Abstract
The Sinorhizobium meliloti (S. meliloti) strain CCNWSX0020 displayed tolerance to high levels exposures of multiple metals and growth promotion of legume plants grown in metal-contaminated soil. However, the mechanism of metal-resistant strain remains unknown. We used five P1B-ATPases deletions by designating as ∆copA1b, ∆fixI1, ∆copA3, ∆zntA and ∆nia, respectively to investigate the role of P1B-ATPases in heavy metal resistance of S. meliloti. The ∆copA1b and ∆zntA mutants were sensitive to zinc (Zn), cadmium (Cd) and lead (Pb) in different degree, whereas the other mutants had no significant influence on the metal resistance. Moreover, the expression of zntA was induced by Zn, Cd and Pb whereas copA1b was induced by copper (Cu) and silver (Ag). This two deletions could led to the increased intracellular concentrations of Zn, Pb and Cd, but not of Cu. Complementation of ∆copA1b and ∆zntA mutants showed a restoration of tolerance to Zn, Cd and Pb to a certain extent. Taken together, the results suggest an important role of copA1b and zntA in Zn homeostasis and Cd and Pb detoxification in S. meliloti CCNWSX0020.
Similar content being viewed by others
Introduction
Heavy and transition metal homeostasis is crucial in all biological systems. Transition metals such as zinc (Zn), copper (Cu) and iron (Fe) are essential micronutrient that are required for many physiological processes but are also extremely toxic in excess. Other metals, such as cadmium (Cd), silver (Ag) and lead (Pb), are acutely toxic and represent a major threat for cell survival1,2. Organisms have evolved to contain multiple defense mechanisms to prevent overaccumulation of heavy and transition metals, such as efflux transport, intracellular sequestration, precipitation, bioadsorption and transformation, etc.3,4,5,6.
The P1B-type ATPase subfamily belongs to the P-type ATPases family and couples ATP hydrolysis to transition metal transport across cellular membranes. The P1B-type ATPases are the most widely distributed group and have the largest substrate range7. P1B-ATPases display several structural characteristics that include six to eight transmembrane helices (TMs)8, the signature sequence (CPC, CPH, SPC, PCP) present in the sixth TM (TM6), one or more metal binding domains in the cytoplasmic N-terminal or C-terminal region (N-MBD, C-MBD) and the catalytic phosphorylation site (DKTGT) between TM6 and TM79. The P1B-ATPases play an essential role in transition metal homeostasis. P1B-ATPases have been divided into five subclasses designated P1B-1 to P1B-5 according to a combination of substrate specificity, sequence similarity and conserved metal binding residues present in transmembrane segments9,10. The metal specificity of the five ATPase subfamilies have been extensively studied and it has been shown that the P1B-1-ATPases transport Cu+ and Ag+ 11,12, the P1B-2-ATPases transport Zn2+, Cd2+ and Pb2+ 13,14, the P1B-3-ATPases are suggested to transport Cu2+ 15 and the P1B-4-ATPases transport Co2+, Ni2+ and/or Zn2+ 16. As to P1B-5 ATPase, only the S. meliloti Sma1163 gene encoding for a P1B-5-ATPase that denoted Nia was biochemically characterized indicating Ni2+ and Fe2+ are substrates of Nia17. Moreover, two new subtype of P1B-ATPases (P1B-6, P1B-7) have been recognized recently, but none of these P1B-6 and P1B-7 ATPases were biochemically examined beyond their sequence classification18.
Previous studies on S. meliloti 2011 P1B-1-ATPases showed that five homologous Cu+-ATPases exhibited functional diversity19. These five homologous Cu+-ATPases are divided into three major subgroups, including CopA1-like transporter (CopA1a and CopA1b), CopA2-like ATPase (FixI1 and FixI2) and CopA3-like ATPase (CopA3). CopA1a is a typical Cu+-ATPase catalyzing cytoplasmic Cu+ efflux to prevent overaccumulation of cytoplasmic copper. A mutation of copA1a resulted in a copper sensitive phenotype and an increase in cytoplasmic copper levels. CopA1b displayed 80% homology to CopA1a. However, a mutant of copA1b changed neither copper tolerance nor cytoplasmic copper accumulation. Rather, it changed the differentiation of the mutant strain into bacteroids, the number of viable bacteria (undifferentiated bacteria) in the copA1b mutant strain-induced nodules was increasing faster than differentiated bacteroids. The CopA2-like (FixI) ATPase in S. meliloti 2011, is encoded in an operon together with genes encoding cytochrome oxidase subunits. It has been proposed that FixI1 ATPase may be involved in respiration under microaerobiosis during symbiosis while FixI2 ATPase is required for respiration during all steps of bacterial life. CopA3 looks like a novel Cu+-ATPase. Mutation of copA3 gene did not lead to sensitivity to Cu+ or cytoplasmic copper accumulation in the mutant strain and the gene was regulated by redox stress and was required during symbiosis. The P1B-5 ATPase has not been well characterized. It was shown that S. meliloti 2011 Nia was induced by Fe2+ and Ni2+ and a nia mutant accumulated nickel and iron, suggesting a possible role in Fe2+ and Ni2+ detoxification17.
P1B-2-ATPases (Zn2+/Cd2+/Pb2+ transporter) are less studied in rhizobia but have been well characterized in other bacteria such as ZntA in Escherichia coli and CadA in Staphylococcus aureus. ZntA is not only for Zn2+ efflux but also transports the non-physiological substrates Cd2+ and Pb2+ 13. The expression of the zntA gene is activated via the Zn2+-responsive transcriptional regulator (ZntR)20. CadA is known to encode a Cd2+ efflux ATPase which plays a role in the cadmium resistance of S. aureus21. In S. meliloti 1021 the SMc04128 gene encodes a P1B-2-type ATPase. A transposon insertion mutant of SMc04128 showed sensitivity to high concentrations of Zn2+ and Cd2+ and slightly increased sensitivities to Cu2+, Pb2+, Ni2+ and Co2+, which indicated that SMc04128 plays a role in the defense of S. meliloti 1021 against these heavy metals22. Moreover, it has been reported that a gene named cadA in Mesorhizobium metallidurans isolated from a zinc-rich mining soil also encodes a PIB-2-type ATPase involved in cadmium and zinc resistance. The cadA gene was induced by zinc and cadmium and a cadA-deleted strain failed to grow at high zinc concentrations23.
