Abstract
Gene regulatory networks involved in flowering time and photoperiodic responses in legumes remain unknown. Although the major maturity gene E1 has been successfully deciphered in soybean, knowledge on the functional conservation of this gene is limited to a certain extent to E1 homologs in legumes. The ectopic expression of Phvul.009G204600 (PvE1L), an E1 homolog from common bean, delayed the onset of flowering in soybean. By contrast, the ectopic expression of Medtr2g058520 (MtE1L) from Medicago truncatula did not affect the flowering of soybean. Characterization of the late-flowering mte1l mutant indicated that MtE1L promoted flowering in Medicago truncatula. Moreover, all transgenic E1, PvE1L and MtE1L soybean lines exhibited phenotypic changes in terms of plant height. Transgenic E1 or PvE1L plants were taller than the wild-type, whereas transgenic MtE1L plants produced dwarf phenotype with few nodes and short internode. Thus, functional conservation and diversification of E1 family genes from legumes in the regulation of flowering and plant growth may be associated with lineage specification and genomic duplication.
Similar content being viewed by others
Introduction
Soybean [Glycine max (L.) Merr], an important source of plant protein and oil, plays a vital role in ensuring food security. Soybean yield generally depends on the production of seeds to be used as crop. Transition from vegetative stage to reproductive stage is critical in seed production in spermophytes and flowering time is influenced by a series of environmental and endogenous factors. Many genes in the model plant Arabidopsis thaliana, participate in several flowering pathways1,2,3, e.g. photoperiod, gibberellin, ambient temperature, vernalization, aging and autonomous pathways. The photoperiod pathway controls flowering time in response to seasonal changes in day length. Light (day-length) in plants is perceived by photoreceptors in leaves and downstream signals are regulated by the circadian clock. Florigen is induced and transported into the shoot apex to trigger flowering by inducing the expression of floral meristem identity genes. Two photoperiod responsive genes, namely, CONSTANS (CO) and FLOWERING LOCUS T (FT), are vital in regulating photoperiodic flowering. CO encoding the B-box zinc finger protein4 integrates the circadian clock and photoperiod pathways5. By contrast, FT, which encodes a phosphatidylethanolamine-binding protein, is induced in leaves and transported into the meristem through the phloem6. The CO-FT module controls flowering across many plant species, such as rice7 and barley8.
Genetic analysis revealed 10 flowering and maturity loci (E1 to E9, J) in soybean9,10,11,12,13,14,15,16,17. E1, E3, E4 and E7 are strongly responsive to photoperiod10,11,12,18. E1–E4 were cloned using either candidate gene approach or positional cloning. E2 encodes GmGIa, which is an ortholog of GIGANTEA in Arabidopsis19. E3 and E4 are homologs of phytochrome A (PHYA) and encode the photoreceptors GmPHYA320 and GmPHYA221, respectively. The genome of soybean, an ancient tetraploid, contains genes with multiple copies. These copies may promote neo- or sub-functionalization, which facilitates the evolution of distinct functions in photoperiodic regulation22. Three GmFTs (i.e., GmFT2a, GmFT5a and GmFT4) regulate flowering in soybean23,24,25. GmFT2a corresponds to previously identified flowering locus E917. In addition, overexpression of several CO homologs, i.e., GmCOL1a, GmCOL1b, GmCOL2a and GmCOL2b26,27, influences flowering. Overexpression of GmCOL1a could also down-regulate E1 expression27. However, the precise role of soybean CO homologs in flowering must be verified through reverse genetic approaches or complementation test. Several FT-like genes have been identified in other legumes. Among these genes, FTb2 in pea (Pisum sativum)28 and MtFTa1 in Medicago (Medicago truncatula)29 regulate flowering. MtFTa1 is also a key regulator of flowering time in Medicago. FT genes possibly possess a conserved function in photoperiodic flowering in legumes. CO homologs have also been identified in legumes, such as common bean (Phaseolus vulgaris)30, pea31, Medicago32,33 and Lotus japonicus34. Most CO-like genes in legumes are related to Arabidopsis CO-like genes than to AtCO and these genes demonstrate an expression pattern distinct from that in Arabidopsis35. Although several CO homologs have been identified in Medicago, no solid evidence proves that CO homologs play a critical role in photoperiodic flowering in this species32,33. Therefore, the function of CO-like genes in regulating flowering in legumes remains unclear, whereas the main flowering time pathway in some leguminous species, e.g. pea, has been proven to be distinct from the CO-FT module in Arabidopsis.
Soybean is an important model plant used to elucidate photoperiodism in legumes36. E1 is a major gene associated with flowering time and maturity and is located at the pericentromeric region; E1 is intron-free and encodes a protein containing a putative bipartite nuclear localizing signal (NLS) and a domain distantly related to the plant-specific B3 domain (B3-like domain)37. Several members of the B3 superfamily directly or indirectly regulate flowering. AtRAV1 in Arabidopsis also influences flowering time38. TEMPRANILLO1 and TEMPRANILLO2 genes can negatively regulate FT expression and floral transition in Arabidopsis through direct binding of the B3 domain to a consensus bipartite sequence element in the FT 5′-untranslated region39. In addition, several genes containing the B3 domain, such as AUXIN RESPONSE FACTOR 140 and LEAFY COTYLEDON2, regulate plant growth and development41. The four major recessive alleles of E1 identified in soybean are e1-as (a single missense point mutation), e1-fs (a 1 bp deletion leading to frame-shift), e1-nl (~130 kb deletion comprising the E1 gene) and e1-b3a (three SNPs and 2 bp deletion in the middle of the B3 domain)42. e1-as is apparently a leaky allele and partially suppresses flowering in soybean37,42, contrary to the functional e1-fs and e1-nl. E1 is expressed in a bimodal pattern, with higher expression in long-day (LD) conditions than in short-day (SD) conditions. E1 is a putative transcription factor (TF) that negatively controls GmFT2a and GmFT5a to delay flowering under the background with functional PHYA genes (E3E4) and LD conditions. Zhai et al.42 proved that E1 can positively regulate GmFT4 to control flowering in soybean. Xu et al.43 revealed that, similar to E1, two E1 orthologs, namely, E1La (Glyma.04G156400.1) and E1Lb (Glyma.04G143300.1), can control the onset of flowering. That LjE1L controls flowering was proposed based on the analysis of the polymorphism of Lj5g3v2221340 (LjE1L, an ortholog of E1) among different wild Lotus japonicus species from various geographic locations in Japan44.
