Abstract
Magnetic resonance (MR) spectroscopic imaging has become an important tool in clinical settings for noninvasively obtaining spatial and metabolic information on a molecular scale. Conventional spectroscopic imaging is acquired in the time domain and its clinical application is limited by the long acquisition time, restricted spatial coverage and complex suppression and reconstruction procedures. We introduce a fast MR spectroscopic imaging technique in the frequency domain, termed phase-cycled spectroscopic imaging (PCSI). PCSI uses a balanced steady-state free precession (bSSFP) sequence with an ultra-low flip angle to achieve very high acquisition efficiency with a short repetition time. This approach enables faster frequency sweeping by changing the cycled RF phase and using flexible non-uniform sampling and it greatly reduces the RF energy deposition in tissue. With its intrinsic water and fat suppression, PCSI more closely resembles routine clinical scans because it eliminates the suppression steps. We demonstrate that it is feasible to acquire PCSI spectra in a phantom and in humans and that PCSI provides an efficient spectroscopic imaging method, even for J-coupled metabolites. PCSI may enable spectroscopic imaging to play a larger role in the clinical assessment of the spatial tissue distribution of metabolites.
Similar content being viewed by others
Introduction
Magnetic resonance spectroscopy (MRS) is widely used in medicine, biology, chemistry and physics. Proton MRS has become an important tool in a broad range of clinical applications, enabling the noninvasive detection of subtle metabolic changes in tissues. As the spectrum is measured in either the frequency domain or the time domain, MRS techniques are categorized as either frequency-resolved or time-resolved techniques1,2,3. The early frequency-resolved techniques, which swept a range of frequencies by changing the magnetic field strength or radio frequency (RF), were very slow and could take several hours to acquire a single spectrum1,2. Therefore, these frequency-resolved techniques have now been supplanted by time-resolved techniques, which are faster and can offer much greater sensitivity3. For in vivo time-resolved spectroscopy, the signal has to be restricted to a selected volume in the subject. Surface coils were first used to receive the signal from a limited volume4. By using RF pulses and gradients, the targeted volume can be selected by different techniques5,6,7,8,9, such as PRESS (point-resolved spectroscopy)9, STEAM (stimulated echo acquisition mode)8 and ISIS (image-selected in vivo spectroscopy)5. These advances have made single-voxel spectroscopy (SVS) feasible in clinical settings.
Magnetic resonance spectroscopy imaging (MRSI) simultaneously acquires both metabolic and anatomic spatial information, thereby providing metabolic mapping that is of great interest to clinicians10,11. However, MRSI has not been widely accepted as a routine clinical tool, mainly because its data acquisition is very time consuming and the post-processing procedures are generally performed offline, automatically or manually by an MR technician. For example, it may take more than 30 min to acquire an image with a 32 × 32 matrix using the PRESS sequence. Multivoxel MRSI with an 8 × 8 or 16 × 16 matrix is used in routine clinical applications. In research studies, fast MRSI methods have been proposed for reducing the acquisition time12,13,14,15,16,17,18. In addition, the strong water signal overpowers the tiny metabolite signal (which is approximately 10,000 times weaker). Water suppression methods, such as chemical-shift selected pulses (CHESS)19 or MEGA20, are normally applied in MRSI. The lipid signal in the skull has to be suppressed to prevent it from contaminating the spectra obtained inside the brain. Lipid suppression can be achieved by several different strategies: outer volume suppression (OVS)21,22,23,24, the inversion recovery (IR) method25,26, or k-space data post-processing27. The most commonly used approach in clinical studies is OVS, which uses spatial presaturation bands to prevent spectral contamination by peripheral lipid and water signals21,22,23,24. The precise placement of the many OVS bands is challenging and time consuming and it requires special technician training and skills.
We propose a novel frequency-resolved method called phase-cycled spectroscopic imaging (PCSI), which uses the bSSFP sequence with an ultra-low flip angle (less than 1°). This PCSI method may overcome the abovementioned problems to achieve very high acquisition efficiency with a short repetition time (TR). PCSI is the first pulsed frequency-resolved method; it uses RF pulses instead of continuous-wave RF as was used in the early frequency-resolved methods1,2. The bSSFP sequence with small flip angles (e.g., 4° or 5°) has been used for functional magnetic resonance imaging (fMRI)28,29, but the application of the bSSFP with ultra-low flip angles (<1°) has not been reported previously. The PCSI method uses ultra-small RF pulses and a phase-cycled frequency-sweep technique to make data acquisition faster, more efficient and more flexible. Here, we demonstrate the feasibility of the PCSI method by conducting a conceptual validation with simulations and provide proof-of-principle testing in phantoms, volunteers and a patient with a heterogeneous brain tumor. Our technique may provide a basis for developing new low-energy and high-sensitivity MRI techniques.
Results
Ultra-low flip angle steady state
The magnetization profiles of the steady-state magnetization were generated for different flip angles, as shown in Fig. 1a. As the flip angle decreased to less than 1°, the profile became a single sharp peak. In Fig. 1b, the real part of the magnetization profile is a pure absorption shape and is much narrower than the magnitude profile. This real magnetization profile can serve as a perfect frequency response function for spectroscopic imaging. Using this response function, we can excite only a very narrow range of frequencies (approximately 1.9 Hz) and simultaneously suppress the signal at other frequencies. Therefore, we can acquire an MR image at a specific frequency point using this steady state with an ultra-low flip angle.
Simulated transverse magnetization profiles of a balanced SSFP sequence obtained using equation [S7].
(a) The magnitude profiles of four selected flip angles. (b) Enlarged magnetization profiles of the absolute, real and imaginary components with α = 0.24°. (c) The real profiles shift along the frequency direction as the cycled RF phase φ is changed.
