Abstract
Flour beetles of the genus Tribolium Macleay (Coleoptera: Tenebrionidae) are important stored product pests in China and worldwide. They are often found or are intercepted in grain depots, flour mills, and entry-exit ports, etc. Traditionally, Tribolium species are identified according to the morphological characteristics of the adult. However, it is almost impossible to rapidly identify adult fragments and non-adult stages based on external morphological characteristics. Molecular techniques for the rapid and accurate identification of Tribolium species are required, particularly for pest monitoring and the quarantine of stored products pests. Here, we establish DNA barcoding, species-specific PCR, and real-time PCR techniques for the identification of six stored-product pest Tribolium species including T. castaneum, T. confusum, T. destructor, T. madens, T. freemani and T. brevicornis. We detected the mitochondrial DNA cytochrome oxidase subunit I (COI) barcodes for Tribolium from 18 geographic populations and 101 individuals, built a Tribolium DNA barcode library, and designed species-specific primers and TaqMan probes for the above six Tribolium species. The three techniques were applied to identify Tribolium collected from stored samples and samples captured from quarantine ports. The results demonstrated that three techniques were all able to identify the six species of Tribolium both rapidly and accurately.
Similar content being viewed by others
Introduction
Flour beetles of the genus Tribolium Macleay, 1825 (Coleoptera: Tenebrionidae) decrease the quality and quantity of stored products by introducing insect fragments and excrement1,2 as well as causing contamination with potential adverse effects to human health3. The genus Tribolium has 36 described species4, ten of which are stored products pests5. T. castaneum and T. confusum are major pests of processed grain and cereal products with cosmopolitan distribution, including P.R. China6, USA7 and Europe8, and five species of Tribolium (T. destructor, T. audax Halstead (1969), T. madens, T. freemani, and T. anaphe Hinton (1948)) are potentially serious pest species. The other Tribolium species are less harmful and include T. brevicorne, T. parallelus (Casey 1890), and T. thusa Hinton (1948).
Traditional methods for the identification of Tribolium species using the morphological characteristics of adults are common and widely used9. However, an individual Tribolium is small and many species are similar in appearance, particularly the non-adult stages including eggs, pupae and larvae. In routine stored-grain insect pest monitoring and port quarantine, it is difficult to identify species on the basis of external morphological characteristics alone. Recently, the rapid and accurate molecular identification of insect pests has become popular10,11,12. Therefore, the molecular identification of Tribolium species is an appropriate alternative to conventional taxonomy based on morphological characteristics11; moreover species-specific primers for T. castaneum and T. confusum have already been designed based on internal transcribed spacer (ITS) fragments encoded by rDNA and on mitochondrial cytochrome oxidase subunit I 1 . General PCR and PCR-RFLP analysis have been used to discriminate T. destructor from other species13. PCR-RFLP has been developed to distinguish Tribolium flour beetles based on a partial 28S rRNA gene sequence14. However, there are currently no correlative studies on the molecular identification of Tribolium species using DNA barcoding technology, species-specific PCR and real-time PCR.
This study focuses on six Tribolium species that are stored-product pests. The aim of the study was to establish molecular techniques to identify Tribolium species accurately, rapidly and practically using DNA barcoding, species-specific PCR, and real-time PCR.
Materials and Methods
Tribolium specimens
Cultures of six Tribolium species from 101 adult specimens including T. castaneum, T. confusum, T. destructor, T. madens, T. freemani and T. brevicornis were used in this study. Specimens were collected from the P. R. China (Henan, Guangxi, Guangdong, Xinjiang), the Czech Republic (Prague, Kyjov, Herink, Rakovník), France (Bordeaux), Croatia (Osijek), and the United States (Kansas). Original collection locations are listed in Table 1. These laboratory strains were reared in darkness at 27 °C and 75% relative humidity on a diet of powdered wheat germ. Voucher specimens, including adults, larvae and pupae, were kept in 100% ethanol and stored at −80 °C. Detailed specimen information is shown in Table 1.
