Abstract
Streptomycetes are filamentous bacteria that produce numerous valuable compounds, including the majority of clinically used antibiotics. At an industrial scale, most of these compounds are produced in bioreactors. Growth of streptomycetes under these conditions is characterized by the formation of complex mycelial particles, whose sizes follow a bimodal distribution. Given the correlation between specific productivity and morphology, this size heterogeneity poses a potential drawback in industry. Recent work indicates that mycelial morphology is controlled by a number of genes that encode proteins required for the synthesis of cell surface-associated glycans. Using a quantifiable system based on fluorescent markers, we here show that these glycans mediate aggregation between germlings and young mycelia, yielding mycelial particles that originate from many different individuals. We also demonstrate that at later time points aggregation between distinct particles is no longer detectable. Notably, the absence of the corresponding glycan synthases yields mycelia that are homogeneous in size, identifying mycelial aggregation as a driving factor towards size heterogeneity. Given that aggregation is widespread within streptomycetes and can also occur between different Streptomyces strains, our work paves the way to improve Streptomyces as a cell factory for the production of known metabolites, but possibly also to discover new ones.
Similar content being viewed by others
Introduction
Streptomycetes are filamentous bacteria that are renowned for their ability to produce several types of bioactive compounds. Standing out among these are a plethora of antibiotics, but also numerous antiviral compounds, anticancer agents and molecules that suppress the immune system1,2. Due to their saprophytic lifestyle, streptomycetes also show a remarkable capacity to produce and efficiently secrete various hydrolytic enzymes that allow them to degrade almost any naturally occurring polymer3. The extensive industrial use of streptomycetes does not come as a surprise when these factors are taken into account.
In contrast to most other bacteria, streptomycetes grow by forming thread-like cells called hyphae4. These hyphae extend at their tip, while new hyphae emerge via subapical branching of pre-existing ones. This mode-of-growth leads to the construction of complex networks of interconnected hyphae called mycelia. When grown in liquid, these mycelia display markedly different morphologies5. Some strains, including the industrial workhorse Streptomyces lividans, predominantly form dense mycelial particles called pellets. In contrast, other strains form so-called mats, which are loosely entangled and scarcely dense mycelial networks5. Notably, a strong correlation exists between morphology and productivity in streptomycetes5,6,7. While growth as pellets is preferred for the production of antibiotics, it is suboptimal for the production of enzymes. For efficient enzyme production mycelial mats are more suitable, which relates to the fact that nutrients are more easily accessible to all hyphae. In fact, hyphae in the central part of pellets often suffer from nutrient stress leading to a phase of controlled cell death8,9. To further complicate matters, recent work from our lab demonstrated that liquid-grown Streptomyces cultures are heterogeneous and contain at least two populations of mycelial particles that differ in size10. This size heterogeneity was observed across a range of different streptomycetes and growth media, suggesting that it has a genetic basis.
In the past few years, several genetic determinants involved in mycelial architecture have been identified in S. lividans. These include among others the recently discovered mat genes11, as well as cslA, glxA and dtpA12,13,14. The common feature of these genes is that they encode proteins that are involved in the synthesis of extracellular glycans. Failure to synthesize these glycans abolishes pellet formation and leads to mycelia with an open, mat-like morphology5. This indicates that extracellular glycans play a pivotal role in shaping Streptomyces pellets, although the underlying mechanism is not known yet. One of the outstanding questions is whether these glycans solely mediate hypha-hypha interactions within individual particles or whether they also mediate aggregation of hyphae between distinct mycelia. The latter process, hereinafter referred to as aggregation, generates larger mycelial structures composed of different individuals. Aggregation has been well characterized in filamentous fungi, such as the ascomycete Aspergillus niger15. Here, aggregation was shown to be a two-step process that starts immediately after inoculation with the clustering of conidia. A second aggregation step coincides with the onset of germination, during which groups of germinated conidia aggregate to give larger particles. In streptomycetes, the clumping of mycelial particles has been noticed in Streptomyces aureofaciens16, although the extent of aggregation and factors involved in this process have remained obscure.
