Abstract
Large scale association studies have identified the single nucleotide polymorphism rs3803662 associated with breast cancer risk. However, the sample size of most studies is too small. Here, we performed this meta-analysis to make the result more convincing. Relevant articles published up to 2016 were identified by searching the PubMed database. 13 studies, involving a total of 29405 participants, were included in the meta-analysis. Odds Ratios (ORs) with 95% confidence intervals (CIs) was calculated with random or fixed effects model. All data analyses were analyzed by Review Manger 5.3 software. In Caucasian subgroup: Dominant model (TT + CT vs CC): OR = 1.17 (1.06, 1.29), Recessive model (TT vs CT + CC): OR = 1.25 (1.13, 1.39) and Allele frequency (T vs C): OR = 1.15 (1.08, 1.22). The present meta-analysis suggests that rs3803662 polymorphism is significantly associated with breast cancer risk in Caucasian women and we did not find the association in Asian women.
Similar content being viewed by others

Introduction
Breast cancer, as a multifactorial disease, is the most common cancer in the world1. The major influences on breast cancer appear to be environmental and genetic factors2,3. Previous studies indicate that genetic influences account for about 27% of the breast cancer risk4. A few genes including BRCA1, BRCA2 and ATM have been known to be associated with the risk of breast cancer5. In the past 30 years, the incidence of breast cancer has been increasing rapidly in many countries. According to the data of International Agency for Research on Cancer (IARC), there are additional 1.68 million breast cancer patient in 2012, which is 25% of the women with tumor6.
It’s reported that genetic factors play an important role in the development of breast cancer. With the completion of the human genome project (HGP) in 2001, single nucleotide polymorphisms (SNPs) come into view as the essential factor in the development of diseases 7,8,9,10. In addition to the highly penetrant (BRCA1, BRCA2 and TP53) and moderately penetrant (CHEK2, BRIP1, ATM and PALB2) genetic variants, breast cancer has been associated with low penetrant risk (FGFR2, TNRC9, MAP3K1 and LSP1)11. About 5% of breast cancer incidence is attributable to high-penetrance mutations12. Therefore, it’s important to evaluate the association between low penetrant and breast cancer risk.
Recent studies have identified the single nucleotide polymorphism rs3803662 associated with breast cancer risk7,13,14,15,16,17,18,19,20,21,22,23,24. In 2010, Rulla M Tamimi et al. and TV Gorodnova et al. found significant association in Swedish and Russian, respectively15,17. In 2011, Martha L. Slattery et al. found the association in Hispanic subsequently22. In 2014, Isabel Elematore et al. considered that rs3803662 was associated with breast cancer risk in Chilean24. In 2016, Yaning He et al. also find the association in Chinese Han population23. However, the sample size of most studies was too small. One of the aims of Meta-Analysis is leading to a higher statistical power and more robust result. Single study does not have significant result statistically sometimes. It is due to that the sample size was so small that statistical power was low. We can improve statistical power by means of combining some samples with small sample size. Therefore, we conducted a meta-analysis of the previously published studies involving SNP rs3803662 and breast cancer to get a more comprehensive result.
Results
Literature search
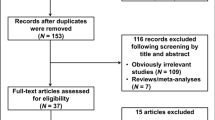
A flow diagram for the study selection process is shown in Fig. 1. A total of 95 articles were identified by the search strategy. 77 articles were removed because they don’t evaluate the association between rs3803662 and breast cancer risk or have sufficient data to calculate the ORs with 95% CIs. Afterwards, 4 articleswere excluded due to that the control group doesn’t meet the Hardy-Weinberg Equilibrium (HWE) and 1 article was excluded because that the study population is male, not female.
The flow diagram showing the study selection process.
A total of 95 articles were identified by the search strategy. Because don’t meet the qualifications, 77 articles were removed and 18 articles remained for further screening. Afterwards, since the control groups don’t meet the Hardy-Weinberg Equilibrium (HWE), 4 articles were excluded. Besides, 1 article was excluded due to that the study population is male, not female. Finally, 13 studies were included in the meta-analysis.