Sinorhizobium meliloti CCNWSX0020 was isolated from Medicago lupulina growing in gold mine tailings in the northwest of China and exhibited higher tolerance towards multiple metals, such as Cu, Zn, Cd and Pb. The heavy metal transporting P1B-type ATPases is vital for heavy metal resistance. However, the role of P1B-type ATPases in S. meliloti CCNWSX00200 remains unknown. There are five genes encoding putative P1B-type ATPases on the S. meliloti CCNWSX0020 genome24. Their predicted signature transmembrane metal binding residues indicated that three genes encoded P1B-1-ATPase (SM0020_05727, SM0020_05912 and SM0020_11415), one (SM0020_22747) encoded a P1B-2-ATPase, another (SM0020_05862) encoded a putative P1B-5-ATPase. These five P1B-ATPases are predicted to be mainly responsible for heavy metals homeostasis and detoxification in S. meliloti CCNWSX00200. To further investigate the function of these five P1B-ATPases we created deletions in these genes and tested different metals tolerance of these deletions. Both deletion of SM0020_11415 (Cu+-ATPase) and deletion of SM0020_22747 (Zn2+-ATPase) displayed sensitivity to Zn2+, Cd2+ and Pb2+. To test whether SM0020_11415 and SM0020_22747 in S. meliloti CCNWSX0020 have similar functions, we investigated these genes expressions in response to different levels of heavy metals exposure and their capability to complement the ∆copA and ∆zntA E. coli mutant strains. The combined results of these studies suggest that SM0020_22747 encodes a classical Zn2+-ATPase which is required for efflux of Zn2+, Cd2+ and Pb2+, whereas SM0020_11415 encoding a Cu+-ATPase surprisingly confers tolerance to Zn, Cd and Pb but not to Cu in S. meliloti CCNWSX00200.
Results
Deletion of P1B-type ATPases made mutant strains more sensitive to a number of heavy metals
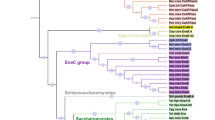
Bioinformatics studies have shown that the genomes of many bacteria including S. meliloti contain a diverse array of genes encoding a number of P1B-ATPases. P1B-ATPases have been associated with the detoxification and tolerance mechanism of heavy and transition metals25. S. meliloti CCNWSX0020, could tolerate up to 1.4 mM CuSO4, 1.0 mM ZnSO4, 3.2 mM Pb(NO3)2, 0.25 mM CdSO4 and 1.0 mM NiSO4 in TY solid medium. A phylogenetic analysis of five predicted P1B-type ATPases from S. meliloti CCNWSX0020 indicated three of them were Cu+-ATPases, one was Zn2+-ATPase and the last belongs to P1B-5-type ATPase (Fig. 1). Furthermore, given the results of sequence homology analysis, the three Cu+-ATPases genes (SM0020_05727, SM0020_05912 and SM0020_11415) displayed the highest similarity with the copA3, fixI1 and copA1b genes which were previously identified in S. meliloti 201119. The SM0020_22747 gene was 99.1% identical to the zntA gene of S. meliloti 1021 while SM0020_05862 showed 98% similarity to the nia gene that encoded a nickel (Ni) and Fe transporter in S. meliloti 201117,22. We therefore selected all five P1B-ATPases that involved in heavy metals resistance of S. meliloti CCNWSX0020. To test our hypotheses, the five P1B-ATPase deletions (∆copA1b, ∆copA3, ∆fixI1, ∆zntA and ∆nia) were characterized using metal-tolerance growth assays in TY liquid medium. The wild type strain and five deletion mutants were cultured in TY supplemented with increasing concentration of CuSO4, ZnSO4, CdSO4, Pb(NO3)2 and NiCl2 (Fig. 2). The ∆zntA mutant showed the greatest sensitivities to 0.2 mM Zn2+ and 0.05 mM Cd2+, while ∆copA1b mutant was slightly more sensitive to high concentration of Zn2+ (0.6 mM) and Cd2+ (0.15 mM) (Fig. 2A,B). Both ∆zntA and ∆copA1b mutants showed sensitivity to high concentration of Pb2+ in different degrees and had no effect on Cu and Ni tolerance (Fig. 2C–E). The CuSO4, ZnSO4, CdSO4, Pb(NO3)2 and NiCl2 metals tolerance of the other three mutants (∆copA3, ∆fixI1 and ∆nia) displayed no difference to the wild type strain (Fig. 2). These results suggested CopA1b and ZntA were involved in Zn, Cd and Pb metals resistance in S. meliloti CCNWSX0020. To further explore the function of CopA1b and ZntA, these two zinc sensitive deletions were further studied.
Phylogenetic analysis of P1B-type ATPases.
Branches indicating proteins in subgroups IB-1, IB-2, IB-3, IB-4 and IB-5 are under different colors. Five P1B-type ATPases genes (SM0020_05727/copA3, SM0020_05862/nia, SM0020_05912/fixI1, SM0020_11415/copA1b, and SM0020_22747/zntA) in Sinorhizobium meliloti CCNWSX0020 are tagged in the unrooted tree.
Influence of deletions in genes encoding different P1B-type ATPases on metal tolerance of Sinorhizobium meliloti CCNWSX0020.
Wild type and mutant strains were grown in TY liquid medium for 48 h in the presence of increasing concentrations of zinc (A), cadmium (B), lead (C), copper (D) and nickel (E). Symbols represent the wild type strain(WT) and mutants ΔcopA3, Δnia, Δfixl1, ΔcopA1b and ΔzntA of S. meliloti CCNWSX0020 (♦,■, ▲, ×, *, ●, respectively). Error bars represent standard deviations of three biological repeats.
CopA1b and ZntA could play a role in zinc, cadmium and lead homeostasis
The ∆copA1b and ∆zntA mutants of S. meliloti CNWSX0020 exhibited sensitivity to Zn2+, Cd2+ and Pb2+ but not to other metals, suggesting CopA1b and ZntA may play a role in Zn, Cd and Pb homeostasis in this strain. To verify the presence of the respective copA1b and zntA genes or either gene alone that was responsible for Zn, Cd and Pb resistance, the two genes were amplified and inserted into the pBBR1MCS-5 vector and transformed into the corresponding mutant and then tested for these metals tolerance. Figure 3 shows the complemented strains (C-copA1b and C-zntA) could restore Zn, Cd and Pb resistance of the mutants by 80%. These results demonstrated that the sensitivity of three metals to the mutants was due to the deletion of copA1b and zntA in S. meliloti CCNWSX0020. Thus we can speculate that copA1b and zntA genes are involved in Zn homeostasis and Cd or Pb detoxification. Surprisingly zntA encodes a Zn2+/Cd2+/Pb2+ transporter whereas copA1b is predicted to encode a Cu+/Ag+ transporter. A mutant containing a zntA deletion did not grow in the presence of low Zn levels as expected. In contrast, the growth of a mutant containing a copA1b deletion was not inhibited by high Cu levels but rather by high concentrations of Zn, Cd and Pb. To better understand the differences between the two genes in responsive to metals exposure, the capabilities of S. meliloti CopA1b and ZntA to complement the E. coli ∆copA and ∆zntA strains were tested. As expected, CopA1b could complement the Cu sensitive phenotype of E. coli ∆copA strain while ZntA could complement the Zn sensitive phenotype of E. coli ∆zntA strain. Meanwhile, CopA1b could restore Zn tolerance of an E. coli ∆zntA strain to some degree whereas ZntA failed to restore Cu tolerance of an E. coli ∆copA strain (Fig. 4). These results suggested that CopA1b and ZntA conferred resistance to Zn, Cd and Pb tolerance and CopA1b also had a capacity for Cu tolerance.