Phase-cycled frequency sweeping
The peak locations of the response function are determined by the cycled RF phase φ in Fig. 1c. The cycled phase corresponds to a certain off-resonance frequency, Δf, determined via the equation Δf = φ/(2π · TR) derived from equation [S12] in the Supplemental Information. By changing the cycled RF phase, the peak location can be shifted to a target frequency within a frequency period, [−1/(2TR), 1/(2TR)], which corresponds to one RF phase cycle. Changing the cycled RF phase is much easier than changing the magnetic field, as was required in early frequency-resolved techniques. Taking advantage of the high acquisition efficiency of the bSSFP sequence, we obtained a PCSI sweep rate of approximately 13 images/s (or 76.8 ms per image), which is much more efficient than the early frequency-sweeping methods (which might have a sweep rate of approximately 1 Hz/s). This high sweep rate enables PCSI to acquire multiple averages in order to improve its signal-to-noise ratio (SNR). After obtaining a series of MR images at different frequency points, we can generate a spectrum in each voxel.
Specific absorption rate
The ultra-small flip angle may greatly reduce the RF energy deposited in tissue, because the specific absorption rate (SAR) is proportional to the square of the flip angle, as follows:

where the RF duty cycle is the ratio of the total RF duration time to the TR. There are three pulses (90°–180° –180°) in each TR (2000 ms) for the PRESS sequence and one RF pulse (e.g., 0.3°) in each TR (2.4 ms) for the PCSI sequence. The SAR ratio of CSI to PCSI is computed to be 2916 by assuming the same duration for RF pulses. Additional RF pulses for the water- and lipid-suppression in CSI, if included, could further substantially increase this SAR ratio. The total RF energy of the PCSI sequence is 2.6 W·s, based on the reported protocol with a flip angle of 0.3°.
Conceptual validation by simulation
We first assessed whether and under what conditions the extremely small metabolite signals can be obtained by using small RF pulses. We performed simulations with multiple small flip angles to validate the theoretical feasibility of PCSI and investigate the importance of the choice of RF flip angle. In the simulations, a spectrum was created with four peaks: water, choline (Cho), creatine (Cr) and N-acetyl aspartate (NAA), which were assumed to be delta functions with magnitudes of 10,000, 0.6, 0.8 and 1.2, respectively. Figure 2a shows the original spectrum with a water peak at 200 Hz. Figure 2b shows the simulated spectrum with an optimal flip angle, α = 0.24°, computed using equation [S11] with TR = 2.4 ms. This simulated spectrum was generated by convolution of the real response function in Fig. 1b and the spectrum in Fig. 2a. All three metabolic peaks could easily be observed (Fig. 2b), having heights proportional to the prescribed values. Figure 2c shows the simulated spectrum with α = 1°, in which the three small metabolic peaks are barely detectable. The three metabolic peaks are not appreciable in Fig. 2d with α = 3°. These simulations demonstrate that it is theoretically feasible to obtain a spectrum by using PCSI and that the metabolic peaks become smaller and eventually disappear as the flip angle increases. Therefore, an ultra-low flip angle is crucial if a spectrum is to be acquired by PCSI.
Simulation of a spectrum using the PCSI method.
(a) The assumed metabolite signals of NAA (1), Cr (2), Cho (3) and water (4) on a log scale. (b) The simulated real signal obtained using equation [S7] with TR = 2.4 ms, α = 0.24°. The inset shows the enlarged plot with three metabolite peaks. (c) The simulated real signal with α = 1°. (d) The simulated real signal with α = 3°. In the simulations, the water peak was assumed to reside at 200 Hz.
Phantom study
A proof-of-principle study was performed on a phantom filled with a brain-mimicking solution that contained the metabolites NAA, Cr and Cho. PCSI data were acquired using a modified bSSFP sequence, in which the cycled RF phase was changed for each measurement by using a non-uniform frequency-sampling scheme. Figure 3a shows PCSI signals in a region of interest (ROI), including nine voxels (18.75 × 18.75 × 15 mm, or 5.3 cm3) at the center of the phantom. The peaks on either side are water peaks, which wrap around as a result of the periodic response function. Phase correction based on the water peaks was performed to convert the real component of each complex signal to a pure absorption shape. The inset enlargement in Fig. 3a clearly demonstrates the three metabolite peaks in the real signal. This signal can be converted to the spectrum, as shown in Fig. 3b after post-processing (see the Methods section for details). Figure 3c shows a spectrum from a single voxel (6.25 × 6.25 × 15 mm, or 0.59 cm3) at the same location as that in Fig. 3b. The single-voxel spectrum is similar to the spectrum in Fig. 3b from a larger ROI but has slightly more noise. For comparison, Fig. 3d shows a spectrum from a single voxel (20 × 20 × 20 mm, or 8 cm3) obtained by using the conventional time-resolved SVS sequence. The two spectra shown in Fig. 3b (PCSI) and 3d (SVS) were similar and had consistent peak positions. This phantom study demonstrates that PCSI can require much less time than the time-resolved SVS method to acquire comparable spectra.
MR spectra from a phantom.
(a) The PCSI spectral signal from nine voxels in the center (18.75 × 18.75 × 15 mm) with an inset showing three metabolite peaks. (b) The real and fitted PCSI spectra calculated from the spectral signal in (a), with the location shown in the inset. (c) PCSI spectra from a single voxel (6.25 × 6.25 × 15 mm) at the same location as (b) (shown in the inset). (d) Conventional spectra from a commercial single-voxel spectroscopy sequence; the voxel size is 20 × 20 × 20 mm (shown in the inset). The peaks labeled 1, 2, 3 and 4 are the NAA, Cr, Cho and water peaks, respectively. Ampl, amplitude; Real, real part of the signal; Imag, imaginary part of the signal; Fit, the fitted spectrum. The inset images in (b) and (c) were generated from the PCSI data. The inset image in (d) was a high-resolution T2-weighted image at the same slice position shown in the insets in (b) and (c).