Study characteristics
The primary characteristics of the 13 studies are summarized in Table 1. A total of 29405 participants with 14306 cases and 15099 controls were included in this study. The studies were divided into two subgroups according to the ethnicity of their participants: East-Asian subgroup with 4 studies and Caucasian subgroup with 8 studies and others subgroup with 1 study.
Association between rs3803662 polymorphism and breast cancer risk
The forest plot concerning the association between the rs3803662 polymorphism and the risk of breast cancer is shown in Fig. 2. Figure 2A–D are forest plots of dominant model, recessive model, additive model and Allele frequency, respectively.
In Caucasian subgroup, dominant model (TT + CT vs CC): OR = 1.17 (1.06, 1.29); recessive model (TT vs CT + CC): OR = 1.25 (1.13, 1.39); additive model (TT + CC vs CT): OR = 0.93 (0.84, 1.03); allele frequency (T vs C): OR = 1.15 (1.08, 1.22).
In East Asian subgroup, dominant model (TT + CT vs CC): OR = 0.86 (0.63, 1.18); recessive model (TT vs CT + CC): OR = 0.96 (0.74, 1.25); additive model (TT+CC vs CT): OR = 1.01 (0.87, 1.17); allele frequency (T vs C): OR = 0.95 (0.77, 1.16).
Assessment of publication bias
Funnel plot were carried out to assess publication bias. There are many reasons leading to publication bias: low quality research with small sample, heterogeneity and negative result. Figure 3A–D are funnel plots of dominant model, recessive model, additive model and Allele frequency, respectively. We found publication bias in Fig. 3B (recessive model). In this study, it mainly comes from few researches with small sample and heterogeneity.
Discussion
Our meta-analyses, including 14306 cases and 15099 controls group numbers from 13 case-control studies, explored the association between the rs3803662 polymorphism and the risk of breast cancer. The result indicates that rs3803662 is significant associated with breast cancer risk in Caucasian women. And we did not find the association Asian women.
The SNP rs3803662 is significant associated with breast cancer risk in Caucasian women which is consistent with results of previous studies15,16,17,18,19,20,21,22. But in Asian women, we get inconsistent result. That can be attributed to the following reasons. First, only 4 studies with 5237 cases and 5130 controls were included in the meta-analysis according to the study selection criteria7,13,14,23. The inadequate participant results in that rs3803662 is not significantly associated with breast cancer risk in Asian women statistically. The more the studies were included in meta-analysis, the more accurate the result will be. More independent studies concentrating on the association in Asian women should be added into this analysis and we will focus on the latest research. Second, we hypothesize that genetic background varies among different ethnic populations. Furthermore, this variation leads to change in susceptibility to some cancers. The hypothesis needs a huge amount of samples to validate whether it is true or not.
The SNP rs3803662 lies 8 kb upstream of TNRC9 which is located on the chromosome 16q12 and consists of seven exons25. Rs3803662 has been confined to estrogen receptor-positive tumors26. Though we are not familiar with the function of TNRC9, this gene has been found to be relevant to bone metastasis in breast cancer27. TNRC9 is not only expressed in brain, but also expressed in breast with higher expression level in breast cancer compared to that in normal tissue28,29. And the minor allele of rs3803662 was associated with lower mRNA expression of TNRC9 gene30. James Owain Jones et al. found that TNRC9 maps to a known breast cancer susceptibility locus and hypothesized that TNRC9 could be a candidate tumor suppressor gene in 16q31.
There are some limitations in this study. First, we conducted subgroup analysis according to the ethnicity of participants. Compared with Caucasian subgroup, only 4 studies with 5237 cases and 5130 controls were included in the study. Second, the variation in genetic background may have effects on the susceptibility to breast cancer. More studies focusing on the same population should be included.
In conclusion, rs3803662 polymorphism is significantly associated with breast cancer risk in Caucasian women and we did not find the association in Asian women.
Methods
Search strategy
A comprehensive literature searches for suitable studies published up to 2016 was conducted in the PubMed, EMbase and Web of Science database. Studies that investigated the rs3803662 and breast cancer were included in this meta-analysis. Studies should be published as a full paper. The search was conducted using the following keywords: “rs3803662” and “breast cancer”.
