 6). However, since most Olig2+ progenitors are late-phase progenitors and ready to leave the cell cycle, it is unlikely for them to re-enter the cell cycle and transition to PIP. Whether other VZ progenitors such as Olig2−/BrdU+ cells (probably Ptf1a+) are able to transition to PIPs is still an open question, but no strong evidence exists to support it yet. Alternatively, the observed cerebellar VZ progenitor phenotype15 can be explained by expansion and/or migration of the Gsx1+ PIPs while PCPs are exhausted by PC generation between E11.5 and E13.5. Our data cannot exclude a small-scale transition between different types of progenitors since we do observe a small number of Parvalbumin+ small interneurons (in the ML) that are labeled by Olig2-Cre (Fig. 6L), although this partial labeling of Pax2+ interneurons can also be explained by a partial overlapping expression pattern of the markers rather than a transition. Further investigations are needed to clarify the underlying mechanism.
Conclusion
Our work demonstrates that Olig2 is expressed in the late phase of the VZ progenitor cell cycle in the developing cerebellum and functionally required for normal PC generation from cerebellar VZ progenitors. The “pro-neuronal” characteristic of Olig2 in the cerebellar VZ progenitors distinguishes its role from other CNS regions such as spinal cord where Olig2 regulates specification of both neurons and oligodendrocytes. In addition, our observations from Cre-lox-mediated long-term genetic lineage tracing analyses challenge the “cerebellar progenitor identity transition” model that has been recently proposed. In fact, our data indicate that the majority of Olig2+ cells in the cerebellar VZ are either PC-committed progenitors or newborn PCs, and they unlikely switch their progenitor identity to give rise to cerebellar interneurons, at least, not in a large scale.
Methods
Mouse lines and genotyping
The Olig1-Cre23 and Olig2-Cre36 mice were crossed with the Z/EG2137, ROSA26;tdTomato (Ai14)38, or ROSA26;LacZ39 reporter mice, to examine promoter-specific Cre activity. The Olig123 and Olig23 single gene knockout mice were obtained by crossing heterozygous mice. Genotyping of the mice were performed as previously described3,23,40. The Institutional Animal Care and Use Committee of West China 2nd University Hospital at Sichuan University approved the use of the animals and the experiments. All procedures were carried out in accordance with the approved guidelines.
RNA extraction and quantitative reverse transcriptase PCR (qRT-PCR)
Total RNA was isolated from micro-dissected E18.5 cerebellar tissue samples using TRIzol® reagent, according to the manufacturer’s instructions (Invitrogen). qRT-PCR analysis of gene expression was performed as previously described40. The relative gene expression levels were normalized to that of the housekeeping gene GAPDH. The primer sequences of the target genes are shown in Table 1.
In vivo BrdU pulse-labeling
Pulse-labeling of dividing cerebellar progenitors was carried out by intraperitoneal injection of BrdU (5-bromo-2′-deoxyuridine, Sigma) into timed pregnant mice at 100 mg/kg body weight. Mice were sacrificed 0.5, 2, 8 or 14 hours after injection. BrdU staining was performed by incubating frozen sections at 37 °C in 2 M hydrochloric acid (HCl) solution for 30 minutes before standard immunohistochemistry.
Acute cerebellar cultures
The acute cerebellar cultures were prepared as described40. Briefly, embryonic cerebella were micro-dissected with meningeal membranes removed. Tissues were dissociated and plated at a density of 100,000 cells/coverslip. Cells were then fixed after 2 hours incubation at 37 °C with 5% CO2 and subjected to immunostaining.