Complementation of ΔcopA E. coli copper sensitive phenotype and ΔzntA E. coli zinc sensitive phenotype by heterologously expressed S. meliloti Cu+-ATPase (C-copA1b) and Zn2+ −ATPase (C-zntA).
All strains were grown to exponential phase in LB liquid medium. Five 10-fold dilutions were carried out and spotted on the LB agar plates from left to right, in the presence of the indicated concentrations of CuSO4 and ZnSO4.
Expression of copA1b and zntA could be induced by different types of heavy metals
To further investigate CopA1b and ZntA in S. meliloti CCNWSX0020, the gene expressions of zntA and copA1b under different metal stresses were examined using qRT- PCR. The expression profiles of copA1b and zntA genes in wild type, ∆copA1b and ∆zntA mutants in response to Cu, Ag, Zn, Cd and Pb showed some unexpected results. The expression of copA1b was strongly up-regulated by Ag (234-fold) and Cu (37-fold) exposure respectively (Fig. 5A), while the expression of zntA was significantly induced by Zn (254-fold), Cd (330-fold) and Pb (231-fold) respectively (Fig. 5B). In addition, a deletion of copA1b significantly inhibited zntA gene expression by nearly 50% (P < 0.01) compared to the wild type strain in response to the above three metals (Fig. 5B). These observations implied that ZntA was a typical Zn2+/Cd2+/Pb2+ transporter, whereas CopA1b contribute to Zn, Cd and Pb tolerance as a classical Cu+-ATPase but not contribute to Cu tolerance of S. meliloti CCNWSX0020.
Gene expression analysis.
Expression of copA1b (SM0020_11415) and zntA (SM0020_22747) under copper, silver, zinc, cadmium and lead stress (A,B). (C) Expression of genes in the vicinity of copA1b (SM0020-11415) under copper, silver and zinc stress. Wild type, ∆copA1b and ∆zntA mutants of S. meliloti CCNWSX0020 strains at OD600 of 1.0 were incubated with 0.6 mM CuSO4, 0.05 mM AgNO3, 0.4 mM ZnSO4, 0.1 mM CdSO4 and 1.5 mM Pb(NO3)2 for 30 min. Samples were then processed for qPCR analysis and normalized against the ribosomal 16 S rRNA. Error bars represent standard deviations of three biological repeats. **P < 0.01.
Since ∆copA1b strain had an unexpected phenotype further studies of the genetic region in the vicinity of copA1b were performed. SM0020_11410 gene locus was located directly upstream of copA1b (SM0020_11415) and predicted to be in an operon with copA1b. SM0020_11410 encodes a transcriptional regulator which is highly homologous with CueR belonging to the MerR family26. CueR is copper sensor and induces the expression of the Cu+-translocating P-type ATPase CopA in response to Cu+, Ag+ or Au+ 27,28. The copA1b promoter contains a 7-7-7 bp inverted repeat (ACCTTCC-CATTATTT-GGAAGGA) between −35 and −10 similar to the E. coli copA promoter. This predicted gene SM0020_11410 product might bind to the inverted repeat sequence and activate copA1b gene expression in the presence of Cu and Ag as a homolog of CueR. Figure 5C showed that SM0020_11410 displayed a 15-, 130-, 4-fold induction by Cu+, Ag+ and Zn2+, respectively. A deletion of SM0020_11410 led to a slight sensitivity to Cu and suppressed the SM0020_11415 gene expression (data not shown), suggesting that SM0020_11410 really regulated the expression of copA1b (SM0020_11415).
There are some other genes in the vicinity of copA1b (SM0020_11415) that may also be part of this operon. SM0020_11405 and SM0020_11425 both encode unknown proteins, SM0020_11420 encodes a potential periplasmic copper chaperone and SM0020_11430 encodes an ArsR family transcriptional regulator. These genes in the vicinity of copA1b might work together against heavy metal stress. The expression of the SM0020_11405 (unknown protein) was inducible by Ag+ and Cu+, whereas SM0020_11430 (ArsR family) was inducible by Cu+ and Zn2+. In addition, the gene expression level of SM0020_11420 (copper chaperone) and SM0020_11425 (unknown protein) was very low under Cu, Ag and Zn stress (Fig. 5C).
Deletions of copA1b and zntA led to increased intracellular concentrations of zinc, lead and cadmium but not copper
A possible explanation for the sensitivity of ∆copA1b and ∆zntA mutants toward Zn, Cd and Pb is that higher levels of these metal ions were accumulated in the mutant cells. This would imply that CopA1b and ZntA play roles in expelling these surplus metals from cytoplasm. To test this hypothesis, total internal metal content of wild type and mutant cells were measured by furnace atomic absorption spectroscopy (AAS). The cells were grown in TY medium individually supplemented with 0.4 mM ZnSO4, 2.0 mM Pb(NO3)2, 0.05 mM CdSO4 and 0.8 mM CuSO4. The ∆zntA mutant which was hypersensitive to Zn and Cd accrued significantly (P < 0.01) higher amounts of intracellular Zn (~3 fold), Cd (~8 fold) and Pb (~2.5 fold) compared with the WT (Fig. 6). The ∆copA1b mutant had a smaller but significant effect on the metal content since the loss of copA1b led to an increased accumulation of Zn by 1 fold, Cd by 1.5 fold (P < 0.05) and Pb by 2 fold (P < 0.01) relative to the WT (Fig. 6). The increase in the accumulation of Zn, Cd and Pb in the ∆copA1b and ∆zntA mutants suggested that CopA1b and ZntA play a role in the efflux of Zn2+, Cd2+ and Pb2+ ions. In addition, the accumulation of intracellular Cu was quite high in the ∆copA1b and ∆zntA mutants as well as in the WT strain (Fig. 6). This result demonstrated that CopA1b was not involved in Cu export and S. meliloti CCNWSX0020 accumulates high levels of intracellular Cu.
Analysis of intracellular metal concentrations in the wild type and two zinc sensitive mutants of S. meliloti CCNWSX0020.
The wild type strain, ∆copA1b and ∆zntA mutant strains were cultured in the presence of 0.4 mM ZnSO4, 2.0 mM Pb(NO3)2, 0.05 mM CdSO4 and 0.8 mM CuSO4. Cells were harvested and washed with metal-free buffer, then were dried at 65 °C and total internal metal concentrations were measured by atomic absorption spectrophotometer. Error bars represent standard deviations of three biological repeats. *P < 0.05, ***P < 0.001.