Figure 4 shows a comparison between a short-TE SVS spectrum and a uniformly-sampled PCSI spectrum on the phantom. Small J-coupled metabolite signals, such as myo-inositol (mI) and glutamate (Glu), are clearly visible in both spectra. Figure 5 shows two spectra from a uniform PCSI with a 64 × 32 matrix and a higher in-plane resolution (3.75 × 3.75 mm). The metabolites with larger signals, such as NAA, Cr and Cho, can be easily identified in the spectrum, even from a single voxel. In comparison, J-coupled metabolites, such as Glu and mI, can be identified in the averaged spectrum from nine voxels.
Spectra of a phantom obtained using single-voxel spectroscopy (SVS) and PCSI methods.
(a) The SVS PRESS spectrum with a short TE of 30 ms; the voxel size was 20 × 20 × 20 mm. (b) The PCSI spectrum from an ROI with three voxels (each voxel was 6.25 × 6.25 × 15 mm). J-coupled metabolites, such as glutamate (Glu) and myo-inositol (mI), can be identified in both spectra.
Human studies
Spectroscopic imaging of the human brain is more challenging than phantom imaging. The field inhomogeneity of the human brain is greater than that of the phantom and can greatly diminish or even destroy metabolite signals. Also, the strong lipid signal in humans can degrade the quality of the spectrum. As a practical proof of principle of PCSI in human brain, volunteer and patient studies were performed using the same non-uniform–sampling protocol as was used in the phantom study.
Figure 6a shows an in vivo PCSI spectrum from an ROI including 9 voxels (18.75 × 18.75 × 15 mm, or 5.3 cm3), in which three metabolite peaks can easily be identified and fitted. In comparison, a spectrum from a voxel (20 × 20 × 20 mm, or 8 cm3) obtained by using the SVS sequence is shown in Fig. 6b, in which the inset shows the location of the ROI to be similar to that in Fig. 6a. Both spectra show consistent peak positions and relative peak heights after alignment. Figure 6c shows a PCSI spectrum from a single voxel (6.25 × 6.25 ×15 mm, or 0.59 cm3) at the edge of the brain where field inhomogeneity becomes severe. Figure 6d shows a spectrum from an ROI at the same position that includes nine voxels (18.75 × 18.75 × 15 mm, or 5.3 cm3). Although the two spectra appear similar, the spectrum in the larger ROI has smaller peaks than does the single-voxel PCSI spectrum because of the high field inhomogeneity near the edge. Both spectra had less noise than did the spectrum in Fig. 6a, because the locations in Fig. 6c,d were closer to the head coil where the SNR is higher. The greater peak width in the PCSI spectra was mainly due to greater field inhomogeneity, because PCSI acquired data from a whole slice, whereas SVS acquired data from only a single voxel.
MR spectra from a healthy human volunteer.
(a) PCSI spectra from the nine voxels (3 × 3 voxels with dimensions 18.75 × 18.75 × 15 mm) shown in red in the inset. (b) Conventional spectra from a commercial single-voxel spectroscopy sequence. The voxel size is 20 × 20 × 20 mm; the inset shows the ROI. (c) PCSI spectra from a single voxel (6.25 × 6.25 × 15 mm) at the periphery of the brain (shown in red in the inset). (d) PCSI spectra from nine voxels (3 × 3 voxels, with dimensions 18.75 × 18.75 × 15 mm) at the same location shown in the insert in (c). The inset images in (a,c,d) were generated from the PCSI data. The inset image in (b) was a high-resolution T2-weighted image acquired at the same slice position. ROI, region of interest; Real, real part of the signal; Fit, the fitted spectrum.
Parametric maps of the heights of the three-metabolite peaks and of the Cho/NAA ratio were generated for two repeated measurements, as shown in Fig. 7. The spectra were computed for each voxel (6.25 × 6.25 mm) by averaging the voxel and its four neighbors to improve its SNR. The two sets of maps were then interpolated and overlaid on a high-resolution T2-weighted image (voxel size: 0.52 × 0.52 mm) for visual comparison. The PCSI method demonstrates a good robustness for separate measurements.
As a final practical proof of principle, we compared the conventional chemical-shift imaging (CSI) parametric maps with PCSI maps for a patient with a multifocal anaplastic astrocytoma (a WHO grade 3 tumor) (Fig. 8). There were four lesion foci (1–4) visible in one T2-weighted image, in which lesion 1 corresponds to a surgical cavity and lesions 2 through 4 represent tumor foci. Figure 8 shows the importance of full coverage of the whole slice because of the heterogeneity of the tumors. It took approximately 4 min longer for CSI to acquire parametric maps with only 10 × 10 voxels than for PCSI to acquire maps with 32 × 32 voxels. The main tumor (lesion 1) can be clearly identified in the Cho/NAA ratio maps obtained with either method. However, the conventional CSI maps covered only small parts of lesions 2 and 3, which clearly demonstrate characteristic tumor profile features in the PCSI Cho/NAA map.
Parametric maps of three metabolites (NAA, Cr and Cho) and of the Cho/NAA ratios from a patient with a multifocal anaplastic astrocytoma.
Maps from the conventional CSI sequence with 10 × 10 voxels are in the upper row. The corresponding PCSI parametric maps are in the lower row. A T2-weighted image (far-right end of the lower row) shows the lesion foci (1–4); lesion 1 corresponds to a surgical cavity and lesions 2 through 4 represent tumor foci. The colored parametric maps were overlaid on a T2-weighted image. The dashed squares were manually drawn on PCSI maps and the T2-weighted image for comparison purposes.