Immunohistochemistry, immunocytochemistry and fluorescence microscopy
Cerebellar frozen and vibratome sections were prepared as previously described40,41,42. Tissue sections or fixed cell cultures were incubated with monoclonal antibodies against BrdU (rat IgG, 1:200, Accurate), Olig2 (mouse IgG, 1:200, Millipore), APC (mouse IgG, clone CC-1, 1:200, Calbiochem) and NeuN (mouse IgG, 1:400, Millipore); polyclonal antibodies against β-Galactosidase (β-gal) (chicken IgY, 1:2000, Abcam), BLBP (rabbit IgG, 1:500, Abcam), Tbr1 (rabbit IgG, 1:400, Proteintech), Pax2 (rabbit IgG, 1:50, Proteintech), Pax6 (rabbit IgG, 1:100, Proteintech), GABA (rabbit IgG, 1:2000, Sigma), Parvalbumin (rabbit IgG, 1:800, ImmunoStar), Olig2 (rabbit IgG, 1:500, Millipore or guinea pig IgG, 1:500, a gift from Dr. Ben Novitch at UCLA), GFAP (chicken IgY, 1:1000, Aves or rabbit IgG, 1:2000, DAKO), Calbindin (rabbit IgG, 1:500, ImmunoStar), Calretinin (rabbit IgG, 1:500, ImmunoStar), cleaved caspase-3 (rabbit IgG, 1:1000, Cell Signaling), and DCX (guinea pig IgG, 1:1000, Millipore), followed by appropriate species-specific secondary antibodies (Molecular Probes). DAPI (10 μg/ml, Sigma) was included in the secondary antibody incubations to label nuclei. The tissue sections or cultured cells were then mounted in mounting medium (Zhong Shan-Golden Bridge Biotech, P.R. China) and analyzed by conventional (Nikon Eclipse Ti) or confocal (Zeiss LSM 5 duo) fluorescence microscopy.
Measurements and statistical analysis
For cell culture staining, in Fig. 2B,C, numbers of Calbindin+ cells were counted in multiple image fields of cultures with a similar cell density from at least 3 different animals for both controls and mutants; in Fig. 2D,E, percentage of Pax6+ cells were counted and calculated from 100 to 500 DAPI+ cells in multiple image fields of cultures from at least 3 animals for both controls and mutants. For section staining, in Fig. 2A, the number of Calbindin+ cells per cerebellar section were counted from 3 animals for both controls and mutants; in Fig. 3, the number of Pax2+ cells per cerebellar section were counted from 3 animals for both controls and mutants; in Fig. 4B, percentage of BrdU+ cells were calculated from more than 600 DAPI+ cells on multiple sections of at least 3 animals for both controls and mutants; in Fig. 4D, a total of more than 600 BrdU+ cells were counted, and percentage of Lhx1/5+ in BrdU+ cells were calculated; in Fig. 5C,D, percentage of marker-positive cells were calculated from more than 600 total cells on multiple sections of at least 3 animals. All data were presented as means ± standard deviation except for Fig. 5C,D where only means were presented. Statistical analysis was performed in Microsoft Excel using Student’s t-test.
Additional Information
How to cite this article: Ju, J. et al. Olig2 regulates Purkinje cell generation in the early developing mouse cerebellum. Sci. Rep. 6, 30711; doi: 10.1038/srep30711 (2016).
References
Lu, Q. R. et al. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317–329 (2000).
Zhou, Q., Wang, S. & Anderson, D. J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25, 331–343 (2000).
Lu, Q. R. et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 (2002).
Zhou, Q. & Anderson, D. J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73 (2002).
Petryniak, M. A., Potter, G. B., Rowitch, D. H. & Rubenstein, J. L. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55, 417–433 (2007).
Furusho, M. et al. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol 293, 348–357 (2006).
Leto, K., Rolando, C. & Rossi, F. The genesis of cerebellar GABAergic neurons: fate potential and specification mechanisms. Front Neuroanat 6, 6 (2012).
Maricich, S. M. & Herrup, K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol 41, 281–294 (1999).
Pascual, M. et al. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci USA 104, 5193–5198 (2007).
Hoshino, M. et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47, 201–213 (2005).
Kim, E. J., Battiste, J., Nakagawa, Y. & Johnson, J. E. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci 38, 595–606 (2008).
Lundell, T. G., Zhou, Q. & Doughty, M. L. Neurogenin1 expression in cell lineages of the cerebellar cortex in embryonic and postnatal mice. Dev Dyn 238, 3310–3325 (2009).
Florio, M. et al. Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development 139, 2308–2320 (2012).
Seto, Y., Ishiwata, S. & Hoshino, M. Characterization of Olig2 expression during cerebellar development. Gene Expr Patterns 15, 1–7 (2014).
Seto, Y. et al. Temporal identity transition from Purkinje cell progenitors to GABAergic interneuron progenitors in the cerebellum. Nat Commun 5, 3337 (2014).