In this study we show that aggregation is restricted to the very early stages of growth, leading to the formation of mycelial networks that originate from different individuals. A qualitative and quantitative image analysis approach using fluorescent reporter strains demonstrated that aggregation depends on cslA, glxA and matAB. Excitingly, the absence of these genes yields mycelial particles that originate from individual spores and which, as a consequence, leads to a homogeneously distributed population. We also show that aggregation in streptomycetes can occur between different wild-type species. Taken together, our data provides the basis to enhance the industrial exploitation of these prolific bacteria.
Results
A quantifiable system to analyse particle aggregation in Streptomyces lividans
To study aggregation in liquid-grown cultures, fluorescent strains of S. lividans 1326 were created that constitutively express eGFP or mCherry by placing these genes under control of the gap1 promoter of S. coelicolor. The reporter strains were co-cultivated in TSBS or NMMP medium, resulting in the formation of pellets10. When spores of both fluorescent strains were mixed, pellets obtained after 24 h of growth were invariably composed of green and red fluorescent hyphae (Fig. 1A,B). Aggregates of fluorescent germlings were already visible 6 h after inoculation (Fig. 1C,D), indicating that aggregation is an early event (see below). Quantification of aggregation using the COPAS, a flow cytometer for large particles17, revealed that >99.8% of pellets contained both red and green fluorescent hyphae, irrespective of the used medium (Fig. 1E, Table 1).
Analysis of aggregation of fluorescent S. lividans strains.
Spores of strains expressing eGFP (S. lividans pGreen) or mCherry (S. lividans pRed) were mixed at the onset of growth in NMMP (A,C) or TSBS (B,D). After 24 h of growth, pellets are composed of both types of fluorescent hyphae (A,B). Aggregates of fluorescent germlings are already detected 6 hours after inoculation (C,D). (E) Quantitative analysis of aggregation using particle sorting. Each plot represents the fluorescence intensities of pellets in the red (X-axis) and green (Y-axis) channel. The top two panels show the green:red ratios of pellets from the eGFP-expressing strain in NMMP (left) and TSBS (right) medium, while the middle two panels reveal the green:red ratios of pellets from the strain expressing only mCherry. The bottom panels represent the fluorescence intensities of pellets from the co-cultivations of both fluorescent strains, which indicate that almost all pellets express both reporters. Scale bars represent 100 μm (A,B) or 20 μm (C,D).
Aggregation is mediated via germlings
The early stages of aggregation of S. lividans were monitored using light microscopy (Fig. 2). In the first 2 h after inoculation, only individual spores were detected in the medium (Supplementary Fig. S1). Germination of spores after 4 h coincided with small aggregates that increased in size in time. Germ tubes were consistently visible in these aggregates (see magnification Fig. 2 and Supplementary Fig. S2), implying that they are required for aggregation. In agreement, individual spores that had not initiated germination were not part of aggregates (Supplementary Fig. S3).
Microscopy analysis of early stages of growth and aggregation.
Wild-type spores of S. lividans were inoculated in TSBS medium. Only individual spores (arrowheads) are observed 2 h after inoculation (A). Aggregation becomes visible after 4 h of growth (B) coinciding with the appearance of germ tubes (see also panel F). Further aggregation and growth leads to the formation of particles that increase in size over time (C–E). Panel (F) shows a magnification of the aggregate visible in panel (B). Arrows indicate germ tubes. Scale bars represents 20 μm (A–E) or 5 μm (F).
To more closely study whether aggregation specifically involves germlings or can also occur between mature mycelia, co-cultivations were performed of the red and green fluorescent strains that had been grown separately for 2, 4, 6, 8, 10 or 12 h. Analysis of the mixed cultures 24 h after the inoculation of spores indicated that virtually all pellets were composed of red and green fluorescent hyphae if the strains were mixed after 2, 4, 6 and 8 hours of growth (Fig. 3, Table 1). Conversely, when the strains were mixed after 10 or 12 h, aggregation became increasingly rare, as most pellets were only red or green fluorescent (Table 1). Notably, while pellets contained intertwined red and green fluorescent hyphae when the cultures were mixed after 2 h of growth, a patched pattern of green and red mycelial parts was observed in pellets when the cultures were mixed after 8 h of growth (Supplementary Fig. S4). To exclude that compounds accumulating in the culture medium reduced aggregation at late time points, we mixed washed mycelia of the fluorescent strains in fresh medium after 12 hours of growth. After washing, 3.5 ± 0.6% of particles aggregated in NMMP medium, while in TSBS medium 16.1 ± 6.1% of particles contained both fluorescences (Supplementary Fig. S5). Given that these values are similar to those observed without washing, these data indicate that aggregation is not hampered by compounds accumulating in the culture broth.