Fossil and molecular dating approaches indicated that legume originated ~60 million years ago (Mya)45,46 followed by speciation. The major legume species belong to two clades, namely, Hologalegina and Papilionoideae (Millettioid/Phaseoloid). Hologalegina, which includes Medicago, chickpea (Cicer arietinum) and L. japonicus, originated ~51 Mya, earlier than Millettoid/Phaseoloid (~45 Mya), which includes common bean, soybean and pigeon pea (Cajanus cajan)45. Moreover, Medicago and soybean underwent two rounds of whole genome duplication (WGD) events; the older WGD occurred at ~58 Mya before Glycine/Medicago split45 and the recent WGD in soybean occurred at ~13 Mya47. The duplicated genes underwent sub- or neo-functionalization after the WGD and some of these genes became pseudogenes22. Many highly syntenic regions were identified and characterized in Medicago, L. japonicus, common bean and soybean48,49,50. The soybean genome exhibits higher conservation with the common bean genome than those with Medicago and L. japonicus. Therefore, whether E1 homologs from legume species exert a function similar to or different from that of E1 as a flowering repressor in soybean remains unknown.
In this study, we analyzed the protein structure and phylogenetic relationships among E1 family genes identified in legumes. We also retrieved sequences of E1 homologs in common bean and Medicago and performed functional analysis by overexpressing the homologs in soybean, Arabidopsis and rice. Functional conservation and diversification among E1 family genes of leguminous species were consistent with the divergence time of genes and lineage species in this plant group.
Results
Gene and protein structures of the E1 family
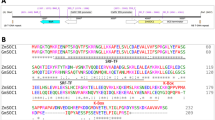
Eight gene products that are highly homologous [amino acid (aa) identity >60%] to E1 genes were retrieved from Phytozome, NCBI, or Legume Information System (LIS). The eight genes, namely, Phvul.009G204600 (PvE1L) from common bean, C. cajan_45915 (CcE1L1) and C. cajan_26468 (CcE1L2) from pigeon pea, Ca_21849 (CaE1L) from chickpea, Medtr2g058520 (MtE1L) from Medicago, LjE1L from L. japonicas and E1La and E1Lb from soybean, along with E1, are referred to as E1 family hereafter. Phylogenetic analysis of E1 family proteins in legume showed that all nine proteins divided into two main groups, namely Group I and Group II (Fig. 1a). The Group I consists of E1, E1La, E1Lb, PvE1L, CcE1L1 and CcE1L2, whereas the Group II consists of LjE1L, MtE1L and CaE1L. We followed the annotation of Xia et al.37, that is, E1 encodes a 174-aa protein, which was supported by the result of rapid amplification of cDNA ends (RACE). Although Glyma.06G207800 was annotated at the E1 locus in the version of Wm82.a2.v1 at Phytozome with 60 aa longer than E1, this finding has not been supported by experimental data. RACE analysis revealed that E1La and E1Lb encode 192-aa proteins43, which are 19 aa longer than that predicted by Xia et al.37. Both proteins shared high similarity (85%) to E1 at the aa level. Six other E1 homologs, namely, PvE1L, CcE1L1, CcE1L2, CaE1L, MtE1L and LjE1L, putatively encoded proteins containing aa residues ranging from 173 (PvE1L) to 184 (MtE1L) and sharing 62% (MtE1L) to 91% (PvE1L) aa identity with E1. All E1 family genes were predicted to contain no intron but a putative bipartite NLS at 34–55 aa residue in the consensus sequence; this bipartite NLS is composed of the KKRK and RRR basic domains at either ends separated by 14 aa residues. Moreover, all genes were predicted to contain a B3-like domain (Fig. 1b). The alignment showed that most residues were highly conserved (Fig. 1b). In addition, three-dimensional (3D) structures of all E1 family proteins were predicted to possess a structure similar to that of the DNA-binding protein RAV1 (SWISS-MODEL template library ID: 1wid.1.A). We predicted that all nine E1 family proteins contain the seven basic beta sheets and loops, along with 1–3 helices. However, the third beta sheet in CcE1L2 was predicted to separate into two beta sheets (Fig. 2). Moreover, six protein structures (E1, E1La, E1Lb, PvE1L, CcE1L1 and CaE1L) of the E1 family possessed only one helix of a sequence stretch “FVRRR”. By contrast, MtE1L or LjE1L contained an extra helix of “ESDL” or “ASDL” at the position of “DSDL” in most proteins. CcE1L2 also possessed extra two helices, namely, “DSDL” and “AYLVKKQI” (Figs 1b and 2).
Phylogenetic relationship and sequence alignment of E1 family genes.
(a) Phylogenetic tree of E1 family genes. The full length amino acid sequences of E1 family proteins were aligned using Clustal W and the phylogenetic tree was constructed using the Neighbor-joining method in MEGA 6.0 (Bootstrap = 1000). Two main groups, Group I and Group II, corresponding to genes from Millettioid/Phaseoloid and Hologalegina clade marked in red and blue. (b) Amino acid sequence alignment of nine E1 family genes from legumes. The B3-like domain is underlined in black. Putative bipartite nuclear localization signals (NLS) are shown in dotted black boxes. The residues sequences shown in black and red boxes form putative extra helices in MtE1L and LjE1L, respectively. The residues in the yellow box correspond to the conserved helix of nine E1 family proteins. All putative helices are shown as blue arrows.
Putative 3D protein structures of E1 family genes.
Putative 3D protein structures of nine E1 family genes from legumes. The structures of E1, E1La, E1Lb, PvE1L, CcE1L1 and CaE1L are displayed in the dotted red frame, whereas those of MtE1L and LjE1L are shown in the dotted blue frame with the extra helix shown in the green box. The structure of CcE1L2 is shown in the dotted purple frame with extra helices shown in the green box or blue circle. The separated position of one helix is shown in the black oval. The structures of the nine proteins are superimposed on the figure shown in the right-hand.