Zordan, P., Croci, L., Hawkes, R. & Consalez, G. G. Comparative analysis of proneural gene expression in the embryonic cerebellum. Dev Dyn 237, 1726–1735 (2008).
Morales, D. & Hatten, M. E. Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. J Neurosci 26, 12226–12236 (2006).
Zhao, Y. et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA 104, 13182–13186 (2007).
Fink, A. J. et al. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci 26, 3066–3076 (2006).
Hatten, M. E. & Heintz, N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci 18, 385–408 (1995).
Nakagawa, S., Watanabe, M. & Inoue, Y. Prominent expression of nuclear hormone receptor ROR alpha in Purkinje cells from early development. Neurosci Res 28, 177–184 (1997).
Meijer, D. H. et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci 13, 819–831 (2012).
Xin, M. et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci 25, 1354–1365 (2005).
Zhao, X. et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612–626 (2010).
Grimaldi, P. et al. Origins and control of the differentiation of inhibitory interneurons and glia in the cerebellum. Dev Biol 328, 422–433 (2009).
Dalgard, C. L., Zhou, Q., Lundell, T. G. & Doughty, M. L. Altered gene expression in the emerging cerebellar primordium of Neurog1−/− mice. Brain Res 1388, 12–21 (2011).
Lee, S. K., Lee, B., Ruiz, E. C. & Pfaff, S. L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev 19, 282–294 (2005).
Mizuguchi, R. et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757–771 (2001).
Novitch, B. G., Chen, A. I. & Jessell, T. M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789 (2001).
Scardigli, R., Schuurmans, C., Gradwohl, G. & Guillemot, F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31, 203–217 (2001).
Chakrabarti, L. et al. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat Neurosci 13, 927–934 (2010).
Ligon, K. L. et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53, 503–517 (2007).
Ali, F. et al. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277 (2011).
Buffo, A. & Rossi, F. Origin, lineage and function of cerebellar glia. Prog Neurobiol 109, 42–63 (2013).
Mecklenburg, N. et al. Cerebellar oligodendroglial cells have a mesencephalic origin. Glia 59, 1946–1957 (2011).
Schuller, U. et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008).
Novak, A. et al. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis 28, 147–155 (2000).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21, 70–71 (1999).
Kuang, Y. et al. Dicer1 and MiR-9 are required for proper Notch1 signaling and the Bergmann glial phenotype in the developing mouse cerebellum. Glia 60, 1734–1746 (2012).
Li, H. et al. Spatiotemporal heterogeneity of CNS radial glial cells and their transition to restricted precursors. Dev Biol 271, 225–238 (2004).
Li, H. & Grumet, M. BMP and LIF signaling coordinately regulate lineage restriction of radial glia in the developing forebrain. Glia 55, 24–35 (2007).
Acknowledgements
We thank members of Dr. Hedong Li’s laboratory for their technical support and helpful discussions. This work is supported by grants from New Jersey Commission on Spinal Cord Research, the National Natural Science Foundation of China (30971633 and 31171045), the Department of Science and Technology of Sichuan Province (Young Scientific Innovation Team in Neurological Disorders grant 2011JTD0005) and the Program for Changjiang Scholars and lnnovative Research Team in University (PCSIRT) (IRT0935) to HL; grants from the US National Institutes of Health R01NS072427 and R01 NS078092 and the National Multiple Sclerosis Society (NMSS-4727) to QRL.
Author information
Authors and Affiliations
Contributions
J.J. and H.L. designed research; J.J., Q.L., Y.Z., Y.L., M.J., L.Z., X.H., C.P. and T.Z. performed research; J.J., Q.L., Y.Z., M.J., T.Z., Q.R.L. and H.L. analyzed data; and H.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ju, J., Liu, Q., Zhang, Y. et al. Olig2 regulates Purkinje cell generation in the early developing mouse cerebellum. Sci Rep 6, 30711 (2016). https://doi.org/10.1038/srep30711
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30711
This article is cited by
-
Revisiting the development of cerebellar inhibitory interneurons in the light of single-cell genetic analyses
Histochemistry and Cell Biology (2024)
-
Elimination of glutamatergic transmission from Hb9 interneurons does not impact treadmill locomotion
Scientific Reports (2021)
-
Transcriptome programs involved in the development and structure of the cerebellum
Cellular and Molecular Life Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.