DNA extraction, PCR and COI sequencing
Total genomic DNA at least three Tribolium adults from different geographic population was extracted from the thorax of Tribolium adults using a commercial TIANamp Genomic DNA kit (TIANGEN, China) according to the manufacturer’s protocol. A pair of universal forward LCO1490 (5′-GGTCAACAAA TCATAAAGATATTGG-3′) and reverse HCO2198 (5′-TAAACTTCAGGGTGACCA AAAAATCA-3′) primers were used for COI amplification15. PCR was performed based on methods by Wang et al.11 and was modified for half volume reactions containing 12.5 μl MasterMix with loading dye, 10 μl sterilized distilled water, 1.5 μl extracted DNA (approximately 20 ng μL−1), and 0.5 μl forward and reverse primers (10 μM). The PCR protocol included an initial denaturing step at 94 °C for 3 min, followed by 35 cycles of 94 °C for 1 min, 52 °C for 1 min and 72 °C for 1 min with a final extension at 72 °C for 10 min. The reactions were performed on a Veriti TM 96-well Thermal Cycler (ABI, USA). The amplified DNA fragments were resolved on a 1.0% (w/v) agarose gel (1 × Tris Acetate-EDTA buffer), stained with ethidium bromide and visualized with a UV light (Gel Logic 212 PRO, Carestream Health, Inc.). DNA purification and bidirectional sequencing using the same amplification primers was commercially performed by Sangon Biotech (Shanghai) Co., Ltd. and Beijing Aoke Biotechnology Co., Ltd.
Sequence assembly and analysis
Contig Express program was used to produce contigs from the forward and reverse reads of each COI amplicon and correct each read by looking at the chromatogram. Primer sequences from each contig were removed within the Contig Express program also. DNAMAN 7.0.2 software was used for DNA multiple sequences alignment. 110 amplicon sequences were aligned, among which 9 from GenBank and 101 from laboratory. Haplotypes were identified using the DnaSP v.5.1 software16. Pairwise genetic distances for COI genes were computed with the Kimura 2-parameter method (K2P). Neighbour-joining (NJ) phylogenetic trees were constructed in MEGA 6.017, and distance histograms were generated with the online version of automatic barcode gap discovery (ABGD)18. All of the identified haplotypes were submitted to GenBank.
Specific primer design, selection and sensitivity test
According to the variability of the partial COI gene sequences from six Tribolium species, suitable areas for designing species-specific primers were identified with Bioedit (version 7.2.0), and species-specific primers for Tribolium identification were designed with Primer Premier 5.0. Primer pairs were evaluated according to eight factors: (1) length between 18 bp to 30 bp; (2) absolute value of Delta G less than 9; (3) 3′-end contains one or more specific bases; (4) no distinct hairpin structure; (5) GC% from 30% to 70%; (6) primers for distinguishing different species; (7) false priming less than 100%; and (8) optimal annealing temperatures. All of the primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Specificity testing with each primer pair in the PCR assays was performed using 18 selected samples (Table 1). PCR amplification in a final reaction volume of 25 μl consisted of 12.5 μl MasterMix with dye, 10.5 μl ddH2O, 0.5 μl specific forward primer, 0.5 μl specific reverse primer and 1 μl template DNA. The PCR cycler conditions used were an initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 54 °C for 30 s and 72 °C for 30 s with a final extension at 72 °C for 10 min. After separation by 1.5% agarose gel electrophoresis and staining in ethidium bromide, the products were confirmed under UV light (Gel Logic 212 PRO, Carestream Health, Inc.) and were sequenced in both directions by Beijing Aoke Biotechnology Co., Ltd.
Sensitivity testing with the selected six Tribolium species-specific primers was determined in PCR runs with a series of samples using decreasing DNA concentrations with the same primer concentration. The DNA concentrations used were 100, 10, 1, 0.1, 0.01 and 0.001 ng μl−1.