Expansion pattern and microsynteny of the E1 family genes
E1 family genes, except for CcE1L1 and CcE1L2, were annotated on six chromosomes in different genomes (Supplementary Fig. S1). E1La and E1Lb possibly originated from segmental duplication because both genes were annotated on chromosome 4 and located at approximately 10 Mb apart (Supplementary Fig. S1). To examine whether E1 family genes are involved in segmental duplication, we retrieved the syntenic blocks of individual E1 homologs from the Plant Genome Duplication Database (PGDD). Nonsynonymous (Ka) and synonymous (Ks) substitution rates were then used to estimate date of divergence. The Ka/Ks ratio of the segmental pairs in soybean varied from 0.1736 to 0.2089 (ratio < 1) (Table 1), indicating that E1, E1La and E1Lb underwent negative or purifying selection. The soybean genome underwent two WGD rounds, which occurred ~58 and ~13 Mya47. The divergence time between E1 and E1La/E1Lb can be traced back to 10.6 and 8.8 Mya, respectively (Table 1), which is possibly associated with the recent WGD in soybean. The divergence time between E1 and PvE1L was also estimated to have occurred 14.1 Mya. By contrast, the divergence time between E1 and MtE1L could be traced back to 62.9 Mya (Table 1). Among the E1 family members in non-soybean legumes, PvE1L displayed the closest genetic relationship with E1. In addition, estimated divergence times between E1 and CcE1L1/CcE1L2 are 28.3 and 28.1 Mya, respectively. Similarly, the divergence time between E1 and LjE1L/CaE1L are 60.8 and 88.5 Mya, respectively (Table 1). The evolution rates may not be inaccurate because of differences among species or even among the distinct locations of genes in a chromosome. Nevertheless, these predicted results revealed the sequential evolutionary relationship of these genes. The E1 family genes can be divided into two main groups, Group I and Group II, based on the estimated divergence times (Supplementary Table S1), corresponding to the Millettiod/Phaseoloid and Hologalegina branch, respectively51. More conserved syntenic blocks were also observed between the region of the E1 and that of E1La or E1Lb than that between the region of E1 and that of PvE1L, MtE1L, or CaE1L (Supplementary Fig. S2). These results are consistent with expected decline in conservation with taxonomic distance. We could not retrieve any E1 homolog in L. japonicus and C. cajan from the CoGe server for syntenic analysis.
Effects of E1 and PvE1L overexpression on flowering time and plant growth in soybean
Xia et al.37 reported that the expression of an additional copy of E1 driven by its native promoter in an early-flowering cultivar Kariyutaka (E1) can significantly delay flowering. In this study, we investigated the functional complementation of the E1 gene under recessive e1-as allele genetic background. We simultaneously transformed the PvE1L gene into the soybean to test whether PvE1L can control the onset of flowering. All transformations were performed using the soybean cultivar Dongnong 50 (WT-DN) carrying the e1-as allele.
Two independent E1 complementation lines (E1#L16 and E1#L18) and one PvE1L overexpression line (PvE1L#L2) were obtained, We painted the leaves with 160 mg L−1 glufosinate to test herbicide resistance and the resistant individuals were segregated from the non-resistant individuals in an approximately 3:1 ratio (Supplementary Table S2). Herbicide-resistant T1 and T2 plants of transgenic E1 and PvE1L lines exhibited late-flowering phenotype, Statistical analysis of flowering time data of herbicide-resistant T3 plants of transgenic lines revealed that two E1 transgenic lines and PvE1L#L2 flowered significantly later than the WT-DN plants in both LD (Figs 3a and 4a–c) and SD (Figs 3b and 4d–f) conditions. The WT-DN plants flowered at 35.3 days after emergence (DAE) under LD conditions and 24.2 DAE under SD conditions. In addition, E1#L16 and E1#L18, which are two E1 transgenic lines, flowered at 48.7 and 43.2 DAE under LD conditions and at 29.7 and 27.8 DAE under SD conditions. These results indicate that E1 overexpression delays the onset of flowering. The PvE1L transgenic line also flowered at a significantly later time (44.2 DAE and 33.2 DAE under LD and SD conditions, respectively) compared with the WT-DN plants (35.3 and 24.2 DAE under LD and SD conditions, respectively) (Figs 3c and 4g). PvE1L overexpression had no effect on the expression of native e1-as in PvE1L#L2 (Fig. 4h). Moreover, the expression levels of GmFT2a and GmFT5a were significantly reduced, whereas GmFT4 expression was significantly induced (Figs 3d,e and 4i,j) in transgenic lines when E1 (Fig. 3f) and PvE1L (Fig. 4k) were overexpressed under LD and SD conditions. These results are consistent with previous findings25,37. E1 complementation test results further proved that E1 is a flowering repressor in soybean. PvE1L also demonstrated a conserved physiological function as a flowering repressor. Interestingly, the PvE1L gene was annotated in a physical position, where two most significant QTLs (df9.1 and df9.2) for flowering time were mapped52 (Supplementary Fig. S1); Thus, PvE1L possibly suppresses flowering by regulating the expression of FT-like genes in common bean. In this study, we propose that PvE1L plays a key role in modulating flowering time in common bean, although this assumption requires more actual evidence confirmation.
Overexpression of E1 in soybean delayed flowering under LD and SD conditions.
Flowering phenotype of wild-type Dongnong 50 (WT-DN) and two E1 transgenic lines under LD (a) and SD (b) conditions. Annotation of cultivar or lines is indicated underneath. Close-up images that correspond to each pot show the axils of trifoliate leaves at approximately 35 and 24 DAE under LD and SD conditions, respectively. Two E1 transgenic lines (E1#L16 and E1#L18) did not display flowers or flower buds under LD conditions. (c) Days to flowering in WT-DN plants and E1 transgenic lines. Herbicide-resistant T3 plants of two transgenic lines were grown for flowering time investigation. Values represent the average of six replications +s.d. Similar results were observed in three separate experiments. Statistical significance was determined using Dunnett’s post hoc test after a significant one-way ANOVA. (d,e) Expression of GmFTs (GmFT2a, GmFT5a and GmFT4) in the third fully expanded trifoliate leaves of WT-DN and E1 transgenic lines under LD and SD conditions, respectively. (f) Expression of E1 in E1 transgenic lines. Relative expression levels were analyzed by quantitative RT-PCR and normalized to TUA5. Values represent the average of three biological replicates +s.d. *, ** and ***indicate significant differences between transgenic lines and WT-DN plants at P < 0.05, P < 0.01 and P < 0.001, respectively.
Ectopic overexpression of PvE1L in soybean delayed flowering under LD and SD conditions.
Flowering phenotype between WT-DN lines and the PvE1L (PvE1L#L2) transgenic issue under LD (a–c) and SD (d–f) conditions, respectively. (a,d) WT-DN plants exhibiting flowers and pods at the axils of trifoliate leaves at approximately 35 and 24 DAE under LD and SD conditions, respectively. Close-up images of (b) WT-DN plant with flowers and (c) one PvE1L transgenic plant with flower buds under LD contidions. Close-up images of (e) WT-DN plants with a pod (approximately 8 mm in length) and (f) PvE1L#L2 with flower buds at the axils of trifoliate leaves under SD conditions. (g) Days to flowering in WT-DN line and PvE1L#L2. Herbicide-resistant T3 plants of PvE1L#L2 were grown for flowering time investigation. Values represent the average of six replicates + s.d. P-value was determined using two-tailed Students’ t-test. Similar results were observed in three separate experiments. (h) Expression of native e1-as in WT-DN and PvE1L overexpression lines under LD and SD conditions. (i,j) Expression of GmFTs (GmFT2a, GmFT5a and GmFT4) in the third fully expanded trifoliate leaves of WT-DN and PvE1L transgenic lines under LD and SD conditions, respectively. (k) Expression of PvE1L in WT-DN and transgenic plants. Relative expression levels were analyzed by quantitative RT-PCR and normalized to TUA5. Values represent the average of three biological replicates +s.d. *, ** and ***indicate significant differences between transgenic lines and WT-DN plants at P < 0.05, P < 0.01 and P < 0.001, respectively.