Because of the limited coverage of CSI maps, most lesion foci, including the left frontal surgical resection cavity (lesion 1) are inadequately evaluated. A slight increase in Cho is suggested in the portions of lesions 2 and 4 covered by the CSI maps. The corresponding PCSI parametric maps also show slightly increased Cho in lesions 2 and 3, decreased NAA concentrations in all lesion foci and resultant robust increases of Cho/NAA in lesions 2 and 3 and a modest increase of Cho/NAA in lesion 4. The reason for the increased Cho/NAA ratio in lesion 1 is unclear; it may indicate residual or recurrent tumor, but this has not been confirmed.
Discussion
We have demonstrated the PCSI method, a new, flexible and efficient way to perform MR spectroscopic imaging in the frequency domain. PCSI uses a new frequency-sweep technique and a non-uniform sampling scheme that reduce the total acquisition time. PCSI also offers intrinsic water and fat suppression, thereby reducing operator dependence and greatly simplifying the scanning procedures. By using an ultra-low flip angle, PCSI greatly reduces the SAR in human subjects. In addition, PCSI may be suitable for measuring J-coupled metabolites because of its short effective TE and synchronized phases. PCSI has great potential to enable faster and simpler clinical applications of MRSI.
Imaging speed
Conventional time-resolved MRSI is limited in its clinical applications because of the long acquisition times. Fast time-resolved MRSI methods have been proposed to accelerate the acquisition12,13,14,15,30,31,32,33,34. Of these methods, EPSI or PEPSI12,14, spiral MRSI13 and SI-CONCEPT with circular echo-planar trajectories34 are the most efficient; they work by interleaving spatial k-space readouts of echo-planar, spiral, circular encoding with the spectral acquisition to speed up the acquisition and cover more volume. However, these fast MRSI techniques usually have a limited spectral width and require more complex reconstruction procedures to correct strong off-resonance effects caused by echo-planar or spiral encoding. The PCSI method provides a different means of speeding up acquisition by using adaptive sampling in targeted ranges of the frequency domain. We compared these methods based on the parameters related to speed and spectral width, which are summarized in Table 1. When SNR is not considered in this comparison, PCSI is the most efficient of the five methods compared, based on the acquisition time per slice per average. Figure S6 shows that PCSI can generate well-defined spectra for a phantom or human, even with only one average. More averages can be used to improve the SNR when necessary.
All of the above methods can be further accelerated by using a parallel imaging or compressed sensing technique35,36,37,38. Acceleration has been demonstrated to achieve additional speed factors in phase and slice directions when parallel imaging techniques are used12,16,17,18,39. In the future, parallel imaging or compressed sensing techniques may be used with speed factors of 2 or more in conjunction with PCSI to acquire higher-resolution spectroscopic images when the SNR is sufficient.
The PCSI method has great flexibility for sampling the frequency domain. In this study, we demonstrated a non-uniform sampling scheme that covered only three metabolite peaks (NAA, Cr and Cho), which decreased the total acquisition time by a factor of 2.5. If we cover only a single metabolite, the acquisition time can be further reduced by a factor of 3 to 5 to achieve the total acquisition in less than 1 min. PCSI may thus become suitable for studying the dynamic changes of certain metabolites.
J-coupled metabolites
J-coupled metabolites (e.g., Glu and mI) have low signals in the conventional spectrum. With decreasing TE, the spectral signal increases considerably and makes the measurement of J-coupled metabolites feasible40,41,42. The PCSI method has a very short TE (1.2 ms). The PCSI signal is from a steady state mixed with shorter and longer TE signals. Even though the effective TE may be longer than the actual TE, the effective TE in PCSI may still be shorter than the minimum TE (30 ms) of PRESS SVS on the scanner. In Fig. 4, the mI peak in the PCSI spectrum is much higher than that in the SVS spectrum. Figure 4 clearly demonstrates that the PCSI method has some advantages with regard to higher signals in J-coupled metabolites.
Besides peak splitting of coupled metabolites, J coupling also causes phase evolutions that lead to peak and baseline distortion, thereby making measurements more difficult in a conventional time-resolved spectrum43,44. The PCSI method acquires data in the frequency domain. All metabolite peaks are acquired separately and should have consistent phases, because PCSI synchronizes the phase of each peak by changing RF cycled phase to match its frequency. The synchronized phases for all the frequencies in one spectrum may further increase the signal of coupled metabolites, which may make PCSI an ideal tool for measuring J-coupled metabolites. This will be investigated in future studies.
Other advantages
PCSI offers other advantages over conventional MRSI. First, PCSI is less prone to problems such as unreliable spectra at the periphery of the brain due to OVS or a limited coverage of PRESS excitation. Second, by using an ultra-low flip angle, PCSI reduces the SAR in humans thousands of times compared to conventional spectroscopy sequences. PCSI also does not require spatial and water suppression, leading to an even greater reduction in the SAR19,21. Finally, the precise manual placement of OVS bands is often highly operator dependent and is poorly reproducible, even by the same operator, resulting in greater inter-subject variability45. With its intrinsic water and fat suppression, PCSI avoids the above issues, reduces subjective variability and greatly simplifies the scanning procedures, thereby more closely resembling a routine clinical imaging procedure.
Factors influencing PCSI
The T1/T2 ratio and flip angles are the main factors influencing PCSI. The relative heights of the peaks in Figs 3, 4 and 6 were somewhat different for PCSI and SVS, possibly reflecting the different mechanisms of data acquisition. The PCSI signal is from the bSSFP sequence, which is a steady state with mixed magnetizations across a broad range of TE values. The relative heights are determined mainly by factors such as T1/T2 and flip angles according to the signal equation [S8]. By comparison, the SVS signals are from a spin-echo sequence and the relative heights are determined by T2 and TE.
The distribution of metabolite signals in Fig. 8 is not entirely consistent between chemical shift imaging (CSI) and PCSI, possibly as a result of T1/T2, flip angle and B0 field inhomogeneity. In addition, there are still small differences between repeated measures in Figs 7 and S7. More robust results may be achieved by further optimizing the acquisition scheme with more dense sampling around the targeted peaks (e.g., with a step of 0.5° or less).