TaqMan probe and real-time PCR primer design, selection and sensitivity test
According to the partial COI gene sequences of six Tribolium species, suitable areas for specific primers were identified by Bioedit (version 7.2.0). TaqMan probes and real-time PCR primers were designed with Beacon Designer 8.12. TaqMan probes were evaluated according to seven factors: (1) length between 18 bp and 30 bp; (2) C% more than G%, otherwise complementary use; (3) GC content between 30% and 80%; (4) no G bases at the 5′ end; (5) avoided repeats of the same type of bases, especially four G bases encoded together; (6) probe should be as close as possible to the primers; and (7) no complementary secondary structures or primers.
Real-time PCR primers were evaluated according to six factors: (1) the length of a pair of primers was no more than four bases, with single base primers between 18 bp and 30 bp; (2) GC content between 40% and 60%; (3) no A bases as the first base on the 3′ end; (4) avoided repeat of the same type of base, especially four G bases encoded together; (5) did not use three G or C bases in a row at the 3′ end; (6) no complimentary primers.
Real-time quantitative PCR reactions were processed in 96-well plates in the PCR amplifier (ABI7500) using commercial Premix Ex Taq (rr390A) according to the manufacturer’s protocol. PCR amplification in a final reaction volume of 20 μl contained 10 μl Premix Ex Taq (Probe qPCR, 2X), 0.4 μl specific forward primer, 0.4 μl specific reverse primer, 0.8 μl TaqMan probe, 7.4 μl ddH2O, and 1 μl template DNA. The PCR cycler conditions used were an initial denaturation at 95 °C for 30 s, followed by 35 cycles of 95 °C for 5 s, 60 °C for 34 s and with a final extension at 72 °C for 10 min.
Sensitivity testing of the selected TaqMan probes for the six Tribolium species was determined in PCR runs with a series of samples using decreasing DNA concentrations with the same primer concentration. The DNA concentration series was 100, 10, 1, 0.1, 0.01 and 0.001 ng μl−1. Three replicates of each treatment were tested.
Results
DNA barcoding, species-specific PCR and real-time PCR accurately identified six stored-product pest Tribolium species including T. castaneum, T. confusum, T. destructor, T. madens, T. freemani and T. brevicornis.
DNA barcoding, genetic divergence and phylogenetic analysis
A 658-bp long region of the mtDNA COI gene was amplified from 101 individual Tribolium beetles (Table 1) using a set of universal COI primers. Alignment of these sequences, and nine additional Tribolium sequences from GenBank (Table 2), revealed six haplotypes were identified for T. castaneum, one haplotype was observed for T. confusum, T. destructor, T. madens, T. freemani and T. brevicornis. The sequences of common haplotype from different geographic populations have been submitted to GenBank, obtain 24 accession number (Table 1). The DNA multiple sequence alignment using DNAMAN 7.0.2 software showed that four bases A, G, C, T average content of these sequences, 30.04%, 22.45%, 16.22%, 31.29% respectively, A + T content 61.33%. Base composition of Tribolium sequences in line with insect mitochondrial genes.
K2P model calculation results using MEGA 6.0 showed that intra- and inter-specific genetic distance exists obvious difference. The inter-specific K2P divergence of the six Tribolium species averaged 19.61%, ranging from 16.34% to 22.28%, such as genetic distance of T. brevicornis and T. freemani reached 22.28%, T. castaneum and T. freemani only 16.34%. The intra-specific K2P divergence ranged from 0.00% to 1.86%, with an average of 0.5% (Table 3). Specimens of T. castaneum were characterized by K2P divergence values up to 1.86%, but less than 2.0%. Intra-specific divergences of other Tribolium species were all less than 1.0%. All inter-specific divergence values were greater than intra-specific values, more than 39 times. A favourable DNA barcode should have a higher divergence among species than within species. In the case of COI, the suggested standard divergence threshold value is ten times (10×) the mean intra-specific variation20.