Deletions of copA1b and zntA decreased antioxidant enzyme activity
P1B-ATPases also have a function in providing metals for assembly of periplasmic metalloproteins since some heavy and transition metals are essential component of many free-radical detoxifying enzymes, like catalase, peroxidase and superoxide dismutase29,30. So the capability of ∆copA1b and ∆zntA to cope with redox stress was tested. Although the H2O2 resistance on the agar plates of ∆copA1b and ∆zntA mutants showed no big change with S. meliloti wild type strain (data not shown), the CAT, POD and total SOD activities were dramatically changed. When all three strains were treated with H2O2, the CAT and POD activities increased obviously while the total SOD activity displayed no change (Fig. 7). However, compared to S. meliloti wild type strain, the CAT and POD activity levels of ∆copA1b and ∆zntA mutants showed apparent decline to a different degree under H2O2 treatment condition whereas the enzyme activities showed no major differences under H2O2-free condition (Fig. 7A,B). The total SOD activity of ∆copA1b and ∆zntA mutants also decreased slightly in comparison to a wild type strain whether under H2O2 treatment or not (Fig. 7C). These results suggest that deletions of CopA1b and ZntA have an influence on the activity levels of these antioxidant enzymes.
The effect of deletions of copA1b and zntA on the antioxidant activities of CAT (A), POD (B) and SOD (C) under H2O2 stress. The wild type strain, ∆copA1b and ∆zntA mutant strains were cultured to exponential phase and then treated or untreated with 500 μM H2O2 for 30 min. Cells were harvested and crude bacterial lysates were subsequently prepared for spectrophotometric analyses. Error bars represent standard deviations of three biological repeats. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Sinorhizobium meliloti CCNWSX0020 is a multiple heavy metals resistant bacterium isolated from root nodules of M. lupulina growing on mine tailings in the northwest of China31. The genome of S. meliloti CCNWSX0020 has been sequenced and some copper resistance genes have been analyzed in the previous studies24,32,33. It has been reported that merR encoding an MerR family transcriptional regulator displayed significantly decreased copper resistance in a merR-interrupted mutant of S. meliloti CCNWSX0020 and the expression of two genes (SM0020_05727 and SM0020_05862) encoding putative P1B-type ATPases were found to be down-regulated under Cu+/Zn2+/Pb2+/Cd2+ stresses in this merR mutant32. These results suggested that the P1B-type ATPases might be involved in heavy metal resistance of S. meliloti CCNWSX0020. To test this hypothesis, the five P1B-type ATPases in S. meliloti CCNWSX0020 were further studied. The phylogenetic analysis of these P1B-type ATPases suggested that three of them are predicted to be Cu+-ATPase, one to be a Zn2+-ATPase and the last one to be a P1B-5-type ATPase which may be a Ni2+-ATPase.
According to the analysis of deletion mutants the five genes encoding P1B-type ATPases obtained upon double homologous recombination, a ∆zntA mutant was shown to be hypersensitive to low concentrations of Zn and Cd compared to other mutants and the wild type, indicating the vital role of ZntA in Zn and Cd resistance in S. meliloti CCNWSX0020. As pointed out in previous studies, ZntA in S. meliloti CCNWSX0020 contains a conserved phosphorylation motif (DKTGT), a CXXC (CASC) motif in N terminal and typical conserved residues of P1B-2 ATPases in trans-membrane8 helices including a CPC motif in TM6, a T(X)5QN(X)7K motif in TM7 and a DXG(X)7N motif in TM8 (see Supplementary Fig. S1)9,34. The well-characterized ZntA from E. coli mediates the efflux of Zn2+, Cd2+ and Pb2+ 13. Amino acid alignment of ZntA from S. meliloti CCNWSX0020 showed 98.04% identity to the SMc04128-encoded P1B-2-type ATPase in S. meliloti 1021 which plays a crucial role in the defense of S. meliloti against high concentrations of Zn and Cd22. The S. meliloti CCNWSX0020 ∆zntA mutant is highly sensitive to low concentration of Zn and Cd but only slightly sensitive to high concentration of Pb, suggesting that ZntA in S. meliloti CCNWSX0020 is essential to the resistance against these metal ions. RT-PCR showed that the expression of zntA in S. meliloti CCNWSX0020 was induced by heavy metals with the following order of effectiveness: Cd2+ > Zn2+ > Pb2+. In addition, a S. meliloti CCNWSX0020 ∆zntA mutant displayed an increased intracellular accumulation of Zn, Pb and Cd. These results strongly suggest ZntA in S. meliloti CCNWSX0020 to be a typical Zn2+-ATPase having a crucial role in the efflux of Zn, Cd and Pb.
Previous studies have reported the presence of five Cu+-ATPase genes on the S. meliloti 2011 genome and analyzed the functional diversity of these five homologous Cu+-ATPases19. The authors divided these five Cu+-ATPases into three subgroups including CopA1-like ATPases (CopA1a and CopA1b), CopA2-like ATPases (FixI1 and FixI2) and CopA3-like ATPases. Based on sequence alignment, three genes (SM0020_05727, SM0020_05912 and SM0020_11415) encoding Cu+-ATPases on the S. meliloti CCNWSX0020 genome were very similar to CopA3, FixI1 and CopA1b, respectively. The deletions of putative Cu+-ATPases (∆copA1b, ∆copA3 and ∆fixI1) had no effect on Cu resistance in agreement with results obtained from mutants of the genes encoding homologous Cu+-ATPases in S. meliloti 2011. However, one important difference from S. meliloti 2011 was that a ∆copA1b deletion in S. meliloti CCNWSX0020 displayed sensitivity to high concentrations of Zn, Cd and Pb. In addition, we could not identify a typical Cu+-translocating P1B-ATPase such as CopA1a in S. meliloti 2011 on the genome of S. meliloti CCNWSX0020. Moreover, cells of S. meliloti CCNWSX0020 accumulated quite high amounts of Cu in both the wild type strain and the different mutant strains. This could indicate that copper resistance in S. meliloti CCNWSX0020 is not due to efflux but rather increased copper binding in cells most likely in the periplasm.
Based on sequence alignment, S. meliloti CCNWSX0020 CopA1b was predicted to be a Cu+-translocating PIB-ATPases and CueR located upstream of copA1b was responsible for Cu and Ag-dependent induction of copA1b expression. Moreover, copA1b could confer copper tolerance to a copper sensitive E. coli ∆copA strain (Fig. 4). These results suggested that CopA1b itself has a capability for copper tolerance and/or efflux. However, ∆copA1b mutant in S. meliloti CCNWSX0020 did not lead to copper sensitivity or increased copper accumulation. Perhaps the lack of copper sensitive phenotype in the ∆copA1b mutant was masked by functional redundancy with other copper transporters or copper resistance determinants. Based on a previous study in S. meliloti CCNWSX0020, some genes responsible for copper homeostasis could be identified. These genes include lpxXL (SM0020_18047) encoding the LpxXL C-28 acyltransferase, omp (SM0020_18792) encoding a hypothetical outer membrane protein, cueO (SM0020_18797) encoding a periplasmic multicopper oxidase and merR (SM0020_29390) encoding a regulatory activator32. Therefore, we compared expression of these four genes with expression of the genes encoding these three Cu+-ATPase (SM20020_05727/copA3, SM0020_05912/fixI1, SM0020_11415/copA1b) under Cu stress. The data showed that omp and cueO were highly induced by Cu to 646- and 243-fold, respectively (see Supplementary Fig. S2). Previous study showed that the omp mutant was hypersensitive to Cu33 and our data showed omp was highly induced by Cu, revealing Omp to play a crucial role in the copper resistance mechanism of S. meliloti CCNWSX0020. In addition, the capability of S. meliloti CopA3 to complement the E. coli ∆copA strain was also tested. The result showed that the CopA3 did not restore copper tolerance of E. coli ∆copA, indicating there is no functional redundancy of CopA3 with CopA1b (data not shown).