In addition, overexpression of E1 and PvE1L in soybean also resulted in several morphological changes, e.g., plant height and number of nodes (Supplemental Figs S3a–d and S4a–d). WT-DN plants in LDs were 66.3–73.1 cm tall with 18.3–19.0 nodes, whereas the E1#L16, E1#L18 and PvE1L#L2 were significantly taller (27%, 24% and 15% increase, respectively) than WT-DN plants. This characteristic was due to the increase in the number of nodes (3.7, 3.3 and 2.4 nodes more, respectively) because internode length was not significantly different from WT-DN plant (Supplemental Figs S3c–e and S4c–e). Although all lines appeared shorter than under LD conditions, similar trends in plant height and node number between transgenic E1/PvE1L lines and WT-DN were observed under SD conditions (Supplemental Figs S3 and S4).
Functional characterization of MtE1L in Medicago and soybean
Two individual mutant lines, namely, NF16583 (NF1) and NF20110 (NF2), containing Tnt1 retrotransponson-tagged insertions occurring at different positions in MtE1L, were obtained from the M. truncatula Mutant Database (http://medicago-mutant.noble.org/mutant/). Four homozygous mutant plants, namely, NF1-1 and NF1-2 from NF1 line and NF2-2 and NF2-4 from NF2 line, were verified by PCR analysis (Supplementary Fig. S5). Sequencing confirmed the insertion positions to be at 218 and 259 bp in the open reading frame of NF1 and NF2, respectively (Fig. 5a). In this study, seedlings from the homozygous mutant lines, NF1-1 and NF2-2, were used to investigate flowering time. No MtE1L expression was detected in both homozygous mutants (Fig. 5b), suggesting that the expression or function of MtE1L in these mutant lines was interrupted by Tnt1 insertion. Moreover, the two homozygous mutant lines flowered at 42.0 (NF1-1) and 39.7 (NF2-2) DAE, later than that of WT-Mt (34.7 DAE) (Fig. 5c,d). Similar phenotype of flowering time between the two mutant lines and WT-Mt was observed in the second independent experiment. Thus, both mutant lines demonstrated late flowering phenotype. Two F2 populations of NF1-2-4 (♂) × R108 (♀) and NF2-4-4 (♂) × R108 (♀) were constructed to further confirm whether the mutation was the causal factor that led to the variation in flowering time. Fifteen plants of NF1-2-4 and thirteen plants from NF2-4-4 derived F2 plants were genotyped and phenotyped in the first trial. The corresponding segregation ratios of wild-type: heterozygous: homozygous are 4:7:4 and 3:6:4, both of which were approximately consistent with the 1:2:1 of Mendelian inheritance. The flowering time of homozygous mutants was significantly later than those of WT-Mt or heterozygous mutants in both F2 populations (P < 0.05, univariate ANOVA)(Supplementary Fig. S6). This phenomenon indicated that both mutations in MtE1L were significantly associated with flowering time.
Medicago mte1l mutants were late-flowering.
(a) Two independent Tnt1 mutations occurring at different positions in MtE1L. (b) Semi-quantitative RT-PCR analysis of MtE1L expression in wild-type cultivar R108 (WT-Mt) and two mutants lines under LD conditions. (c) Flowering phenotype of WT-Mt and two mte1l lines, NF1-1 and NF2-2, in LDs. Seedlings were vernalized for 10 days and then grown under LD conditions. Close-up of WT-Mt plant showing flowers at approximately 35 DAE. Two homozygous mutant lines did not exhibit flowers. (d) Number of days to flowering in WT-Mt and two homozygous mutant lines. Values represent the average of twelve replicates +s.d. Similar results were observed in two separate experiments. Statistical significance was determined using Dunnett’s post hoc test after a significant one-way ANOVA. ***indicates significant difference between mutant lines and WT-Mt plants at P < 0.001.
These results suggested that MtE1L may regulate flowering in Medicago. To investigate whether ectopic overexpression of MtE1L in soybean affects flowering time, we transformed MtE1L into Dongnong 50. Two soybean transgenic lines (MtE1L#L6 and MtE1L#L14) showed high MtE1L expression, whereas no MtE1L expression was detected in WT-DN lines (Fig. 6a). Meanwhile, progenies of both transgenic lines were segregated when tested for herbicide resistance. The resistant to non-resistant individuals were consistent with 3:1 of Mendelian inheritance (Supplementary Table S2). Two transgenic MtE1L soybean lines flowered at 36.3–37.2 and 24.0–24.3 DAE under LD and SD conditions not significantly different from that of the WT-DN plants (36.5 and 24.2 DAE under LD and SD conditions) (Fig. 6b–d). In addition, the expression levels of native e1-as, GmFT2a, GmFT5a and GmFT4 in transgenic plants were consistent with those in WT-DN plants both under LD and SD conditions (Fig. 6e–g). The significant difference of WT-DN plants in flowering time between LD or SD conditions revealed that the soybean cultivar Dongnong 50 was sensitive to photoperiod. Previous study showed that e1-as represents a partial functional allele that controls flowering in soybean42. Thus, the ectopic overexpression of MtE1L in soybean did not affect flowering in soybean. Taken mutant analysis and transgene in soybean together, we propose the MtE1L gene is a flowering promoter in Medicago, contrary to the E1 or PvE1L genes as flowering repressor.
Ectopic overexpression of MtE1L in soybean exerted no noticeable effect on flowering time under LD and SD conditions.
(a) Semi-quantitative RT-PCR analysis of MtE1L in two transgenic lines under LD and SD conditions. (b,c) Flowering phenotype of WT-DN and two MtE1L transgenic lines (MtE1L#L6 and MtE1L#14). All lines flowered at ~37 and 24 DAE under LD and SD conditions, respectively. (d) Days to flowering in WT-DN plants and MtE1L transgenic lines. Herbicide-resistant T3 plants of two transgenic lines were grown for flowering time investigation. Values represent the average of approximately six replicates +s.d. Similar results were observed in three separate experiments. Statistical significance was determined using Dunnett’s post hoc test after a significant one-way ANOVA. Expression of (e) native e1-as and (f,g) GmFTs (GmFT2a, GmFT5a and GmFT4) in the third fully-expanded trifoliate leaves of MtE1L transgenic lines and WT-DN plants under LD and SD conditions. Relative expression levels were analyzed by quantitative RT-PCR and normalized to TUA5. Values represent the average of three biological replicates +s.d.