The results of applying the ABGD algorithm to the COI data set are presented in Fig. 1. Distance values show a gap between the intra-specific and the inter-specific distances (Fig. 1a). The data set was partitioned into six groups when the prior assumption of maximum intra-specific divergence was set as high as 0.46% (Fig. 1b).
The NJ tree grouped the six morphologically identified Tribolium species based on the COI gene sequences as well as the outgroup species P. depressus (GenBank submission number KM450509) and P. subdepressus (KM452267) (Fig. 2). The resulting trees showed a clear clade of six Tribolium species distinct from the outgroup clades. There was high bootstrap support (100%) for the terminal branches at the species level.
Specific primer design, selection and sensitivity test
One hundred and one partial COI gene sequences (658 bp) from six adult species in the genus Tribolium were used to develop an accurate and fast method for identifying these six Tribolium species (Fig. 3). The specificity tests of the designed primers were performed by uniplex PCR. Six primer pairs were selected for the reliable identification of six Tribolium species and are listed in Table 4. These primer pairs were designed to identify different Tribolium species regardless of life stage. The results clearly demonstrated that each primer pair produced a species-specific band without any nonspecific bands (Fig. 4).
(a) Specific primers for T. destructor. (b) Specific primers for T. brevicornis. (c) Specific primers for T. madens. (d) Specific primers for T. freeman. (e) Specific primers for T. castaneum. (f) Specific primers for T. confusum. M: D2000 DNA Marker; 1: T. destructor (Prague); 2: T. brevicornis (York); 3: T. madens (Kansas); 4: T. freemani (Kansas); 5: T. castaneum (Prague); 6: T. castaneum (Rakovník); 7: T. castaneum (Osijek); 8: T. castaneum (Bordeaux); 9: T. castaneum (Kansas); 10,11: T. castaneum (guangxi); 12,13: T. castaneum (Xinjiang); 14: T. castaneum (Guangdong); 15: T. castaneum (Henan); 16: T. confusum (Prague); 17: T. confusum (Bordeaux); 18: T. confusum (Herink); 19: T. confusum (Kyjov); 20: T. confusum (Kansas); 21: Negative control (ddH2O).
Sensitivity of a selected specific primer set for each of the six species was determined using one sample from each species. In all of the species, DNA concentrations of 100, 10, 1, and 0.1 ng μl−1 resulted in strong intensity bands (Fig. 5).
TaqMan probe and real-time PCR primer design, selection and sensitivity test
Six TaqMan probe and primer pairs were selected for the reliable identification of six Tribolium species and are listed in Table 5. These primer and probe sets were designed to identify different Tribolium species regardless of their life stage. The results clearly demonstrated that each primer and probe set produced a species-specific band without any nonspecific bands (Fig. 6).
In the six Tribolium species, template concentrations lower than 0.01 ng μl−1 noticeably decreased the intensity of the visualized bands. Finally, the lower limit for detection was set at 0.01 ng μl−1 for T. destructor and T. freeman and 0.001 ng μl−1 for T. castaneum, T. confusum, T. destructor and T. madens (Fig. 7).
Discussion
Molecular biology techniques have been increasingly applied to species identification. In our study, a series of experiments based on DNA barcoding, species-specific primers were performed to identify six species of the genus Tribolium. The results showed that DNA barcoding technology, species-specific PCR and real-time PCR are useful for the rapid and accurate identification of six Tribolium species. Our data demonstrated that regardless of whether the specimen is an adult, larva or pupa of Tribolium, it is possible to extract sufficient DNA for DNA barcoding, species-specific PCR and real-time PCR for identification of the sample. We speculate that the universal primers for the mtDNA COI gene, the species-specific primers, and the TaqMan probe and primers sets for Tribolium can feasibly identify all of the ontogenetic stages.