The ∆copA1b mutant was sensitive to Zn, Cd and Pb and led to increased intracellular Zn, Cd and Pb concentrations, demonstrating that although copA1b in S. meliloti CCNWSX0020 is predicted to encode a Cu+-ATPase, it is involved in Zn, Cd and Pb homeostasis. Previous studies have reported that P1B-ATPases have a high specificity for substrate they transport, for example Cu+-ATPase transports monovalent heavy metal ions (Cu+, Ag+) and Zn2+-ATPase transports divalent heavy metal ions (Zn2+, Cd2+)9,10. Sequence comparisons and functional characterization have underlined the importance of the difference between Cu+-ATPases and Zn2+-ATPases in the presence of unique and conserved trans-membrane amino acid residues which could contribute to substrate specificity, such as Tyr/Asn of TM7, Pro/Met/Ser of TM8 in Cu+-ATPase while Leu/Lys of TM7 and Asp/Val/Ala of TM8 in Zn2+-ATPase34,35,36. Furthermore, in agreement with most Cu+-ATPases, CopA1b in S. meliloti CCNWSX0020 contains two CXXC(CASC) motifs in the N terminal and typical conserved residues of P1B-1 ATPases in trans-membrane helices including a CPC motif in TM6, a YN(X)4P motif in TM7, a MXXSS motif in TM8 (see Supplementary Fig. S1)9,34. It is probable that the presence of the same N-terminal CASC motif in CopA1b and ZntA in S. meliloti CCNWSX0020 could bind Zn2+/Cd2+/Pb2+ ions. However, the function of the N-terminal CXXC motif in P1B-type ATPase remains controversial, as two N-terminal CXXC motifs in E. coli CopA have distinct functions (the MBD1 functions as metallochaperones and the MBD2 has a regulatory role in CopA activity)37 while CXXC motif in Streptococcus pneumoniae CopA is able to bind a dicopper center and might be responsible for delivery of Cu+ to the TM metal-binding sites38. To study the function of N-terminal CXXC motif of CopA1b and ZntA in S. meliloti CCNWSX0020, we constructed these two N-terminal deletions. The results showed that the zntA N-terminal deletion was slightly sensitive to high zinc concentration while copA1b N-terminal deletion did not affect copper and zinc tolerance in S. meliloti and that neither N-terminal deletions of CopA1b nor ZntA could restore copper or zinc tolerance of E. coli ∆copA and ∆zntA mutant trains (data not shown). These indicate that the presence of N-terminal domain of CopA1b and ZntA is essential for their complete transportation function. In addition, the S. meliloti CopA1b confers a slight increase in zinc tolerance of E. coli ∆zntA mutant (Fig. 4). Therefore, it is very possible that the similar N-terminal domain of CopA1b to ZntA could bind Zn2+/Cd2+/Pb2+ ions but these metal ions binding by CopA1b N-terminal domain is not essential for cellular Zn2+/Cd2+/Pb2+ resistance, just played a primary role in cytoplasmic Zn2+/Cd2+/Pb2+ sequestration or delivery to the transmembrane site of CopA1b for cellular efflux. Notably, similar to copA1b, SMc04128 gene in S. meliloti 1021 encoded a Zn2+- ATPase and transposon insertion mutant of SMc04128 was not only highly sensitive to Zn2+ and Cd2+ but also slightly increased sensitivities to Cu2+, Pb2+, Ni2+ and Co2+ 22. This phenomenon has attracted our attention and it is likely that Cu+-ATPase and Zn2+-ATPase in S. meliloti might not have strict specificity for the heavy and transition metal ions they transport.
Moreover, P1B-1-ATPases were also shown to be involved in protein maturation and delivering metal cofactors into metalloenzymes39,40. Previous work demonstrated that both functions are important for bacterial virulence25,41. A novel host immune defense against bacterial invaders was identified and involved intoxication by transition metals (such as copper and zinc) in the phagosome to kill bacteria42,43,44. This mechanism could also be identified in response to protozoa45. P1B-ATPases in bacteria are required for transition metal efflux and assembly of metalloproteins which are essential for bacterial survival in extreme oxidative environments46. So we speculate that although CopA1b in S. meliloti CCNWSX0020 is not responsible for copper tolerance, it may incorporate copper into metalloenzymes (such as Cu/Zn-SOD) that protect against metal or oxidative stress. In line with our speculation, the deletion of copA1b led to a decrease in CAT, POD and total SOD activity level (Fig. 7), so a copA1b deletion could result in sensitivity to high concentration of Zn, since ∆copA1b mutant might lack enough capability to cope with the oxidative stress caused by high Zn. However, Cu and Zn usually served as cofactor for Cu/Zn-SOD not for CAT and POD, but the deletions of copA1b and zntA also decreased the CAT and POD activity levels. It is speculated that the reduced CAT and POD activity might result from the periplasmic metal disturbances caused by deletions of copA1b and zntA.
In conclusion, in S. meliloti CCNWSX0020 ZntA is a typical Zn2+-ATPase playing a crucial role in the efflux of Zn, Cd and Pb while copA1b encoding a Cu+-ATPase is involved in tolerance to Zn, Cd and Pb. Moreover, both of CopA1b and ZntA are potentially being involved in assembly and maturation of metalloenzymes crucial for tolerance to heavy metal and oxidative stress.
Materials and Methods
Bacterial strains, plasmids and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains (E. coli DH5α, E. coli GR16 and E. coli RW3110) were grown in Luria-Bertani (LB) medium at 37 °C. Sinorhizobium meliloti CCNWSX0020 was grown at 28 °C in TY medium (5 g tryptone, 3 g yeast extract and 0.7 g CaCl2·2 H2O per liter)47. Media were supplemented with the following antibiotics as required: 100 μg/mL ampicillin (Amp), 50 μg/mL kanamycin (Km), 100 μg/mL gentamicin (Gm) (Table 1).