The MtE1L transgenic lines exhibited a dwarf phenotype with fewer nodes and shorter internode length compared with that of WT-DN plants under LD or SD conditions (Supplemental Fig. S7a,b). Transgenic MtE1L lines in LD conditions were significantly shorter (25% to 32% decrease) than WT-DN plants (73.4 cm tall). This characteristic was contributed by a combination of 2.7–3.3 fewer nodes and 0.6–0.7cm shorter in internode length than WT-DN plants possessing 19.0 nodes with 4.0 cm long of internode length (Supplemental Figs S7c–3e). All lines under SD conditions were shorter than that under LD conditions, but similar variations in plant height, node number and internode length between MtE1L transgenic lines and WT-DN were observed (Supplemental Fig. S7).
Effect of ectopic overexpression of E1 on flowering in Arabidopsis and rice
Three Arabidopsis E1 overexpression lines (E1#L3, E1#L10 and E1#L11) and three vector-only transgenic lines (VC#L2, VC#L4 and VC#L7) were obtained (Supplementary Fig. S8a,b). All three E1 overexpression lines demonstrated high levels of E1 expression (Supplementary Fig. S8b). In addition, E1 overexpression lines flowered at 28.4–29.7 DAE, which was not significantly different from those of WT-At or VC lines under LD conditions (28.7–29.5 DAE) (Supplementary Fig. S8c). E1 overexpression lines, namely, WT-At and VC lines, also displayed similar number of rosette leaves (13.5–14.9 on average) when flowering (Supplementary Fig. S8d). The ectopic overexpression of E1 during flowering time apparently did not cause phenotypic changes in Arabidopsis. Similar results were observed in a separate experiment. Expression profiles of AtFT genes and AtCO were not significantly different between WT-At and all transgenic lines (Supplementary Fig. S8e).
Two rice E1 overexpression transgenic lines, namely, E1#L2 and E1#L4, were obtained (Supplementary Fig. S9a). All lines, including wild-type rice (WT-Os) and two E1 transgenic lines, required 52.1–52.8 days to flower under SD conditions (Supplementary Fig. S9b,c). Similar phenotypes of flowering time of all lines were observed in an independent experiment. Similar to that observed in Arabidopsis, the expression profiles of OsHd1 and OsHd3a, which are respective AtCO and AtFT orthologs in rice, were not significantly different between WT-Os and transgenic rice lines (Supplementary Fig. S9d).
Discussion
The legume family consists of crops that serve as primary sources of plant proteins for food and feed. Flowering is one of the major agronomic traits of legumes and has been the focus of fundamental and applied studies. Flowering regulation pathway in soybean is distinct from that in model plants, such as Arabidopsis and rice. Moreover, E1 is a key gene in the gene regulatory network of flowering in soybean. The product of this gene is a negative regulator of the photoperiod response pathway. However, the exact molecular function and evolutionary history of the E1 gene remains to be elucidated.
Most TF proteins consist of two domains, namely, a DNA-binding domain that binds to the promoter of target genes and a transcription activation (or suppression) domain that activates or suppresses transcription of target genes. E1 has been proven that E1 as a negative regulator of the flowering pathway37. However, the mechanism of action of the E1 family genes remains unclear. TFs are often associated with conserved functions across distantly related species. However, E1 family proteins appear to be only effective in closely related species. Given that E1 family genes are only recovered from legume species, the failure to detect any phenotype caused by overexpression of E1 in Arabidopsis and rice suggests that the exogenous E1 gene is independent of the regulation networks of photoperiodic flowering in Arabidopsis and rice. E1 function within legume might be, conserved between soybean and common bean, only diverging for ~14 Mya, which is a rather short distance based on evolutionary perspective. In this study, ectopic overexpression of PvE1L gene in soybean resulted in similar flowering and morphologic changes to transgenic E1 lines. The extent by which the PvE1L gene contributes in the control of flowering time in common bean needs further investigation. No flowering phenotype was observed, when MtE1L was overexpressed in soybean. This result suggests that the E1 network may differ between the two species that diverged for ~60 Mya. Alternatively, more soybean varieties should be tested to verify the result from this particular cultivar. Nevertheless, E1 could evidently perform distinct functions in the same family of plants. Therefore, E1 represents a TF that evolved rapidly.
In this study, we recovered a total of nine E1 family genes from six legume species. E1 is a single copy gene in four species and duplicated in soybean and pigeon pea, suggesting that E1 duplication is not a frequent event. Sequence divergence among the orthologs of E1 indicates that the initial E1 gene can be tracked back prior to the legume speciation. The two E1 family genes in pigeon pea have separated for ~3.9 Mya (Supplementary Table S1). Three E1 copies, namely, E1, E1La and E1Lb were found in soybean. These copies were derived from two duplication events. The first duplication event engendered E1 and the ancestor of E1La and E1Lb, which occurred between 8.8 Mya to 10.6 Mya. The second duplication event led to the generation of E1La and E1Lb, which occurred at ~3 Mya because of segmental duplication in chromosome 4. If neither E1 duplication events were due to genome wide duplication, the amplification of E1 gene has been suggested to present a selective advantage in soybean. The differentiation in expression patterns, in conjunction with the combination of functional, partially functional and non-functional copies of the E1 gene, may have enabled soybean to adapt in various geological locations.
These genes apparently perform different functions in flowering, although E1 family genes in legume seem to have the same origin. E1, E1La and E1Lb43 in soybean suppress flowering, whereas MtE1L in Medicago appears to promote flowering. The distinct function was probably due to the presence of an extra helix in the proteins (Fig. 2). Interestingly, the extra helix was present in MtE1L from M. truncatula, but not in E1. The association between E1 structure and flowering physiology might be an interesting topic for further studies. From a parsimony perspective, the evolution of different forms of TFs from the same ancestral protein is efficient for “turning on” or “turning off” the same set of target genes to fulfill the photoperiod requirement in different plants or in different seasons of the same plant. The two E1 copies in pigeon pea that only diverged for ~3.9 Mya support this notion. Interestingly, the predicted product from CcE1L2 contained the extra helix, whereas that from CcE1L1 was devoid of the extra helix. Moreover, Ka/Ks value between the gene pair was almost 1 (0.94, Supplementary Table S1), implying the lack of purifying selection between two genes or evolution of different functions. We infer that both copies may exert different functions considering the distinct structures between the two concurrent copies of E1 homologs in pigeon pea. Nevertheless, this result provides a good example to investigate the functional diversity in relation to structural variation.