The COI gene provides a robust DNA barcode for identifying the six species of stored-product Tribolium with non-overlapping genetic distances between intra- and inter-specific samples (Fig. 1a). The genetic distance between sequences provides an approach for ‘DNA barcode’ evaluation19. A favourable DNA barcode should have a higher divergence between species than within species. For COI, the suggested standard divergence threshold value is ten times (10X) the mean intra-specific variation20. In this study, the ratio between species is thirty nine times (39X) the within species variation. The NJ tree organized all of the six species determined by morphology by forming robust clades.
The PCR assay with species-specific primers clearly demonstrated that each primer pair produced a species-specific band without any nonspecific bands (Fig. 4). Compared with DNA barcoding technology, species-specific PCR does not require sequencing, only routine laboratory techniques such as DNA extraction, PCR and electrophoresis, so this method is convenient for any quarantine laboratory. The same PCR assay with species-specific primers can be performed within 3 h using unknown Tribolium species DNA.
Six probe and primer sets were designed and selected for the real-time PCR method, which was also able to distinguish the six Tribolium species. Compared with traditional PCR, the real-time PCR method can be assessed directly without melting curve analysis. The amount of fluorescence generated during the reaction directly reflects the number of amplicons in real-time gene copies. However, this method also has drawbacks, e.g., the relatively high price of real-time instruments, reaction kits and TaqMan probes.
Future work should focus on collecting more samples, screening commonly used molecular markers and developing multiplex PCR. In this study, three Tribolium species, T. destructor, T. brevicornis and T. madens, are only distinct populations geographically. Meanwhile, T. parallelus, T. thusa, and especially T. audax Halstead in North America and T. anaphe Hinton in central Africa4 have not been collected, although attempts to acquire these samples were made. Next, internally transcribed spacer (ITS) can possibly be used for Tribolium species identification, despite the COI gene providing a robust DNA barcode for identifying the six species of Tribolium stored-product pests. Finally, species-specific primers and real-time PCR have the potential to be implemented in multiplex PCR.
Additional Information
How to cite this article: Zhang, T. et al. DNA barcoding, species-specific PCR and real-time PCR techniques for the identification of six Tribolium pests of stored products. Sci. Rep. 6, 28494; doi: 10.1038/srep28494 (2016).
References
Nowaczyk, K. et al. Molecular techniques for detection of Tribolium confusum infestations in stored products. J. Econ. Entomol. 102, 1691–1695 (2009).
Lagisz, M., Wilde, K. E. & Wolff, K. The development of PCR-based markers for molecular sex identification in the model insect species Tribolium castaneum . Entomol. Exp. Appl. 134, 50–59 (2010).
Stejskal, V. & Hubert, J. Risk of occupational allergy to stored grain arthropods and false pest-risk perception in Czech grain stores. Ann. Agric. Environ. Med. 15, 29–35 (2008).
Angelini, D. R. & Jockusch, E. L. Relationships among pest flour beetles of the genus Tribolium (Tenebrionidae) inferred from multiple molecular markers. Mol. Phylogenet. Evol. 46, 127–141 (2008).
Nakakita, H. Rediscovery of Tribolium freemani Hinton: a stored product insect unexpected to entomologists for past 100 years. Jpn. Ag. Res. Q. 16, 239–245 (1983).
He, P. H., Zhang, T. et al. Species of stored grain insects and mites trapped by corrugated board method on the surface of grain bulk in granary. J. Henan University of Technology. 36, 50–54 (2015).
Hagstrum, D. & Subramanyam, B. Stored-product Insect Resource. AACC Press. St. Paul. p. 509 (2009).
Stejskal, V., Hubert, J., Aulickyv, R. & Kucerova, Z. Over-view of present and past and pest-associated risks in stored food and feed products: European perspective. J. Stored. Prod. Res. 64, 122–132 (2015).
Hinton, H. E. A synopsis of the genus Tribolium Magley, with some remarks on the evolution of its species-group (Coleoptera, Tenebrionidae). Bull. Entomol. Res. 39, 13–55 (1948).