Quantitative analysis of time-dependent aggregation.
Fluorescence profiles of 24-hour-old pellets obtained from co-cultures of S. lividans pRed and S. lividans pGreen that had initially been grown separately for 2, 4, 6, 8, 10, or 12 h. Each plot represents the fluorescence intensities of pellets in the red (X-axis) and green (Y-axis) channel determined by particle analysis. The growth media were NMMP (top) and TSBS (bottom). Note that aggregation is strongly reduced when the fluorescent strains have initially been grown separately for more than 8 h.
Genes involved in glycan synthesis are required for germling aggregation
Given the crucial role of extracellular glycans in pellet morphology, we studied whether these polymers also mediate germling aggregation. We therefore created reporter strains of the S. lividans cslA, glxA and matAB mutants, all of which are affected in glycan biosynthesis11,12. Aggregation of the cslA and glxA mutants was strongly affected in both NMMP and TSBS, as indicated by the presence of particles that only showed green or red fluorescence (Fig. 4, Supplementary Figs S6–S9). Quantification revealed that 20.2 ± 3.6% and 3.5 ± 0.7% of the particles of the cslA mutant contained both red and green fluorescent hyphae in NMMP and TSBS medium, respectively. These values were comparable to those of the glxA mutant, where 23.0 ± 3.9% (TSBS) and 0.57 ± 0.2% (NMMP) of all particles were aggregated. Aggregation in the matAB mutant was also greatly reduced (Fig. 4), with only 4.8 ± 1.7% (NMMP) and 2.3 ± 0.2% (TSBS) of particles displaying both red and green fluorescent hyphae. These results indicate that proteins involved in glycan biosynthesis play a crucial role in the aggregation of germlings.
Aggregation depends on glycan synthases.
Microscopy images of particles obtained 24 h after mixing spores of red- or green-fluorescent derivatives of the wild-type (WT), cslA, glxA or matAB mutant strains. Note that the majority of particles only have either green or red fluorescence in the absence of cslA, glxA, or matAB in both NMMP (top) and TSBS (bottom). Scale bar represents 100 μm (A,E) or 200 μm (B–D, F–H).
Considering that the majority of aggregation occurs within the first 12 h of growth (see above), we asked whether expression of the genes involved in glycan biosynthesis was changed after 12 h of growth. We therefore isolated RNA from mycelia grown in NMMP and TSBS medium after 12 and 24 h and performed RT-PCR using primers directed against cslA, glxA, matA and matB. Expression of these genes was detected in both media at both time points (Supplementary Fig. S10). This implies that the reduced aggregation after 12 hours is probably not caused by changes in the abundance of these glycan synthases.
Germling aggregation leads to size heterogeneity in a medium-dependent manner
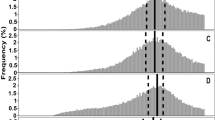
Mycelia of the wild-type strain and its cslA, glxA and matAB mutant derivatives were analysed using automated image analysis to assess how aggregation affects the size distribution of mycelial particles. This approach enabled the analysis of particles that are too small for detection with the COPAS (see Methods). Mathematical analysis revealed that the size distributions of all strains grown for 12 h in NMMP was better explained by assuming the presence of at least two populations of particles that differ in size (Fig. 5A, Table 2). The size distribution of the wild-type strain was very similar to that of the cslA and matAB mutants, in which the number of small particles exceeded that of the bigger particles, as deduced from the skewed participation fractions (Table 2). In contrast, the glxA mutant in NMMP medium was more heterogeneous in size, with 60.7% of the particles being part of the population of small particles. Notably, the degree of heterogeneity in the size of wild-type pellets was considerably higher in TSBS medium compared to NMMP (Fig. 5B). This is concluded from the fact that in this case 38% and 62% of the particles belonged to the population of small and large particles, respectively (Table 2). In contrast, TSBS-grown cultures of the cslA, glxA and matAB deletion strains revealed a dramatic reduction in particle size heterogeneity. In all cases, the population of small particles accounted for the majority of mycelia in these cultures, with participation fractions of 93%, 99% and 84% for the cslA, glxA and matAB deletion strains, respectively (Table 2). Taken together, these results demonstrate that aggregation leads to heterogeneity in a medium-dependent manner.