Bioinformatic analysis
The known P1B-ATPase protein sequences of most bacterial genomes used in our study were obtained from UniProtKB (http://www.uniprot.org/uniprot/)48. The whole set of bacterial P1B-ATPase sequences were aligned using ClustalW249 and the phylogenetic tree visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The neighboring genes of the genes encoding P1B-ATPases were obtained from the draft genome sequence of S. meliloti CCNWSX0020 which had been reported with the accession number AGVV00000000.1 in GenBank24.
Generation of deletion mutants in genes encoding P1B-type ATPases
An in-frame, tagged P1B-ATPase deletion mutant of S. meliloti CCNWSX0020 was constructed by a method involving crossover PCR50 and the suicide vector pK18mobsacB, which cannot replicate in S. meliloti CCNWSX002051. The total genomic DNA of S. meliloti CCNWSX0020 was extracted according to the protocol of Wilson and Carson52. The plasmid pK18mobsacB-∆copA1b was used to construct the S. meliloti CCNWSX0020 copA1b deletion mutant. A 683 bp upstream and a 655 bp downstream fragment of copA1b were amplified using primer pairs copA1b-F1/copA1b-R1 and copA1b-F2/copA1b-R2, respectively. The upstream and downstream PCR products were ligated by crossover PCR with primer pairs copA1b-F1/copA1b-R2. The resulting fragment was cloned into pMD18-T vector (TaKaRa) that introduces XbaI/HindIII sites generating vector pMD18-∆copA1b. The plasmid pMD18-∆copA1b was digested with XbaI and HindIII and the ∆copA1b fragment was inserted into the XbaI/HindIII site of the suicide vector pK18mobsacB to produce pK18-∆copA1b. The plasmid pK18-∆fixI1, pK18-∆copA3, pK18-∆zntA, pK18-∆Nia were constructed in a similar manner and all primers used in this study are listed in Table S1.
The deletion was obtained through double homologous recombination. In the first step, the generated plasmid pK18-∆copA1b was transferred into S. meliloti CCNWSX0020 by triparental mating which included S. meliloti CCNWSX0020 (Ampr) as the recipient, E. coli DH5α cells containing pK18-∆copA1b (Kmr) as the donor and E. coli DH5α cells containing pRK2013 as helper cells. The selective SM agar medium (0.2 g MgSO4·7 H2O, 0.1 g CaCl2, 0.5 g KNO3, 0.5 g K2HPO4, 0.1 g NaCl, 10 g mannitol, 75 mg pantothenic acid, 75 mg biotin, 75 mg thiamine and 15 g agar per liter)53 with kanamycin and ampicillin was used for screening the first recombination events. Since pK18-∆copA1b is unable to replicate in S. meliloti CCNWSX0020, kanamycin-resistant clones should have integrated the plasmid into the chromosome by homologous recombination via one of the copA1b flanking regions. In addition S. meliloti CCNWSX0020 contains ampicillin resistance whereas E. coli could not grow in SM medium. In the second step to select for a second recombination event, clones resistant to both kanamycin and ampicillin were grown in TY liquid medium with ampicillin for 24 h and plated onto TY plates containing 10% (w/v) sucrose. As 10% sucrose was lethal to single crossover clone, sucrose-resistant clones must be the wild type strain or the deletion mutant by a second crossover event. Double crossover recombinants were confirmed by PCR using copA1b-F1 and copA1b-R2 as primers. Finally, the correct PCR product was sequenced.
Other tagged P1B-ATPase deletion mutants of S. meliloti CCNWSX0020 were constructed in the similar manner as described above for the copA1b deletion mutant. The resulting strains were designated as ∆copA1b, ∆fixI1, ∆copA3, ∆zntA and ∆nia (Table 1).
Heavy and transition metal sensitivity assays of the five mutants
Sinorhizobium meliloti CCNWSX0020 and each of the five deletion mutants were grown to midexponential phase in TY liquid medium at 28 °C with shaking at 150 rpm and cell suspensions were prepared at the same OD600 of 1.0 (optical density at 600 nm). Then 1% of the cell suspensions were added to fresh TY medium supplemented with different concentration of CuSO4, ZnSO4, CdSO4, NiCl2 and Pb(NO3)2. The cells were again incubated with shaking at 150 rpm for 48 h and growth was monitored at OD600. The data were shown as the means of biological triplicates ± SD.
Complementation of ∆copA1b and ∆zntA mutants
To complement the copA1b mutant, the entire copA1b gene including the regulatory region was amplified with primers C-copA1b-F/C-copA1b-R using S. meliloti CCNWSX0020 genomic DNA as template. The PCR product was digested with SmaI/XbaI and inserted into broad-range plasmid pBBR1MCS-5 to generate pBBR-copA1b. The sequence of this construct to be used in complementation was verified by automated DNA sequencing, transformed into E. coli donor strain DH5α and delivered into the ∆copA1b mutant via triparental conjugation. Single clones carrying pBBR-copA1b were selected on TY plates containing gentamicin. The presence of the copA1b gene in the mutant strain was confirmed by PCR. The complementation of a zntA deletion was performed in a similar fashion.
Metal sensitivity assays of S. meliloti CCNWSX0020 wild type, ∆copA1b mutant and ∆zntA mutant and the corresponding complementations (C-copA1b and C-zntA) were performed on TY solid medium with different concentrations of CuSO4, ZnSO4, CdSO4 and Pb(NO3)2. Cells were grown to exponential phase in TY liquid medium and then diluted to an OD600 of 0.1. Four 10-fold dilutions were carried out and spotted on the agar medium. Each experiment was repeated three times.
Heterologous expression of S. meliloti copA1b and zntA in E. coli ∆copA and ∆zntA strains
Complementation plamids pBBR-copA1b and pBBR-zntA were transformed into E. coli ∆copA strain and E. coli ∆zntA strain via triparental conjugation, respectively. Single clones carrying pBBR-copA1b or pBBR-zntA were selected on LB plates containing gentamicin. The presence of the copA1b or zntA gene in the E. coli ∆copA and ∆zntA strains was confirmed by PCR.
Copper and zinc sensitivity assays of E. coli ∆copA and ∆zntA strains and the corresponding complementation (C-copA1b-E∆copA, C-copA1b-E∆zntA, C-zntA-E∆copA and C-zntA-E∆zntA) were performed on LB solid medium. Cells were grown to exponential phase at 37 °C in LB liquid medium and then diluted to an OD600 of 0.1. Five 10-fold dilutions were carried out and spotted on the agar medium with indicated concentrations of CuSO4 and ZnSO4. Each experiment was repeated three times.
Heavy and transition metal accumulation assay
Sinorhizobium meliloti CCNWSX0020 wild type and mutant strains were grown at 28 °C in TY liquid medium until cells reached exponential phase at the same OD600 of 0.8. Then cells were incubated for another 24 h with shaking at 150 rpm after TY medium had been supplemented with 0.8 mM CuSO4, 0.4 mM ZnSO4, 0.05 mM CdSO4 and 2.0 mM Pb(NO3)2. Then cells were harvested by centrifugation at 8000 g for 10 min. The intracellular accumulated heavy and transition metals were measured by furnace atomic absorption spectroscopy (Varian SpectrAA 880/GTA 100) as described previously31. The data were shown as the means of biological triplicates ± SD.