The transition from vegetative to reproductive growth is a complex process that requires coordination among numerous genes, as well as the redistribution of resources between reproductive and vegetative organs. Consequently, late flowering is logically associated with increased plant height (see Supplementary Figs S3 and S4). Whether the reduced plant size accompanying early flowering is a result of reduced available resources for growth or an intentional inhibition of growth occurs to ensure flower development is a key question in the relationship of flowering development and growth. No flower phenotype was observed when MtE1L was overexpressed in soybean. This finding is unexpected because the relevant gene seemed to function in Medicago. More surprisingly, the dwarf phenotype was observed in the overexpression lines. This observation clearly indicates that the retarded growth was not a consequence of early flowering. Instead, this type of growth might be a part of the MtE1L regulation. The role and mechanism in flowering and plant development of MtE1L and other members of the E1 family merit further studies. Taken together, putative molecular networks of photoperiodic flowering regulation are proposed in legumes according to variations in photoperiodic response (Fig. 7). Networks of photoperiodic flowering regulation are conserved in short-day plants soybean and common bean, in which E1 or PvE1L is highly expressed and suppresses the expression of the florigen genes GmFT2a/5a or PvFTs under LD conditions. However, under SD conditions, the expression of both genes is suppressed and the florigens are upregulated. On the other hand, in long-day plant Medicago, the function of MtE1L is shifted to flowering promoting. However, in the proposed flowering networks in legume, some putative interactions need to be tested or confirmed in the future study, e.g. how PvE1L and MtE1L genes regulate florigen genes in their native species. Nevertheless, the functional diversification of E1 family genes revealed in this study and further characterization of flowering networks will provide insights in natural selection, domestication of important traits as well as genome evolution in legume.
Proposed flowering regulation networks in legumes.
Genes from soybean, common bean and Medicago are marked blue, red and green, respectively. Arrows represent stimulation of gene expression; T-shaped symbols represent inhibition of gene expression; Potential interactions are connected in dotted line. Symbol × represent the negation of promotion/inhibition. The deduced interactions were indicated by question marks.
Methods
Sequence collection, protein structure and phylogenetic analysis
The E1 protein sequence was used to search for E1 homologs at NCBI (http://www.ncbi.nlm.nih.gov/), Phytozome v10.3 (http://phytozome.jgi.doe.gov/pz/portal.html) and Legume Information System (LIS, http://www.legumeinfo.org/). We retrieved general information, such as position in genome, coding sequence (CDS) and protein sequence, of nine highly similar homologs. In silico 3D protein structures of nine E1 family proteins from G. max, P. vulgaris, C. cajan, C. arietinum, M. truncatula and L. japonicas were generated using SWISS-MODEL53 online modeling server and visualized with PyMOL.
The online NCBI conserved domain database (http://www.ncbi.nlm.nih.gov/cdd) was used to extract the protein sequences of the B3-like domain for phylogenetic analysis. Multiple sequence alignments were performed using the Clustal W program with default parameters in MEGA v6.0 with some manual editing. An unrooted neighbor-joining phylogenetic tree was constructed with a 1000 bootstrap repetition. Multiple sequences alignment of nine full-length E1 family protein sequences was generated using Clustal X v1.81 software with default parameters.
Microsynteny between chromosome segments encompassing E1 or its homologs from legumes
The syntenic blocks of each gene were searched against the PGDD to determine whether E1 or its homologs are located in the regions involved in segmental duplication. Ka/Ks and date of duplication occurrence were estimated using a previously reported method54. Pairwise alignments of CDS without the stop codon of duplicated genes were performed using the MUSCLE program in MEGA6.0 with default parameters. Phylogenetic tree was constructed using maximum likelihood method with Ka/Ks value that was calculated online by CODEML program built in Phylemon 2 (http://phylemon2.bioinfo.cipf.es/evolutionary.html). The estimation of the date of the segmental duplication event was performed using the formula, T = Ks/2λ, in which the mean synonymous substitution rate (λ) was set to 6.1 × 10−9 based on previous report for Fabaceae55.
The SynFind and GEvo tools at CoGe online server (https://genomevolution.org/CoGe/index.pl) were used for microsyntenic analysis. The segment (PAC.26299773), which represents a region that contained the E1 sequence, was blasted using BlastZ built in SynFind against the genomes of legumes, namely, soybean, common bean, pigeon pea, chickpea, Medicago and L. japonicus. Each segment containing E1 family gene along with 200 kb flanking sequences at both sides without masking was used to identify genome evolution patterns. These segments were also used to construct the syntenic relationship map at GEvo with default parameters.
Plant materials and growth conditions
Soybean cultivar Dongnong 50 was used to genetically transform E1 and its homologs. Soybean plants were grown in a phytotron under constant temperature of 25 °C and average light fluence rate of 200–300 μmol m−2s−1 under either LD (16 h light/8 h dark) or SD (12 h light/12 h dark) conditions. Furongdidou, a common bean cultivar, was grown under LD conditions and was used to clone the E1 homolog in common bean.
Medicago truncatula cultivar R108 and Tnt1 insertion lines were obtained from the Medicago mutant database (http://medicago-mutant.noble.org/mutant/). Seeds were scarified, germinated overnight at room temperature (25 °C) on wet filter papers and stored in Petri dishes at 4 °C in the dark for 10 days to allow seeds to germinate. Young seedlings were transplanted into the soil and grown in a phytotron under a constant temperature of 22 °C and average light fluence rate of 150 μmol m−2s−1 under LD (16 h light/8 h dark) conditions.
A. thaliana ecotype Columbia (Col-0) was used for genetic transformation and phenotypic analysis. Seeds were surface-sterilized with 70% ethanol and 10% sodium hypochlorite. Then, the seeds were rinsed several times with sterile distilled water and sown onto 1/2 MS medium plates for germination. Seedlings with 2–4 leaves were transferred into the soil and grown in a phytotron under constant temperature of 22 °C and average light fluence rate of 150 μmol m−2s−1 under LD (16 h light/8 h dark) conditions.
Oryza sativa L. ssp. Japonica cv. Longjing 11 was used for genetic transformation and phenotypic analysis. Seeds were harvested, dried at 37 °C and then germinated in sterilized water under SD (10 h light/14 h dark) conditions. The plants were transplanted into soil under constant temperature of 26 °C and average light fluence rate of 200–300 μmol m−2s−1 under SD conditions when the shoots reached a height of approximately 5–6 cm.