Quantitative analysis of particle size heterogeneity in the absence of cslA, glxA and matAB.
Normalized particle size distributions of the S. lividans wild-type strain, the cslA deletion strain, the glxA deletion strain and the matAB deletion strain in NMMP (A) and TSBS (B) after 12 h of growth. The x-axis represents the normalized Feret diameter, while the y-axis represents the relative abundance. The two dashed lines in each plot represent the two normally distributed populations of particles fitted to the actual measured distribution (solid line). Size heterogeneity is strongly diminished in the absence of the cslA, glxA and matAB genes in TSBS medium.
Mycelial aggregation of different streptomycetes yields multi-species pellets
To determine whether aggregation is a common trait in streptomycetes, spores of different wild-type strains, i.e. S. lividans, S. coelicolor, S. scabies and S. albus, were inoculated in NMMP medium, after which the cultures were analysed via light microscopy. Aggregates of germinated spores were detected in all strains 6 h after inoculation (Fig. 6). In the case of S. lividans and S. coelicolor the aggregates appeared rather compact, unlike those of S. scabies and S. albus that had a more open morphology. To assess whether aggregation also occurs between different species, fluorescent wild-type strains of S. lividans, S. coelicolor and S. scabies were co-cultured in NMMP and in TSBS. The strains of S. coelicolor and S. scabies used in this experiment expressed eGFP and mCherry respectively, whereas S. lividans was engineered to produce both fluorescent proteins and was false-colored cyan. Notably, all pellets after 24 h of growth were composed of the three different strains, irrespective of the medium (Fig. 7). These results not only indicate that germling aggregation is common in streptomycetes, but also shows that it occurs between different Streptomyces species.
Formation of multi-species Streptomyces pellets.
Germlings of S. coelicolor M512 (green), S. scabies (red) and S. lividans (false-colored in cyan) aggregate to form multi-species pellets. The image shows a representative pellet obtained after 24 h of growth of the three strain in NMMP medium. Scale bar represents 100 μm.
Discussion
Streptomycetes are among the biggest players in the fermentative production of antibiotics, hydrolytic enzymes and numerous other compounds with biological activity. The hallmark of the submerged growth of these microorganisms, the preferred mode of growth under industrial settings, is the formation of interconnected clumps of mycelium with varying sizes10,18,19,20. The constraints imposed by this heterogeneity are most evident when one takes account of the strong link between morphology and production, which is a well-established concept in the Streptomyces field5,6,7. We here show that the aggregation of germlings is an important factor leading to size heterogeneity. Our findings open new avenues to improve Streptomyces as a cell factory in the biotech industry.
Aggregation in Streptomyces
In this work we present direct evidence for aggregation of independent particles in liquid-grown Streptomyces cultures. Whereas we mainly focus on the model strain S. lividans to provide insight into this phenomenon, we show that aggregation is common to a wide range of streptomycetes and also occurs between the mycelia of different co-cultured Streptomyces species. About two-thirds of all Streptomyces genomes contain the matAB genes11, including all species used in this work, whereas the cslA/glxA operon is present in all streptomycetes21. The broad conservation of these genes across the streptomycetes could explain why aggregation of germlings is so common within this genus. Unlike in filamentous fungi such as Aspergillus niger and Aspergillus fumigatus22,23, in streptomycetes we did not find evidence for aggregation of ungerminated spores. Spores of streptomycetes are covered with amyloid fibrils that render the spore surface hydrophobic24,25. Apparently, this hydrophobicity is not a driving factor for aggregation, as aggregation was only detected after the visible emergence of germ tubes. The proteins that make up these amyloidal structures at the spore surface, called rodlins and chaplins, were previously shown to mediate attachment to abiotic surfaces25,26.