H2O2 sensitivity test and antioxidant enzyme activity assay
Sinorhizobium meliloti CCNWSX0020 wild type strain, ∆copA1b and ∆zntA mutants were grown to exponential phase in TY liquid medium and then diluted to an OD600 of 0.1. Four 10-fold dilutions were carried out and spotted on the agar medium with different concentration of H2O2 for H2O2 sensitivity test.
The exponential phase cells grown in TY liquid medium were treated or untreated with 500 μM H2O2 for 30 min. Then cells were harvested by centrifugation at 8000 g for 2 min and resuspended with enzyme extracting solution. Cell suspensions were lysed by ultrasonic disruption, followed by centrifugation at 8000 g for 10 min. Clear lysates were used for total protein determination and catalase, peroxidase and superoxide dismutase activity assay. Protein concentrations were determined by using the Bradford Bio-Rad protein assay. The catalase (CAT) activity was assayed by measurement of the degradation of H2O2 at a wavelength of 240 nm according to the method of Aebi54. The peroxidase (POD) activity was determined spectrophotometrically at 470 nm by the method of Hammerschmidt, Nuckles and Kuc55. The superoxide dismutase (SOD) activity was assayed by measuring the enzyme’s ability to inhibit the photochemical reduction of nitroblue tetrazolium (NBT) as described previously by Beauchamp and Fridovich56. The data were shown as the means of biological triplicates ± SD.
Real-time qRT-PCR analysis
Sinorhizobium meliloti CCNWSX0020 and two zinc-sensitive mutants grown to exponential phase in TY liquid medium were supplemented with 0.6 mM CuSO4, 0.05 mM AgNO3, 0.4 mM ZnSO4, 0.1 mM CdSO4 and 1.5 mM Pb(NO3)2 and incubated for 30 min at 28 °C. Then cells were harvested and total RNA was extracted. Procedures including DNA elimination, cDNA synthesis and quantitative RT-PCR were performed as described previously32. All these assays were performed in triplicate. Primer pairs used to monitor transcription of genes are listed in Table S1. To standardize results, 16S rRNA was used as an internal standard and the relative levels of transcription were calculated using the 2−∆∆Ct method57.
Statistical analyses
Statistical analyzes were carried out using SPSS 19.0 software (SPSS 208 Inc., Chicago, IL, USA). Paired two-tailed Student’s t-test was performed to determine significant differences among the different experimental treatments. All of the data was analyzed using the Origin Pro v8.0 (Origin Lab, Hampton, USA) to create the figures.
Additional Information
How to cite this article: Lu, M. et al. Zinc Resistance Mechanisms of P1B-type ATPases in Sinorhizobium meliloti CCNWSX0020. Sci. Rep. 6, 29355; doi: 10.1038/srep29355 (2016).
References
Nelson, N. Metal ion transporters and homeostasis. EMBO J 18, 4361–4371 (1999).
Thompson, K. H. & Orvig, C. Boon and bane of metal ions in medicine. Science 300, 936–939 (2003).
Gilotra, U. & Srivastava, S. Plasmid-encoded sequestration of copper by Pseudomonas pickettii strain US321. Curr Microbiol 34, 378–381 (1997).
Ji, G. & Silver, S. Bacterial resistance mechanisms for heavy metals of environmental concern. J Ind Microbiol 14, 61–75 (1995).
Kuroda, K. & Ueda, M. Bioadsorption of cadmium ion by cell surface-engineered yeasts displaying metallothionein and hexa-His. Appl Microbiol Biotechnol 63, 182–186 (2003).
Nies, D. H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27, 313–339 (2003).
Palmgren, M. G. & Nissen, P. P-type ATPases. Annu Rev Biophys 40, 243–266 (2011).
Gaitan-Solis, E., Taylor, N. J., Siritunga, D., Stevens, W. & Schachtman, D. P. Overexpression of the transporters AtZIP1 and AtMTP1 in cassava changes zinc accumulation and partitioning. Front Plant Sci 6 (2015).
Argüello, J. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biol 195, 93–108 (2003).
Argüello, J. M., Eren, E. & González-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248 (2007).
Mandal, A. K., Cheung, W. D. & Argüello, J. M. Characterization of a Thermophilic P-type Ag+/Cu+-ATPase from the Extremophile Archaeoglobus fulgidus. J Biol Chem 277, 7201–7208 (2002).
Rensing, C., Fan, B., Sharma, R., Mitra, B. & Rosen, B. P. CopA: an Escherichia coli Cu (I)-translocating P-type ATPase. Proc Natl Acad Sci USA. 97, 652–656 (2000).
Sharma, R., Rensing, C., Rosen, B. P. & Mitra, B. The ATP hydrolytic activity of purified ZntA, a Pb (II)/Cd (II)/Zn (II)-translocating ATPase from Escherichia coli. J Biol Chem 275, 3873–3878 (2000).
Rensing, C., Mitra, B. & Rosen, B. P. The zntA gene of Escherichia coli encodes a Zn (II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 94, 14326–14331 (1997).
Mana-Capelli, S., Mandal, A. K. & Argüello, J. M. Archaeoglobus fulgidus CopB Is a Thermophilic Cu2+-ATPase functional role of its histidine-rich n-terminal metal binding domain. J Biol Chem 278, 40534–40541 (2003).
Raimunda, D., Long, J. E., Sassetti, C. M. & Argüello, J. M. Role in metal homeostasis of CtpD, a Co2+ transporting P1B4‐ATPase of Mycobacterium smegmatis. Mol Microbiol 84, 1139–1149 (2012).
Zielazinski, E. L. et al. Sinorhizobium meliloti Nia is a P1B-5-ATPase expressed in the nodule during plant symbiosis and is involved in Ni and Fe transport. Metallomics 5, 1614–1623 (2013).
Smith, A. T., Smith, K. P. & Rosenzweig, A. C. Diversity of the metal-transporting P1B-type ATPases. J Biol Inorg Chem 19, 947–960 (2014).
Patel, S. J., Padilla-Benavides, T., Collins, J. M. & Argüello, J. M. Functional diversity of five homologous Cu+-ATPases present in Sinorhizobium meliloti. Microbiology 160, 1237–1251 (2014).
Brocklehurst, K. R. et al. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol 31, 893–902 (1999).
Tsai, K.-J. et al. Membrane topology of the pl258 cadA Cd (II)/Pb (II)/Zn (II)-translocating p-type ATPase. J Bioenerg Biomembr 34, 147–156 (2002).
Rossbach, S. et al. Response of Sinorhizobium meliloti to elevated concentrations of cadmium and zinc. Appl Environ Microbiol 74, 4218–4221 (2008).