One definition of the SNR in spectroscopy is the peak height divided by the noise40. For the PCSI and CSI methods, the SNR was computed from a single voxel (PCSI voxel size: 6.25 × 6.25 × 15 mm; CSI voxel size: 10 × 10 × 15 mm) from a metabolic phantom by using the NAA peak height and the standard deviation of the noise computed on the left side of the Cho peak, because the PCSI method has a limited frequency range for measuring noise. The SNR values for the PCSI and CSI methods were 19.8 and 90.9, respectively. However, it may not be rational to compare SNRs directly in this case, because the left side of the Cho peak may not be an ideal location for measuring noise due to possible metabolite signal at short TE and several factors influence the SNR, such as the voxel size, number of averages, acquisition matrix, sampling density and TE value, which are not identical for the two methods.
The quality of shimming is a key factor in spectroscopic imaging. The typical FWHM ranges from 12 to 20 Hz for in vivo brain shimming40. In this study, PCSI used an auto-advanced shimming to achieve a very sharp water peak with a 6.5-Hz width on the phantom. However, PCSI had a broader peak with a 35.7-Hz width in one human volunteer, compared to the SVS peak width of 17.5 Hz in a much smaller ROI. Therefore, the quality of the PCSI spectrum may be improved by using advanced shimming techniques (e.g., higher-order shimming), which can approach a width of 15 Hz or less in humans46,47.
Limitations and future applications
The PCSI method has some limitations. PCSI has a smaller frequency spectral window than does conventional MRSI and requires prior knowledge of metabolite location to speed up the acquisition using non-uniform sampling. Therefore, the PCSI method is suitable for studying metabolite signals in a known spectral range but may not be optimal for exploring new metabolites. Prior knowledge of the specific spectrum is, therefore, required in order to develop a specialized PCSI protocol for targeted metabolites. However, this may not be a problem in most clinical scenarios. In addition, because of the signal dependence on the flip angle, PCSI is sensitive to B1 inhomogeneity, which may be the main factor causing the variations in the parametric map shown in Fig. 8. Therefore, a body transmit coil should be used instead of a surface transmit coil to reduce the effects of B1 inhomogeneity. If necessary, B1 correction or parallel transmit can be used to further reduce these B1 inhomogeneity effects48.
The PCSI method has great potential for other applications. First, PCSI could be implemented in 3D imaging, which could have a higher SNR and efficiency because each 3D measurement requires only one steady state. Second, the PCSI can be used in multinuclear spectroscopic imaging, such as fluorine and sodium imaging. In addition, whereas time-resolved spectroscopy has a clear advantage in single-voxel spectroscopy, the PCSI method offers a greater advantage in 2D or 3D high-resolution spectroscopic imaging, in which the number of spectral points is much smaller than the number of conventional phase encoding steps. We have successfully acquired 32 × 32 and 64 × 64 images; a higher-resolution spectroscopic image with a 128 × 128 matrix may be obtainable in the future if high-order gradient shimming, superconductor coils and parallel RF transmission techniques can be used with this PCSI technique.
Methods
The theory and simulations are presented in the Supplementary Information.
Protocol design
The protocol parameters must be precisely selected based on prior knowledge of the targeted metabolite signal. In contrast to conventional MRSI, PCSI does not require additional water- and fat-signal suppression. In PCSI, water and fat signals are intrinsically suppressed, because the very sharp response function suppresses signal that occurs far from the selected frequency, as shown in Fig. 1b. To suppress both water and fat, appropriate values of the protocol parameters, such as the TR and flip angle α, had to be selected to optimize the suppression by positioning the water and fat peaks as closely as possible, using the periodic property of the response function. To do this, the water and fat peaks had to be put into two consecutive frequency periods, as shown in Fig. S3a, so that the fat peak could wrap around to the water period as the red peak. The period length should be similar to the water-fat chemical shift for this configuration. The average chemical shift between body fat and water is approximately 3.35 ppm49, or approximately 413 Hz on a 3T scanner. Therefore, a TR of 2.4 ms was selected so that the period of the response function was 417 Hz, the closest value to the water-fat shift of 413 Hz. Period II was selected for the acquisition window because it included all the metabolite and water peaks. The fat peak was in period I and wrapped around to period II as an inverted red peak, as shown in Fig. S3a. In this case, both the water and fat peaks were located at the end of period II. The response function is different in each period, being positive for period II and negative for the two neighboring periods (I and III) in Fig. S3b. This is why the fat peak in period I was inverted and projected onto period II in Fig. S3a. To place the targeted metabolite at the center of the acquisition window, the system frequency was decreased by a certain amount (e.g., 200 Hz) to shift the water peak to one side of period II in Fig. S3a. For TR = 2.4 ms, T1 = 1300 ms and T2 = 250 ms, the optimal α and the maximum amplitude are 0.24° and 0.22, respectively, according to equation [S11].
Accelerated non-uniform frequency sweeping
When uniform sampling was used in the whole phase cycle (a frequency period) with a step of 1°, PCSI required approximately 11 min with an acquisition matrix 32 × 32 (360 measurements, TR = 2.4 ms, 23 averages). By using non-uniform sampling, we can sample densely in the ranges of target metabolites and sparsely in other ranges to reduce the acquisition time. In this study, the positions of three target metabolites, NAA, Cr and Cho, in the spectrum were calculated in order to select the specific ranges for dense sampling. NAA is located near −115°, Cr is near −6° and Cho is near 13°, with the water peak being shifted to 200 Hz on the 3T scanner. Therefore, the dense sampling windows were selected in the ranges (−142°, −100°) and (−27°, 40°) with a step of 1° and the step in other ranges was selected as 10° (Fig. S4). The complete sample range of the cycled RF phase was chosen as −200° to 250°, which led to a total of 143 measurements and a total acquisition time of 4 min 28 s. The acquisition time was decreased by a factor of 2.5 by using this non-uniform sampling method when compared to a uniform sampling scheme with 360 measurements. With more advanced non-uniform sampling schemes, the acquisition time may be further reduced to less than 1 min in the future.