As aggregation of germlings proceeds, aggregates may also connect to one another leading to the formation of larger mycelial particles. Interestingly, the greater part of aggregation takes place between 4 and 10 h after the inoculation of the spores. These aggregation dynamics are in line with observations performed in a pioneering study on S. aureofaciens, which displayed aggregated filaments after approximately 8 h of growth16. Given that aggregation is not impeded by molecules accumulating in the culture medium and that the glycans involved in aggregation are constitutively produced, we assume that this time-restricted process is caused by the size of the interacting particles, which may become too large to remain connected due to increasing collision forces or shear stress imposed upon them.
As shown in this work, for aggregation to occur between different germlings, extracellular glycans associated with the cell surface are of crucial importance. Notably, aggregation in filamentous fungi is also mediated by glycans. In A. fumigatus, for instance, an α-(1–3) glucan becomes exposed upon swelling of spores and mediates conidial aggregation22. In S. lividans, the glycans produced by CslA and MatAB are key contributors mediating aggregation. The most dramatic phenotype in the absence of cslA and the matAB genes is observed in TSBS-grown cultures, where aggregation was largely abolished in both deletion mutants. Conversely, in NMMP-grown cultures aggregated particles were detected, although significantly fewer than for the parental strain. These results suggest that other adhesives may contribute to aggregation in NMMP-grown cultures. Differences in the composition of a so-called extracellular matrix under various conditions is in fact not uncommon and has also been observed in other bacteria during the formation of biofilms27,28. A wide range of molecules normally composes the extracellular matrix of bacteria, which in addition to glycans may contain proteins, lipids and nucleic acids4,29,30,31. In this respect it is interesting to mention that pellet formation resembles the formation of a free-floating biofilm or floc14,32. In agreement with this, a previous report suggested that extracellular DNA is involved in maintenance of pellet morphology in S. coelicolor33. Besides providing the first direct indication of aggregation between distinct particles in streptomycetes, our work also demonstrates that this phenomenon can occur between different Streptomyces species, resulting in the formation of multi-species pellets. These “synthetic” communities may be useful for the discovery of novel metabolites. Several recent examples indicate that co-cultivations may lead to increased levels of known secondary metabolites, but also to the production of novel compounds34,35,36,37. Notably, physical adhesion was found to be essential to elicit the biosynthesis of interaction-specific compounds in co-cultures of Aspergillus nidulans and Streptomyces hygroscopicus34. We anticipate that the use of aggregation between germlings of different streptomycetes may represent a useful strategy to discover novel compounds. We are currently using this approach to study whether novel metabolites can be produced by these synthetic communities.
Factors leading to size heterogeneity
Our results highlight the importance of extracellular glycans for the aggregation of independent particles, which ultimately provides the basis for mycelial size heterogeneity in Streptomyces. Germling aggregation leads to a range of particles that not only increase in size due to growth, but also due to the incorporation of germlings or small mycelia into pre-existing particles. In addition, larger-sized particles have an increased chance to capture new germlings by accidental collision, thereby further skewing the differences between small and large particles. In contrast, particles of the cslA and matAB mutant strains only increase in size due to growth, which may explain why they are more homogeneous in size. The fact that mycelia likely grow faster in rich medium can explain why pellets of the S. lividans wild-type strain are earlier heterogeneous in size in TSBS medium, compared to the minimal NMMP medium. However, we also cannot exclude that other surface-associated molecules that are differentially produced in both conditions may contribute to generating size heterogeneity.
One surprising outcome was the observation that the glxA mutant strain is heterogeneous in size in NMMP medium after 12 h of growth, while all other strains were homogenous. More specifically, the fraction of large particles in the glxA mutant was considerably larger compared to that of the cslA mutant (Fig. 5 and Supplementary Fig. S11). Notably, these larger particles were significantly denser than those being part of the population of small particles (Supplementary Fig. S11). One main difference between the glxA and cslA mutant is the fact that the glxA mutant is still capable of synthesizing the glycan produced by CslA, on which GlxA would normally act. Perhaps this residual glycan, although not processed by GlxA, is still capable of inducing some mycelial aggregation leading to the formation of denser particles that comprise the observed second distinct population.