Maynaud, G. et al. CadA of Mesorhizobium metallidurans isolated from a zinc-rich mining soil is a P IB-2-type ATPase involved in cadmium and zinc resistance. Res Microbiol 165, 175–189 (2014).
Li, Z., Ma, Z., Hao, X. & Wei, G. Draft genome sequence of Sinorhizobium meliloti CCNWSX0020, a nitrogen-fixing symbiont with copper tolerance capability isolated from lead-zinc mine tailings. J Bacteriol 194, 1267–1268 (2012).
González‐Guerrero, M., Raimunda, D., Cheng, X. & Argüello, J. M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol 78, 1246–1258 (2010).
Humbert, M. V., Rasia, R. M., Checa, S. K. & Soncini, F. C. Protein signatures that promote operator selectivity among paralog MerR monovalent metal ion regulators. J Biol Chem 288, 20510–20519 (2013).
Stoyanov, J. V., Hobman, J. L. & Brown, N. L. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol 39, 502–512 (2001).
Stoyanov, J. V. & Brown, N. L. The Escherichia coli Copper-responsivecopA Promoter Is Activated by Gold. J Biol Chem 278, 1407–1410 (2003).
Fridovich, I. Superoxide dismutases: studies of structure and mechanism. Adv Exp Med Biol 74, 530 (1976).
Hersleth, H.-P., Ryde, U., Rydberg, P., Görbitz, C. H. & Andersson, K. K. Structures of the high-valent metal-ion haem–oxygen intermediates in peroxidases, oxygenases and catalases. J Inorg Biochem 100, 460–476 (2006).
Fan, L.-M. et al. Characterization of a copper-resistant symbiotic bacterium isolated from Medicago lupulina growing in mine tailings. Bioresour Technol 102, 703–709 (2011).
Li, Z., Ma, Z., Hao, X., Rensing, C. & Wei, G. Genes conferring copper resistance in Sinorhizobium meliloti CCNWSX0020 also promote the growth of Medicago lupulina in copper-contaminated soil. Appl Environ Microbiol 80, 1961–1971 (2014).
Li, Z., Lu, M. & Wei, G. An omp gene enhances cell tolerance of Cu (II) in Sinorhizobium meliloti CCNWSX0020. World J Microb Biot 29, 1655–1660 (2013).
Zhitnitsky, D. & Lewinson, O. Identification of functionally important conserved trans‐membrane residues of bacterial PIB‐type ATPases. Mol Microbiol 91, 777–789 (2014).
Okkeri, J. & Haltia, T. The metal-binding sites of the zinc-transporting P-type ATPase of Escherichia coli. Lys 693 and Asp 714 in the seventh and eighth transmembrane segments of ZntA contribute to the coupling of metal binding and ATPase activity. Bba-bioenergetics 1757, 1485–1495 (2006).
González-Guerrero, M., Eren, E., Rawat, S., Stemmler, T. L. & Argüello, J. M. Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J Biol Chem 283, 29753–29759 (2008).
Drees, S. L., Beyer, D. F., Lenders-Lomscher, C. & Lübben, M. Distinct functions of serial metal‐binding domains in the Escherichia coli P1B-ATPase CopA. Mol Microbiol 97, 423–438 (2015).
Fu, Y. et al. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat Chem Biol 9, 177–183 (2013).
Preisig, O., Zufferey, R. & Hennecke, H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol 165, 297–305 (1996).
Hassani, B. K., Astier, C., Nitschke, W. & Ouchane, S. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J Biol Chem 285, 19330–19337 (2010).
Argüello, J. M., González-Guerrero, M. & Raimunda, D. Bacterial transition metal P1B-ATPases: transport mechanism and roles in virulence. Biochemistry 50, 9940–9949 (2011).
Botella, H., Stadthagen, G., Lugo-Villarino, G., de Chastellier, C. & Neyrolles, O. Metallobiology of host–pathogen interactions: an intoxicating new insight. Trends Microbiol 20, 106–112 (2012).
Hood, M. I. & Skaar, E. P. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol 10, 525–537 (2012).
Djoko, K. Y., Cheryl-lynn, Y. O., Walker, M. J. & McEwan, A. G. The Role of Copper and Zinc Toxicity in Innate Immune Defense against Bacterial Pathogens. J Biol Chem 290, 18954–18961 (2015).
Stafford, J. L., Neumann, N. F. & Belosevic, M. Macrophage-mediated innate host defense against protozoan parasites. Crit Rev Microbiol 28, 187–248 (2002).
Botella, H. et al. Mycobacterial P1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10, 248–259 (2011).
Mohamad, O. A. et al. Biosorption of copper (II) from aqueous solution using non-living Mesorhizobium amorphae strain CCNWGS0123. Microbes Environ 27, 234 (2012).
Consortium, U. UniProt: a hub for protein information. Nucleic Acids Res 43, D204–D212 (2014).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Link, A. J., Phillips, D. & Church, G. M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179, 6228–6237 (1997).
Schäfer, A. et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994).
Wilson, T. & Carson, J. Rapid, high-throughput extraction of bacterial genomic DNA from selective-enrichment culture media. Lett Appl Microbiol 32, 326–330 (2001).
Hao, X. et al. Copper Tolerance Mechanisms of Mesorhizobium amorphae and Its Role in Aiding Phytostabilization by Robinia pseudoacacia in Copper Contaminated Soil. Environ Sci Technol 49, 2328–2340 (2015).
Aebi, H. Catalase in vitro. Methods Enzymol 105, 121–126 (1984).
Hammerschmidt, R., Nuckles, E. & Kuć, J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiological Plant Pathology 20, 73–82 (1982).
Beauchamp, C. & Fridovich, I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44, 276–287 (1971).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166, 557–580 (1983).
Grass, G. & Rensing, C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol 183, 2145–2147 (2001).
Kovach, M. E. et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant numbers 31370142 and 31270012). We thank Tanya Soule for donating the pRK2013 plasmid.
Author information
Authors and Affiliations
Contributions
G.W. and Z.L. conceived and designed the experiment, M.L., J.L. and Y.W. performed experiments. M.L. wrote the manuscript and C.R. revised the manuscript and assisted with manuscript preparation. All authors have read the manuscript and agree with its content.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lu, M., Li, Z., Liang, J. et al. Zinc Resistance Mechanisms of P1B-type ATPases in Sinorhizobium meliloti CCNWSX0020. Sci Rep 6, 29355 (2016). https://doi.org/10.1038/srep29355
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29355
This article is cited by
-
Impact of lead (Pb2+) on the growth and biological activity of Serratia marcescens selected for wastewater treatment and identification of its zntR gene—a metal efflux regulator
World Journal of Microbiology and Biotechnology (2023)
-
Characterization of a Bacillus megaterium strain with metal bioremediation potential and in silico discovery of novel cadmium binding motifs in the regulator, CadC
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.