Data acquisition
Phantom study
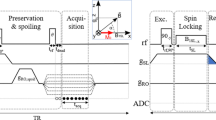
All studies were performed on a Siemens 3T Prisma MRI scanner (Siemens Medical Solutions, Erlangen, Germany). A phantom filled with a brain-mimicking solution containing the metabolites NAA, Cr and Cho was used to investigate the feasibility of the PCSI method. The PCSI data acquisition used a modified bSSFP sequence based on a TrueFISP sequence, in which the cycled RF phase changed for each measurement. A 64-channel head coil was used in the image acquisition to ensure the quality of the data. In the experiments, a single axial slice was selected and shimming with the advance mode was performed to achieve the best field homogeneity. After shimming, the system frequency was manually decreased by 200 Hz to shift the water peak toward the end of the frequency acquisition window. The protocol parameters were as follows: TR = 2.4 ms; TE = 1.2 ms; flip angle α = 0.3°; 40 dummy pulses for the preparation of the steady state; 32 × 32 acquisition matrix; in-plane resolution 6.25 × 6.25 mm; slice thickness 15 mm; bandwidth 1116 Hz; 23 averages; 143 measurements using a non-uniform sampling scheme of the cycled RF phase; and total time approximately 4 min 28 s. A flip angle of 0.3° was used because the actual flip angle may be smaller than the prescribed angle as a result of the electron screening effect.
In addition, a PCSI data set with uniform sampling (a step of 1°, 360 measurements) was acquired in 11 min 12 s. The short TE spectrum was acquired on a phantom by using the SVS protocol with a minimum TE of 30 ms. The PCSI signals of J-coupled metabolites were compared with those in the short-TE SVS spectrum. To test the limits of PCSI, a higher-resolution dataset was acquired with a 64 × 32 acquisition matrix, an interpolated in-plane resolution of 3.75 mm × 3.75 mm and a slice thickness of 15 mm. The total acquisition time was 11 min 12 s. Half of the coil channels were turned off because of the limited memory of the computer used for processing the data. Finally, the above non-uniform protocol was performed three times on the phantom to test its repeatability.
Human studies
Field inhomogeneity is a more serious confounding variable in studies of the human brain than in those performed on a uniform phantom, making shimming a more critical issue. A single tilted axial slice was selected in the superior brains of two healthy volunteers and a patient with multifocal anaplastic astrocytoma of the brain to demonstrate the feasibility of PCSI. The protocol was the same as that used in the phantom study, except for the flip angle and the frequency shift. In the human studies, based on our experience, a flip angle of 0.5° was used to obtain a better signal. The frequency shift was reduced to 190 Hz because of the much broader water peak. Data were acquired from the first volunteer by using a 64-channel head coil (Fig. 6). Data from the second volunteer (Fig. 7) and the patient (Fig. 8) were acquired using a 20-channel head coil to demonstrate the feasibility and reproducibility.
For comparison, spectra from a single voxel were acquired in both phantom and human studies by using the manufacturer’s SVS sequence with PRESS preparation. The protocol TE was 135 ms, the TR was 2000 ms and the voxel size was 20 × 20 × 20 mm. Water suppression was used to improve the SNR of the spectra. The total acquisition time was 4 min 24 s. The conventional CSI data were acquired from a patient with the following protocol parameters: TE/TR = 135/1700 ms; voxel size = 10 × 10 × 15 mm; 10 × 10 acquisition matrix; and total time of 8 min 7 s. These studies were approved by the institutional review board of St. Jude Children’s Research Hospital and written informed consent was obtained from the volunteer, patient, parent, or guardian as appropriate. All the experiments were carried out in accordance with the approved guidelines.
Image reconstruction and post-processing
The measurement data in k-space were saved on the scanner and transferred to a Linux workstation. The image was first reconstructed by using a fast Fourier transform for each channel of the head coil. Phase correction based on the water peak was performed for each voxel to convert the real profile to a pure absorption shape, which ensured that the phases from different coil channels were consistent for each voxel. After phase correction, images from different channels can easily be combined by using a weighted summation based on their magnitude. Because of the field inhomogeneity, the position of the water peak varied for the different voxels in the image. The peaks were shifted in order to align them for subsequent processing. The baseline correction was performed by using polynomial fitting for the magnitude and phase of the profile without the metabolite signal. The profile then subtracted the fitted baseline to obtain the spectrum in units of Hertz. The spectrum was then converted to the final spectrum in parts per million (PPM). To quantify the spectrum, it was fitted using three Lorentzian functions. Normalization was performed by dividing the signal by a factor that was computed using an empirical form: the square root of the cube of the filtered sensitivity map. With the fitted spectrum, the different parameters (e.g., the amplitude and position) related to each peak could be extracted for further processing. After quantification for each voxel, the parametric maps were generated, registered and overlaid on a high-resolution T2-weighted image. The signal-processing procedure is summarized and shown in Fig. S5; in the future, it may allow real-time metabolic maps to be generated directly on the MRI scanner without the need for manual intervention by a technician.
Additional Information
How to cite this article: Guo, J. et al. Fast frequency-sweep spectroscopic imaging with an ultra-low flip angle. Sci. Rep. 6, 30066; doi: 10.1038/srep30066 (2016).
References
Arnold, J. T., Dharmatti, S. S. & Packard, M. E. Chemical effects on nuclear induction signals from organic compounds. J. Chem. Phys. 19, 507, 10.1063/1.1748264 (1951).