Opposite to aggregation, another factor that directly influences size heterogeneity is fragmentation. By fragmentation we mean the detachment of viable mycelial parts from existing particles. This process could be particularly significant for strains such as the cslA and matAB mutants, which no longer form dense pellets and as a consequence are more susceptible to shear stress. Such detached fragments directly lead to a change in the distribution of particles in the culture. By making use of the fluorescent strains described in this paper, we have gathered preliminary evidence for fragmentation (B. Zacchetti and D. Claessen, unpublished results). Whether this process already occurs within the 12-hour timeframe described here remains to be elucidated. We hypothesize that these combined factors provide the basis for the formation of two distinct populations of pellets. We are now building a mathematical model to gain more insight into how these complex processes lead to heterogeneity.
As mentioned previously, a strong link exists between mycelial morphology and production of secondary metabolites and enzymes in streptomycetes. The observation that the deletion of cslA and the matAB genes not only reduces the size of mycelial particles, but also their heterogeneity makes them interesting targets to improve enzyme production. In agreement with this, we recently showed that a S. lividans strain lacking the matAB genes produces more tyrosinase than the wild-type strain11. Given the propensity of many streptomycetes to aggregate and grow heterogeneously, we expect that targeting these processes may be valuable for production purposes in other Streptomyces species.
Methods
Strains and culture conditions
The E. coli and Streptomyces strains used in this study are listed in Supplementary Table S1. E. coli strains were grown at 37 °C in LB medium, supplemented with antibiotics if necessary. For growth of E. coli strains carrying a plasmid with an hygromycin resistance cassette, modified LB medium was used, which contains 1.25 gl−1 NaCl instead of 10 gl−1. Streptomyces strains were grown at 30 °C on solid MS agar plates. For growth in liquid, 250 ml flasks equipped with coils were used, each containing 100 ml TSBS medium (TSB + 10% sucrose) or NMMP medium with glucose as carbon source38. Cultures were inoculated with 106 spores ml−1 (both when single or two strains were used) and were grown at 30 °C in an orbital shaker at 180 rpm.
Constructs and transformation
Constructs and oligonucleotides used in this work are listed in Supplementary Table S2 and Supplementary Table S3, respectively. To create pGreen, the promoter region of gap1 (SCO1947) of S. coelicolor A3(2) M145 was amplified via PCR as a BglII-NdeI 450 bp fragment using primers Gap1-FW and Gap1-RV. This promoter has a strong constitutive activity in S. lividans (Mangiameli and Vijgenboom, unpublished). The promoter fragment was ligated as a BglII-NdeI fragment into pIJ8630, containing the eGFP gene39. To create pRed, the integrative vector pMS82 was used, which carries an hygromycin resistance cassette40. The gap1 promoter was amplified via PCR using primers Gap1-FW* and Gap1-RV, thereby introducing KpnI and NdeI sites. The mCherry gene was amplified as an NdeI-HindIII fragment using vector pRSET-B as a template [R. Tsien] and primers mCherry-FW and mCherry-RV. The two PCR products were ligated in vector pIJ292541, after which the gap1-mCherry fragment was excised with KpnI and HindIII and blunted with T4 polymerase, according to the manufacturer’s protocol. The fragment was subsequently cloned into the EcoRV site of pMS82. Restriction analysis and sequencing confirmed the insertion and orientation of the fragment, with the mCherry coding sequence running in the opposite orientation relative to the hygromycin resistance gene. The reporter constructs were introduced in Streptomyces via conjugation as described38.
RNA Isolation and RT-PCR
Total RNA was isolated in duplicate from mycelia grown for 12 or 24 h in TSBS or NMMP medium using the RNeasy Mini Kit in combination with the RNAprotect Bacteria Reagent (Qiagen), according to the instructions of the manufacturer. Following RNA extraction, 250 ng of RNA was used to generate cDNA followed by PCR amplification using the SuperScript III One-Step RT-PCR System (Invitrogen). The program for cDNA synthesis and amplification was as follows: cDNA synthesis (55 °C, 30 min); 30 cycles of 94 °C for 15 s (denaturation), 60 °C for 30 s (annealing) and 68 °C for 30 s (extension), followed by a final extension for 5 min at 68 °C. Specific primers for the RT-PCR are shown in Supplementary Table S3. As a control, primers directed against the 16S rRNA were used as described previously42.