Huang, C. G., Hsu, J. C., Haymer, D. S., Lin, G. C. & Wu, W. J. Rapid identification of the Mediterranean fruit fly (Diptera: Tephritidae) by loop-mediated isothermal amplification. J. Econ. Entomol. 102, 1239–1246 (2009).
Wang, Y. J. et al. DNA barcoding of five common stored product pest species of genus Cryptolestes (Coleoptera: Laemophloeidae). Bull. Entomol. Res. 104, 671–678 (2014).
Varadínová, Z., Wang, Y. J. & Li, Z. H. COI barcode based species-specific primers for identification of five species of stored-product pests from genus Cryptolestes (Coleoptera: Laemophloeidae). Bull. Entomol. Res. 105, 202–209 (2015).
Zhang, C. W., Xu, L. P., Lu, M., Liang, X. & Li, J. Rapid molecular identification of Tribolium destructor . Agric. Biotechnol. 4, 49–52 (2014).
Zhang, H. S., Feng, Z. J. & Cheng, C. Molecular identification of T. castaneum and T. confusum based on PCR-RFLP analyses of 28S rRNA gene. Environ. Entomol. 36, 171–175 (2014).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G. ABGD, automated barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877 (2012).
Meiklejohn, K. A., Wallman, J. F. & Dowton, M. DNA-based identification of forensically important Australian Sarcophagidae (Diptera). Int. J. Legal Med. 125, 27–32 (2011).
Hebert, P. D. N., Penton, E. H., Burns, J. M., Janzen, D. H. & Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA. 101, 14812–14817 (2004).
Acknowledgements
We thank Prof. Shengfang Zhang (Chinese Academy of Inspection and Quarantine) for the morphological identification of all of the Tribolium species in this study. We thank Dr. Susan Brown (Kansas State University) and Dr. Radek Aulicky (Crop Research Institute Prague) for sample collection. This work was supported by 3 Chinese programs, including the International Science and Technology Cooperation Program (No. 2013DFG32350), the Special Fund for Food Research in the Public Interest of China (No. 201513002-05), and the Research on Data Security and Encryption and Protection of Digital Specimen of Inspection and Quarantine (2015JK002). Zuzana Kucerova and Vaclav Stejskal were supported from the grant RO0414.
Author information
Authors and Affiliations
Contributions
Z.-H.L. designed this research. T.Z., Y.-J.W. and Z.-H.L. conducted experiments and analysis. T.Z., Y.-J.W., Z.-H.L., W.G., D.L., Z.K., V.S. and G.O. wrote the manuscript. Y.C., F.-J.L. and Y.W. provided their feedback on this research. T.Z., Y.-J.W., Z.-H.L.,V.S. and G.O. provided help in reviewing the scientific content and language in this manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, T., Wang, YJ., Guo, W. et al. DNA barcoding, species-specific PCR and real-time PCR techniques for the identification of six Tribolium pests of stored products. Sci Rep 6, 28494 (2016). https://doi.org/10.1038/srep28494
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28494
This article is cited by
-
Effect of high temperature on Wolbachia density and impact on cytoplasmic incompatibility in confused flour beetle, Tribolium confusum (Coleoptera: Tenebrionidae)
BMC Research Notes (2022)
-
Novel molecular identification methods for the larger black flour beetle, Cynaeus angustus (Coleoptera: Tenebrionidae)
Applied Entomology and Zoology (2020)
-
Sequencing of Tuta absoluta genome to develop SNP genotyping assays for species identification
Journal of Pest Science (2019)
-
A protocol for obtaining DNA barcodes from plant and insect fragments isolated from forensic-type soils
International Journal of Legal Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.


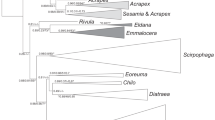







 T. destructor. (b)
T. destructor. (b)  T. brevicornis. (c)
T. brevicornis. (c)  T. madens. (d)
T. madens. (d)  T. freemani. (e)
T. freemani. (e)  T. castaneum. (f)
T. castaneum. (f)  T. confusum.
T. confusum.