Meyer, L. H., Saika, A. & Gutowsky, H. S. Electron distribution in molecules. III. The proton magnetic spectra of simple organic groups. J. Am. Chem. Soc. 75, 4567–4573, 10.1021/Ja01114a053 (1953).
Ernst, R. R. & Anderson, W. A. Application of Fourier transform spectroscopy to magnetic resonance. Rev. Sci. Instrum. 37, 93–102, 10.1063/1.1719961 (1966).
Ackerman, J. J. H., Grove, T. H., Wong, G. G., Gadian, D. G. & Radda, G. K. Mapping of metabolites in whole animals by 31P NMR using surface coils. Nature 283, 167–170, 10.1038/283167a0 (1980).
Ordidge, R. J., Connelly, A. & Lohman, J. A. B. Image-selected in vivo spectroscopy (ISIS): a new technique for spatially selective NMR spectroscopy. J. Magn. Reson. 66, 283–294, 10.1016/0022-2364(86)90031-4 (1986).
Doddrell, D. M. et al. Discrete isolation from gradient-governed elimination of resonances—Digger, a new technique for invivo volume-selected NMR-spectroscopy. J. Magn. Reson. 70, 319–326, 10.1016/0022-2364(86)90017-X (1986).
Aue, W. P., Müller, S., Cross, T. A. & Seelig, J. Volume-selective excitation. A novel-approach to topical NMR. J. Magn. Reson. 56, 350–354, 10.1016/0022-2364(84)90115-X (1984).
Frahm, J., Merboldt, K. D. & Hänicke, W. Localized proton spectroscopy using stimulated echoes. J. Magn. Reson. 72, 502–508, 10.1016/0022-2364(87)90154-5 (1987).
Bottomley, P. A. Spatial localization in NMR spectroscopy in vivo. Ann. N. Y. Acad. Sci. 508, 333–348, 10.1111/j.1749-6632.1987.tb32915.x (1987).
Brown, T. R., Kincaid, B. M. & Ugurbil, K. NMR chemical shift imaging in three dimensions. Proc. Natl. Acad. Sci. USA 79, 3523–3526 (1982).
Maudsley, A. A., Hilal, S. K., Perman, W. H. & Simon, H. E. Spatially resolved high-resolution spectroscopy by 4-dimensional NMR. J. Magn. Reson. 51, 147–152, 10.1016/0022-2364(83)90113-0 (1983).
Posse, S., Otazo, R., Tsai, S. Y., Yoshimoto, A. E. & Lin, F. H. Single-shot magnetic resonance spectroscopic imaging with partial parallel imaging. Magn. Reson. Med. 61, 541–547, 10.1002/mrm.21855 (2009).
Adalsteinsson, E. et al. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn. Reson. Med. 39, 889–898, 10.1002/mrm.1910390606 (1998).
Posse, S., Tedeschi, G., Risinger, R., Ogg, R. & Lebihan, D. High-speed H-1 spectroscopic imaging in human brain by echo-planar spatial-spectral encoding. Magn. Reson. Med. 33, 34–40, 10.1002/mrm.1910330106 (1995).
Duyn, J. H. & Moonen, C. T. W. Fast proton spectroscopic imaging of human brain using multiple spin-echoes. Magn. Reson. Med. 30, 409–414, 10.1002/mrm.1910300403 (1993).
Otazo, R., Tsai, S. Y., Lin, F. H. & Posse, S. Accelerated short-TE 3D proton echo-planar spectroscopic Imaging using 2D-SENSE with a 32-channel array coil. Magn. Reson. Med. 58, 1107–1116, 10.1002/mrm.21426 (2007).
Lin, F. H. et al. Sensitivity-encoded (SENSE) proton echo-planar spectroscopic imaging (PEPSI) in the human brain. Magn. Reson. Med. 57, 249–257, 10.1002/mrm.21119 (2007).
Dydak, U., Weiger, M., Pruessmann, K. P., Meier, D. & Boesiger, P. Sensitivity-encoded spectroscopic imaging. Magn. Reson. Med. 46, 713–722, 10.1002/Mrm.1250 (2001).
Frahm, J. et al. Localized high-resolution proton NMR spectroscopy using stimulated echoes: initial applications to human brain in vivo. Magn. Reson. Med. 9, 79–93, 10.1002/mrm.1910090110 (1989).
Mescher, M., Tannus, A., Johnson, M. O. & Garwood, M. Solvent suppression using selective echo dephasing. J. Magn. Reson. Ser. A. 123, 226–229, 10.1006/jmra.1996.0242 (1996).
Singh, S. & Brody, W. R. Projection presaturation. III. Accurate selective excitation or presaturation of the regions of tailored shape in the presence of short-T1 species. J. Magn. Reson. Ser. B. 101, 52–62, 10.1006/jmrb.1993.1007 (1993).
Duijn, J. H., Matson, G. B., Maudsley, A. A. & Weiner, M. W. 3D phase encoding 1H spectroscopic imaging of human brain. Magn. Reson. Imag. 10, 315–319, 10.1016/0730-725x(92)90490-Q (1992).
Singh, S., Rutt, B. K. & Napel, S. Projection presaturation. II. Single-shot localization of multiple regions of interest. J. Magn. Reson. 90, 313–329, 10.1016/0022-2364(90)90137-X (1990).
Singh, S., Rutt, B. K. & Henkelman, R. M. Projection presaturation: a fast and accurate technique for multidimensional spatial localization. J. Magn. Reson. 87, 567–583, 10.1016/0022-2364(90)90313-X (1990).
Behar, K. L., Rothman, D. L., Spencer, D. D. & Petroff, O. A. C. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn. Reson. Med. 32, 294–302, 10.1002/mrm.1910320304 (1994).