Flow cytometry
5 ml samples were harvested from liquid NMMP or TSBS cultures and fixed in 4% formaldehyde for 20 min on ice. Biomass was pelleted at 1500 rpm for 10 min, washed twice with phosphate-buffered saline (PBS) and stored at −20° until further use. Particles were analysed using a COPAS Plus profiler (Union Biometrica) equipped with a 1 mm nozzle as described10,43, excluding data with an extinction below 25, which correspond to hyphal fragments and debris. All experiments were performed in duplicate and for each replicate at least 10,000 pellets were analysed per sample.
To calculate the number of pellets expressing both eGFP and mCherry, the green to red ratio was calculated of all pellets in the single cultivations of the fluorescent strains. The lowest value in the eGFP-expressing strain and the highest value in the mCherry-expressing strain were used as boundaries between which pellets were considered to express both reporter proteins. For fluorescence measurements, the photomultipliers were set at 800 and 900 Volts to detect the green and red signals, respectively. To compensate for the fluorescence bleed-through, the red signal was multiplied by a factor of 0.04 and subtracted from the green signal. The opposite was done for the red signal, with the green signal being multiplied by a factor of 0.05.
Microscopy
Confocal microscopy was performed using a Zeiss Observer Microscope. Samples were excited with laser light at wavelengths of 488 and 543 nm to detect eGFP and mCherry, respectively. Fluorescence emissions were monitored in the region between 505–545 nm for eGFP, while a 560 nm longpass filter was used to detect mCherry. Capture settings were such that no bleed through could be detected. The pinhole size was adjusted to one airy unit (94 micron) for optimal contrast images which results in 4.5 μm thick coupes. All detailed fluorescence microscopy pictures (i.e. those in Figs 1, 4 and 7) represent Z-projections of stacks with a thickness of 2.2 μm (optimized for optical section thickness).
To quantify aggregation in mutants of S. lividans, whole microscopy wells (1 μ-Slide 2 × 9 well, Ibidi GmbH) containing samples of co-cultivations were imaged as a mosaic of 5 × 5 tiles. The eGFP and mCherry fluorescence emissions were merged to give 8-bit greyscale pictures that were used to detect regions containing fluorescent particles. The mean green and red fluorescence component were subsequently measured in the corresponding regions in RGB pictures. Data were used to quantify the number of particles showing green and/or red fluorescence. Particles with a green to red emission ratio between 1.2 and 0.15 were counted as aggregated, while those with a ratio above 1.2 or below 0.15 were counted as green or red, respectively. For each sample, at least 500 particles were analysed in duplicate. Examples of the images used for quantification are presented as Supplementary Figs S6–S9.
Light microscopy was performed using a Zeiss Axioscope A1 upright microscope with an Axiocam MRc5 camera at a resolution of 37.5 nm/pixel. Automated acquisition of pictures for size analysis of streptomycetes was performed using a brightfield Zeiss Observer microscope.
Size distribution analysis
12 hours after the inoculation of spores, 10 ml samples were harvested from the culture flasks. The biomass was concentrated via centrifugation at 1000 rpm for 30 mins at 4 °C and resuspended in Phosphate Buffer Saline (PBS). Micrographs of samples were obtained via automated image acquisition and analysed using ImageJ (Joost Willemse, Ferhat Büke, manuscript in preparation). For size distribution analysis, the maximum Feret diameter was taken as the value representing the size of particles. For each measurement, 1000 particles were analysed to provide a sufficiently large statistical pool for further analysis. Incorrectly analysed particles (e.g. out-of-focus mycelia) were manually removed from the datasets. The size distribution analysis was performed as described44. Briefly, the datasets representing the normalized maximum Feret diameter of particles were fit by a probability distribution assuming two normal distributions. This model determines five parameters: the participation fraction (p), two means (μ1; μ2) and two standard deviations (σ1; σ2). For each parameter, the 95% confidence interval (CI) estimate was obtained by refitting with the model after bootstrapping (1000 replicates) using the open source Scilab language. Datasets with non-overlapping CIs of the mean and 0.025 < p < 0.975 were considered to be derived from a culture with two populations of pellets.
Additional Information
How to cite this article: Zacchetti, B. et al. Aggregation of germlings is a major contributing factor towards mycelial heterogeneity of Streptomyces. Sci. Rep. 6, 27045; doi: 10.1038/srep27045 (2016).
References
Hopwood, D. A. Streptomyces in nature and medicine: the antibiotic makers (Oxford University Press, USA, 2007).