Genetic transformation in soybean, Arabidopsis and rice
A series of plant overexpression and expression vectors were constructed for genetic transformation. All vectors contained the bar gene for selection. According to the nature of the intron-free E1 family genes, E1, PvE1L and MtE1L were amplified from Harosoy (soybean), Furongdidou (common bean) and R108 (Medicago), respectively, using gene-specific primers to which Xba I and Sac I cloning sites were manually introduced. The fragments of PCR products and pTF101.1-GmFT2a vector23 containing a cauliflower mosaic virus 35S promoter (CaMV 35S) digested by Xba I/Sac I were assembled together to construct pTF101.1-E1, pTF101.1-PvE1L and pTF101.1-MtE1L. These vectors were transferred into Agrobacterium tumefaciens EHA101. Meanwhile, another plant expression vector pMDC123-E137, in which E1 was driven by the E1 native promoter, was transferred into A. tumefaciens EHA105 and subsequently used for rice genetic transformation. Agrobacterium-mediated cotyledon-node explant method was used for soybean genetic transformation56, whereas floral dip method was used for Arabidopsis genetic transformation57, Moreover, Agrobacterium-mediated co-cultivation method was used for rice genetic transformation.
Screening of transgenic plants
Transgenic soybean lines were screened by screening the T0, T1, T2 and T3 transgenic seedlings by daubing 160 mg L−1 glufosinate on unfolded preliminary leaves of seedlings. In addition, E1/PvE1L/MtE1L-specific primers (see Supplementary Table S3) were used for semi-quantitative RT-PCR and real-time RT-qPCR. Herbicide-resistant T3 seedlings were used for phenotypic and molecular analysis.
Seeds from transgenic Arabidopsis lines were surface-sterilized with 70% ethanol and 10% sodium hypochlorite. Then, the seeds were rinsed for 5–6 times with sterile distilled water and sown onto 1/2 MS plates containing 25 μg L−1 glufosinate for testing and germination. T2 lines with more than 95% herbicide-resistant seedlings (approximately 100 seedlings tested for each T2 line) were considered to be homozygous lines. T3 homozygous were confirmed by semi-quantitative RT-PCR with E1-specific primers and used for phenotype and expression analyses.
Seeds from transgenic rice lines were harvested and dried at 37 °C. Then, the seeds were germinated in water under SD (10 h light/14 h dark) conditions. The plants were transplanted into the soil under SD conditions when seedling shoots were about 5–6 cm height. T0, T1 and T2 transgenic lines were verified by semi-quantity RT-PCR with E1-specific primers (see Supplementary Table S3).
Characterization and verification of Medicago mutants
A pair of gene-specific primers (MtE1L-F/MtE1L-R) and a pair of MtE1L-Tnt1 combined primers (MtE1L-F/MtE1L-Tnt1-R) were used to identify Tnt1 insertions using PCR58. PCR products were purified and sequenced. The primers used in genetic screening can be found in Supplementary Table S3. We constructed two populations of NF1-2-4 (♂) × R108 (♀) and NF2-4-4 (♂) × R108 (♀) to further confirm that the mutation was the causal factor determining flowering time. The F2 populations were developed from seeds of self-pollinated F1 plants. Fifteen individuals of NF1-2-4 and thirteen individuals of NF2-4-4 derived F2 populations were used for genotyping and phenotyping. Data were statistically analyzed (univariate ANOVA) for correlation using the GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA).
Phenotypic observation of plants overexpressing E1, PvE1L and MtE1L
Six plants each from WT-DN and T3 herbicide resistant soybean plants were used to investigate flowering time. Six plants from each line were used for growth habit investigation under both LD and SD conditions. The flowering time observed or investigations were performed three times (approximately six plants per instance). Flowering time was recorded for days from emergence to the first open flower at any node on the main stem (R1). WT-At plants and T3 homozygous lines were used to investigate flowering time. Flowering time and total number of rosette leaves were recorded for each line when flowers emerged on the main stem. Approximately 40 plants for each line were recorded and two independent experiments were carried out. WT-Os and T2 overexpression lines in rice were used to investigate heading time. Heading date was recorded when a panicle was just coming out. Eight rice plants for each line were assessed and two independent experiments were carried out. Two-tailed Student’s t-test was used to compare single transgenic line with WT, whereas Dunnett’s post hoc test was used after a significant one-way ANOVA between multiple transgenic lines with WT using GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA).
RNA isolation and quantitative real-time PCR (RT-qPCR) analysis
Total RNA of leaves were extracted as mentioned previously. Reverse transcription was performed with 2 μg of RNA through TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China) according to the manufacturer’s instruction. RT-qPCR was performed on the LightCycle 96 system (Roche, Switzerland) using SYBR Green Master Mixture (TransGen, Beijing, China) according to the manufacturer’s instruction. Soybean TUA5, Arabidopsis AtActin2, or rice OsUbi5 was used as an internal control for normalization. PCR cycling conditions were set up as in the following program: 94 °C for 30 s, followed by 40 cycles of 94 °C for 5 s and 60 °C for 30 s with fluorescence signal collection. Melting and cooling steps were set as default parameters. Relative gene expression levels were calculated using the 2−ΔΔCt method. Three independent biological replicates were obtained and subjected to real-time PCR in triplicate. Raw data were standardized as described previously59. All primers for expression analysis are listed in Supplementary Table S3.
Additional Information
How to cite this article: Zhang, X. et al. Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 6, 29548; doi: 10.1038/srep29548 (2016).
References
Baurle, I. & Dean, C. The timing of developmental transitions in plants. Cell. 125, 655–664 (2006).
Boss, P. K., Bastow, R. M., Mylne, J. S. & Dean, C. Multiple pathways in the decision to flower: enabling, promoting and resetting., Plant Cell. 16 Suppl, S18–31 (2004).
Fornara, F., de Montaigu, A. & Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell . 141, 550, e551–552 (2010).
Mizoguchi, T. et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 17, 2255–2270 (2005).
Suarez-Lopez, P. et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature . 410, 1116–1120 (2001).
Corbesier, L. et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science . 316, 1030–1033 (2007).
Kojima, S. et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105 (2002).
Hemming, M. N., Peacock, W. J., Dennis, E. S. & Trevaskis, B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 147, 355–366 (2008).
Bernard, R. Two major genes for time of flowering and maturity in soybeans., Crop Sci. 11, 242–244 (1971).
Buzzell, R. Inheritance of a soybean flowering response to fluorescent-daylength conditions., Can J.Genet and Cytol . 13, 703–707 (1971).
Buzzell, R. & Voldeng, H. Inheritance of insensitivity to long daylength. Soybean Genet. Newsl . 7, 26–29 (1980).
McBlain, B., Bernard, R., Cremeens, C. & Korczak, J. A procedure to identify genes affecting maturity using soybean isoline testers. Crop Sci. 27, 1127–1132 (1987).