Bydder, G. M. & Young, I. R. MR imaging: clinical use of the inversion recovery sequence. J. Comput. Assist. Tomogr. 9, 659–675, 10.1097/00004728-198507010-00002 (1985).
Haupt, C. I., Schuff, N., Weiner, M. W. & Maudsley, A. A. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn. Reson. Med. 35, 678–687, 10.1002/mrm.1910350509 (1996).
Lee, J., Shahram, M., Schwartzman, A. & Pauly, J. M. Complex data analysis in high-resolution SSFP fMRI. Magn. Reson. Med. 57, 905–917, 10.1002/mrm.21195 (2007).
Miller, K. L. et al. Functional brain Imaging using a blood oxygenation sensitive steady state. Magn. Reson. Med. 50, 675–683, 10.1002/mrm.10602 (2003).
Metzger, G. & Hu, X. P. Application of interlaced Fourier transform to echo-planar spectroscopic imaging. J. Magn. Reson. 125, 166–170, 10.1006/jmre.1997.1114 (1997).
Posse, S., DeCarli, C. & Lebihan, D. Three-dimensional echo-planar MR spectroscopic imaging at short echo times in the human brain. Radiology 192, 733–738 (1994).
Webb, P., Spielman, D. & Macovski, A. A fast spectroscopic imaging method using a blipped phase encode gradient. Magn. Reson. Med. 12, 306–315, 10.1002/mrm.1910120303 (1989).
Mansfield, P. Spatial mapping of the chemical shift in NMR. Magn. Reson. Med. 1, 370–386, 10.1002/mrm.1910010308 (1984).
Furuyama, J. K., Wilson, N. E. & Thomas, M. A. Spectroscopic imaging using concentrically circular echo-planar trajectories in vivo. Magn Reson Med. 67, 1515–1522, 10.1002/mrm.23184 (2012).
Lustig, M., Donoho, D. L., Santos, J. M. & Pauly, J. M. Compressed sensing MRI. IEEE Signal Processing Magazine 25, 72–82, 10.1109/Msp.2007.914728 (2008).
Lustig, M., Donoho, D. & Pauly, J. M. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 58, 1182–1195, 10.1002/Mrm.21391 (2007).
Griswold, M. A. et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn. Reson. Med. 47, 1202–1210 (2002).
Pruessmann, K. P., Weiger, M., Scheidegger, M. B. & Boesiger, P. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med. 42, 952–962 (1999).
Cao, P. & Wu, E. X. Accelerating phase-encoded proton MR spectroscopic imaging by compressed sensing. J. Magn. Reson. Imag. 41, 487–495, 10.1002/jmri.24553 (2015).
Drost, D. J., Riddle, W. R. & Clarke, G. D. Proton magnetic resonance spectroscopy in the brain: report of AAPM MR Task Group #9. Med. Phys. 29, 2177–2197, 10.1118/1.1501822 (2002).
Michaelis, T., Merboldt, K. D., Bruhn, H., Hänicke, W. & Frahm, J. Absolute concentrations of metabolites in the adult human brain in vivo: quantification of localized proton MR spectra. Radiology 187, 219–227 (1993).
Gyngell, M. L. et al. Noninvasive 1H NMR spectroscopy of the rat brain in vivo using a short echo time STEAM localization sequence. NMR Biomed. 4, 150–156, 10.1002/nbm.1940040307 (1991).
Thompson, R. B. & Allen, P. S. Response of metabolites with coupled spins to the STEAM sequence. Magn. Reson. Med. 45, 955–965, 10.1002/Mrm.1128.Abs (2001).
Thompson, R. B. & Allen, P. S. Sources of variability in the response of coupled spins to the PRESS sequence and their potential impact on metabolite quantification. Magn. Reson. Med. 41, 1162–1169, 10.1002/(Sici)1522-2594(199906)41:6<1162::Aid-Mrm12>3.0.Co;2-N (1999).
Posse, S., Otazo, R., Dager, S. R. & Alger, J. MR spectroscopic imaging: principles and recent advances. J. Magn. Reson. Imag. 37, 1301–1325, 10.1002/jmri.23945 (2013).
Zhang, Z. H. et al. Reduction of transmitter B-1 inhomogeneity with transmit SENSE slice-select pulses. Magn. Reson. Med. 57, 842–847, 10.1002/mrm.21221 (2007).
Mo, J., Callot, V., Huang, T.-Y., Poncelet, B. P. & Reese, T. G. In Proceedings of the 12th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, Kyoto, Japan, May 15–21, 2004, 538 (2004).
Pan, J. W., Lo, K. M. & Hetherington, H. P. Role of very high order and degree B0 shimming for spectroscopic imaging of the human brain at 7 Tesla. Magn. Reson. Med. 68, 1007–1017, 10.1002/Mrm.24122 (2012).
Haacke, E. M., Brown, R. W., Thompson, M. R. & Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design, 135 (Wiley-Liss, 1999).
Acknowledgements
This work was supported in part by Cancer Center Support Grant P30 CA-21765 from the National Cancer Institute at the National institutes of Health, Bethesda, MD and by ALSAC, Memphis, TN. The authors thank Dr. Robert Ogg for helpful discussions. The authors wish to acknowledge the valuable contributions of Angela Edwards, MR Technician, for her efforts in acquiring and processing of the CSI data. We acknowledge the editorial assistance of Sharon Naron and Dr. Keith Laycock. We also thank Dr. Samuel Brady for letting us borrow the phantom.
Author information
Authors and Affiliations
Contributions
Concept development, technical implementation and post-processing: J.G. All authors participated in the interpretation of the results and the preparation of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Guo, J., Patay, Z. & Reddick, W. Fast frequency-sweep spectroscopic imaging with an ultra-low flip angle. Sci Rep 6, 30066 (2016). https://doi.org/10.1038/srep30066
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30066
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.