Study selection criteria
Two independent reviewers first screened the titles and abstracts to identify the relevant investigations. Then, full articles were read to include the eligible studies that met the following criteria: (1) used a case-control study design, (2) evaluated the association between rs3803662 and the risk of breast cancer, (3) provided the number of genotypes in case-controls groups for calculating ORs, (4) the control group has to satisfy HWE.
Data extraction
For every eligible study, the following data were extracted by two independent reviewers: name of the first author, publication date, the ethnicity of study population and the number of genotype in case-control group. In addition, the P value of HWE in control group was calculated.
Statistical analysis
All statistical analysis was conducted by STATA version 14.0 (STATA Corporation, College Station, TX, USA). The Odds Ratios (ORs) with 95% confidence intervals (CIs) was calculated to evaluate the association between the rs3803662 polymorphism and breast cancer risk.
The heterogeneity means the variation between different researches in systematic review. It has two major types: the clinical heterogeneity and the heterogeneity of methodology. The former comes from the difference of participants, interventions and outcome among studies. The latter derives from distinction of experimental design. We use Q and I2 statistics to assess the heterogeneity. If I2 < 50% or the P-value of heterogeneity >0.10, we use fixed effects model. Otherwise, the random effects model was selected.
The Begg’s rank correlation test and Egger’s linear regression test were used to assess the publication bias. The genetic models we use here include dominant model, recessive mode, additive model and allele frequency32,33.
Additional Information
How to cite this article: Yang, Y. et al. Association of single nucleotide polymorphism rs3803662 with the risk of breast cancer. Sci. Rep. 6, 29008; doi: 10.1038/srep29008 (2016).
References
Curado, M.-P. et al. Cancer incidence in five continents, Volume IX. (IARC Press, International Agency for Research on Cancer, 2007).
Chen, F. et al. A single nucleotide polymorphism of the TNRC9 gene associated with breast cancer risk in Chinese Han women. Genet Mol Res 13, 182–187, doi: 10.4238/2014.January.10.9 (2014).
Parkin, D. M., Bray, F., Ferlay, J. & Pisani, P. Global cancer statistics, 2002. CA: a cancer journal for clinicians 55, 74–108 (2005).
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark and Finland. New England journal of medicine 343, 78–85 (2000).
Walsh, T. & King, M. C. Ten genes for inherited breast cancer. Cancer Cell 11, 103–105, doi: 10.1016/j.ccr.2007.01.010 (2007).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–386, doi: 10.1002/ijc.29210 (2015).
Han, W. et al. Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiol Biomarkers Prev 20, 793–798, doi: 10.1158/1055-9965.EPI-10-1282 (2011).
Liu, G. et al. Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiology of aging 35, 786–792, doi: 10.1016/j.neurobiolaging.2013.10.084 (2014).
Li, Y. et al. CR1 rs3818361 Polymorphism Contributes to Alzheimer’s Disease Susceptibility in Chinese Population. Molecular neurobiology, doi: 10.1007/s12035-015-9343-7 (2015).
Xiang, Z. et al. Integrating Genome-Wide Association Study and Brain Expression Data Highlights Cell Adhesion Molecules and Purine Metabolism in Alzheimer’s Disease. Molecular neurobiology 52, 514–521, doi: 10.1007/s12035-014-8884-5 (2015).
Harlid, S. et al. Combined effect of low-penetrant SNPs on breast cancer risk. Br J Cancer 106, 389–396, doi: 10.1038/bjc.2011.461 (2012).
Chen, Y. C. & Hunter, D. J. Molecular epidemiology of cancer. Ca A Cancer Journal for Clinicians 55, 45–54 quiz 57 (2005).
Mizoo, T. et al. Effects of lifestyle and single nucleotide polymorphisms on breast cancer risk: a case-control study in Japanese women. Bmc Cancer 13, 5707–5707 (2013).
Jie, L. et al. Genetic variants in trinucleotide repeat-containing 9 (TNRC9) are associated with risk of estrogen receptor positive breast cancer in a Chinese population. Frontiers in Human Neuroscience 7, 924–924 (2014).
Gorodnova, T. V. et al. Distribution of FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 alleles in genetically enriched breast cancer patients versus elderly tumor-free women. Cancer Genet Cytogenet 199, 69–72, doi: 10.1016/j.cancergencyto.2010.01.020 (2010).