Ray, J. D., Hinson, K., Mankono, J. & Malo, M. F. Genetic control of a long-juvenile trait in soybean. Crop Sci. 35, 1001–1006 (1995).
Bonato, E. R. & Vello, N. A. E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet Mol Bio . 22, 229–232 (1999).
Cober, E. R. & Voldeng, H. D. A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701 (2001).
Cober, E. R., Molnar, S. J., Charette, M. & Voldeng, H. D. A new locus for early maturity in soybean. Crop Sci. 50, 524–527 (2010).
Kong, F. et al. A new dominant gene conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535 (2014).
Abe, J. et al. Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci. 43, 1300–1304 (2003).
Watanabe, S. et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics. 188, 395–407 (2011).
Watanabe, S. et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics . 182, 1251–1262 (2009).
Liu, B. et al. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics. 180, 995–1007 (2008).
Gupta, S., Nadarajan, N. & Gupta, D. S. Legumes in the omic era . (Springer, 2014).
Nan, H. Y. et al. GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. Plos One. 9(5), e97669 (2014).
Kong, F. J. et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231 (2010).
Zhai, H. et al. GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. Plos One. 9(2), e89030 (2014).
Wu, F. et al. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. Plos One. 9(1), e85754 (2014).
Cao, D. et al. GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. doi: 10.1093/pcp/pcv152 (2015).
Hecht, V. et al. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod., Plant Cell. 23, 147–161 (2011).
Laurie, R. E. et al. The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224 (2011).
Kwak, M., Velasco, D. & Gepts, P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 99, 283–291 (2008).
Hecht, V. et al. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144, 648–661 (2007).
Pierre, J. B., Bogard, M., Herrmann, D., Huyghe, C. & Julier, B. A CONSTANS-like gene candidate that could explain most of the genetic variation for flowering date in Medicago truncatula. Mol Breeding. 28, 25–35 (2011).
Wong, A. C. et al. Isolation and functional analysis of CONSTANS-like genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Frontiers in Plant Science . 5, 486 (2014).
Ono, N. et al. Genomewide characterization of the light-responsive and clock-controlled output pathways in Lotus japonicus with special emphasis of its uniqueness., Plant Cell Physiol. 51, 1800–1814 (2010).
Liew, L. C., Singh, M. B. & Bhalla, P. L. Unique and conserved features of floral evocation in legumes. J.Integ Plant Bio . 56, 714–728 (2014).
Owen, F. V. Inheritance studies in soybeans. II. Glabrousness, color of pubescence, time of maturity and linkage relations. Genetics. 12, 519–529 (1927).
Xia, Z. J. et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci USA. 109, E2155–E2164 (2012).
Hu, Y. X., Wang, Y. H., Liu, X. F. & Li, J. Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 14, 8–15 (2004).
Castillejo, C. & Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 18, 1338–1343 (2008).
Ulmasov, T., Hagen, G. & Guilfoyle, T. J. ARF1, a transcription factor that binds to auxin response elements. Science. 276, 1865–1868 (1997).
Stone, S. L. et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 98, 11806–11811 (2001).
Zhai, H. et al. Allelic variations at four major maturity E genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. Plos One. 9(5), e97636 (2014).
Xu, M. et al. The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168, 1735–1746 (2015).
Wakabayashi, T. et al. Polymorphisms of E1 and GIGANTEA in wild populations of Lotus japonicus. J Plant Res. 128, 221–221 (2015).
Lavin, M., Herendeen, P. S. & Wojciechowski, M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 54, 575–594 (2005).
Cronk, Q., Ojeda, I. & Pennington, R. T. Legume comparative genomics: progress in phylogenetics and phylogenomics. Curr Opin Plant Biol. 9, 99–103 (2006).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature. 463, 178–183, (2010).
Choi, H.-K. et al. Estimating genome conservation between crop and model legume species., Proc Natl Acad Sci USA. 101, 15289–15294 (2004).
Hougaard, B. K. et al. Legume anchor markers link syntenic regions between Phaseolus vulgaris, Lotus japonicus, Medicago truncatula and Arachis. Genetics . 179, 2299–2312 (2008).
Tsubokura, Y. et al. Characterization of soybean genome based on synteny analysis with Lotus japonicus. Breeding Sci. 58, 157–167 (2008).
Doyle, J. J. & Luckow, M. A. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131, 900–910 (2003).
Blair, M. W., Iriarte, G. & Beebe, S. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theor Appl Genet. 112, 1149–1163 (2006).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. doi: 10.1093/nar/gku340 (2014).
Zhu, Y. et al. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 14, 93 (2014).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000).
Olhoft, P. & Somers, D. L-Cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledonary-node cells. Plant Cell Rep. 20, 706–711 (2001).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Tadege, M. et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347 (2008).
Willems, E., Leyns, L. & Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem. 379, 127–129 (2008).
Acknowledgements
We thank Dr. Kan Wang (Iowa State University of Science and Technology, USA) for providing the soybean transformation vector pTF101.1 and the Agrobacterium strain EHA101. We also thank Professor Jiang N (Michigan State University, USA), Professor Harada K (National Institute of Agrobiological Sciences, Japan) and Professor Cober ER (Agric. and Agri-Food Canada, Eastern Cereal and Oilseed Research Centre) for critical comments. This work was supported by National Key Research and Development Project (grant no. 2016YFD0101902) from the Ministry of Science and Technology; by Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA0801010503); and by Programs (31471518, 31271742 and 31301338) from National Natural Science of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
X.Z. performed most experiments on Arabidopsis and soybean genetic transformation and verification of Medicago mutants, as well as worked on phenotype and expression assays. H.Z., Y.W., Y.Z., H.W., S.L., G.Y., Y.L., L.W., B.H. and C.L. assisted to perform experiments. X.T. worked on rice transformation. Z.X., H.Z. and Q.B. conceived this research. X.Z. and Z.X. wrote this paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, X., Zhai, H., Wang, Y. et al. Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci Rep 6, 29548 (2016). https://doi.org/10.1038/srep29548
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29548
This article is cited by
-
Association mapping in bambara groundnut [Vigna subterranea (L.) Verdc.] reveals loci associated with agro-morphological traits
BMC Genomics (2023)
-
Transcriptome Profile of a Long-Juvenile Soybean Genotype Huaxia-3 Under Short and Long Photoperiod
Plant Molecular Biology Reporter (2022)
-
Identification, genomic organization, and expression profiles of single C2H2 zinc finger transcription factors in tomato (Solanum lycopersicum)
Journal of Applied Genetics (2021)
-
Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days
BMC Plant Biology (2020)
-
Winged bean (Psophocarpus tetragonolobus (L.) DC.) for food and nutritional security: synthesis of past research and future direction
Planta (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.