Butt, S. et al. Genetic predisposition, parity, age at first childbirth and risk for breast cancer. BMC research notes 5, 414 (2012).
Tamimi, R. M. et al. Birth weight, breast cancer susceptibility loci and breast cancer risk. Cancer Causes Control 21, 689–696, doi: 10.1007/s10552-009-9496-7 (2010).
McInerney, N. et al. Low penetrance breast cancer predisposition SNPs are site specific. Breast Cancer Res Treat 117, 151–159, doi: 10.1007/s10549-008-0235-7 (2009).
Shan, J. et al. Genome-Wide Association Studies (GWAS) breast cancer susceptibility loci in Arabs: susceptibility and prognostic implications in Tunisians. Breast Cancer Res Treat 135, 715–724, doi: 10.1007/s10549-012-2202-6 (2012).
Latif, A. et al. Breast cancer susceptibility variants alter risks in familial disease. Journal of Medical Genetics 47, 126–131 (2009).
Antoniou, A. C. et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet 82, 937–948, doi: 10.1016/j.ajhg.2008.02.008 (2008).
Slattery, M. L. et al. Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res Treat 129, 531–539, doi: 10.1007/s10549-011-1498-y (2011).
He, Y. et al. Relationship between five GWAS-identified single nucleotide polymorphisms and female breast cancer in the Chinese Han population. Tumor Biology, 1–6 (2016).
Elematore, I. et al. Association of genetic variants at TOX3, 2q35 and 8q24 with the risk of familial and early-onset breast cancer in a South-American population. Molecular Biology Reports 41, 3715–3722 (2014).
Easton, D. F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093, doi: 10.1038/nature05887 (2007).
Stacey, S. N. et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature Genetics 39, 865–869 (2007).
Smid, M. et al. Genes associated with breast cancer metastatic to bone. Journal of Clinical Oncology Official Journal of the American Society of Clinical Oncology 24, 2261–2267 (2006).
Dittmer, S. et al. TOX3 is a neuronal survival factor that induces transcription depending on the presence of CITED1 or phosphorylated CREB in the transcriptionally active complex. Journal of Cell Science 124, 252–260 (2011).
Udler, M. S. et al. Fine scale mapping of the breast cancer 16q12 locus. Hum Mol Genet 19, 2507–2515, doi: 10.1093/hmg/ddq122 (2010).
Riaz, M. et al. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival and mRNA expression of the nearest genes. Breast Cancer Res Treat 133, 843–851, doi: 10.1007/s10549-011-1663-3 (2012).
Jones, J. O. et al. TOX3 mutations in breast cancer. Plos One 8, e74102–e74102 (2013).
Minelli, C., Thompson, J. R., Abrams, K. R., Thakkinstian, A. & Attia, J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol 34, 1319–1328, doi: 10.1093/ije/dyi169 (2005).
Minelli, C., Thompson, J. R., Abrams, K. R. & Lambert, P. C. Bayesian implementation of a genetic model‐free approach to the meta‐analysis of genetic association studies. Statistics in medicine 24, 3845–3861 (2005).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31301938 and 81300945); Natural Science Basic Research Plan in Shaanxi Province of China (2014JQ3110); Chinese Universities Scientific Fund (2452015077 and 2452016005)
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: M.L. and Y.C.Y. Performed the experiments: Y.Y., W.W. and M.L. Analyzed the data: Y.Y., G.L. and M.L. Contributed reagents/materials/analysis tools: Y.Y. and M.L. Wrote the paper: Y.Y., Y.C.Y. and M.L. All of the authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yang, Y., Wang, W., Liu, G. et al. Association of single nucleotide polymorphism rs3803662 with the risk of breast cancer. Sci Rep 6, 29008 (2016). https://doi.org/10.1038/srep29008
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29008
This article is cited by
-
Meta-analysis of association between TCF7L2 polymorphism rs7903146 and type 2 diabetes mellitus
BMC Medical Genetics (2018)
-
Rs4878104 contributes to Alzheimer’s disease risk and regulates DAPK1 gene expression
Neurological Sciences (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.