Barka, E. A. et al. Taxonomy, physiology and natural products of Actinobacteria. Microbiol Mol Biol Rev 80, 1–43, 10.1128/MMBR.00019-15 (2016).
Vrancken, K. & Anné, J. Secretory production of recombinant proteins by Streptomyces. Future Microbiol 4, 181–188, 10.2217/17460913.4.2.181 (2009).
Claessen, D., Rozen, D. E., Kuipers, O. P., Søgaard-Andersen, L. & van Wezel, G. P. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12, 115–124, 10.1038/nrmicro3178 (2014).
van Dissel, D., Claessen, D. & van Wezel, G. P. Morphogenesis of streptomyces in submerged cultures. Adv Appl Microbiol 89, 1–45, 10.1016/B978-0-12-800259-9.00001-9 (2014).
Wardell, J. N., Stocks, S. M., Thomas, C. R. & Bushell, M. E. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol Bioeng 78, 141–146 (2002).
van Wezel, G. P. et al. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol 72, 5283–5288, 10.1128/AEM.00808-06 (2006).
Manteca, A., Alvarez, R., Salazar, N., Yagüe, P. & Sanchez, J. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl Environ Microbiol 74, 3877–3886, 10.1128/AEM.02715-07 (2008).
Rioseras, B., López-García, M. T., Yagüe, P., Sánchez, J. & Manteca, A. Mycelium differentiation and development of Streptomyces coelicolor in lab-scale bioreactors: Programmed cell death, differentiation and lysis are closely linked to undecylprodigiosin and actinorhodin production. Bioresour Technol 151C, 191–198, 10.1016/j.biortech.2013.10.068 (2013).
van Veluw, G. J. et al. Analysis of two distinct mycelial populations in liquid-grown Streptomyces cultures using a flow cytometry-based proteomics approach. Appl Microbiol Biotechnol 96, 1301–1312, 10.1007/s00253-012-4490-5 (2012).
van Dissel, D., Claessen, D., Roth, M. & van Wezel, G. P. A novel locus for mycelial aggregation forms a gateway to improved Streptomyces cell factories. Microb Cell Fact 14, 44, 10.1186/s12934-015-0224-6 (2015).
Chaplin, A. K. et al. GlxA is a new structural member of the radical copper oxidase family and is required for glycan deposition at hyphal tips and morphogenesis of Streptomyces lividans. Biochem J 469, 433–444, 10.1042/BJ20150190 (2015).
Petrus, M. L. C. et al. The DyP-type peroxidase DtpA is a Tat-substrate required for GlxA maturation and morphogenesis in Streptomyces. Open Biol 6, 10.1098/rsob.150149 (2016).
Petrus, M. L. C. & Claessen, D. Pivotal roles for Streptomyces cell surface polymers in morphological differentiation, attachment and mycelial architecture. Antonie van Leeuwenhoek 106, 127–139 (2014).
Grimm, L. H. et al. Kinetic studies on the aggregation of Aspergillus niger conidia. Biotechnol Bioeng 87, 213–218, 10.1002/bit.20130 (2004).
Tresner, H. D., Hayes, J. A. & Backus, E. J. Morphology of submerged growth of streptomycetes as a taxonomic aid. I. Morphological development of Streptomyces aureofaciens in agitated liquid media. Appl Microbiol 15, 1185–1191 (1967).
de Bekker, C., van Veluw, G. J., Vinck, A., Wiebenga, L. A. & Wösten, H. A. B. Heterogeneity of Aspergillus niger microcolonies in liquid shaken cultures. Appl Environ Microbiol 77, 1263–1267, 10.1128/AEM.02134-10 (2011).
Nieminen, L., Webb, S., Smith, M. C. M. & Hoskisson, P. A. A flexible mathematical model platform for studying branching networks: experimentally validated using the model actinomycete, Streptomyces coelicolor. PLOS One 8, e54316, 10.1371/journal.pone.0054316 (2013).
Reichl, U., King, R. & Gilles, E. D. Characterization of pellet morphology during submerged growth of Streptomyces tendae by image analysis. Biotechnol Bioeng 39, 164–170, 10.1002/bit.260390207 (1992).
Tough, A. J. & Prosser, J. I. Experimental verification of a mathematical model for pelleted growth of Streptomyces coelicolor A3(2) in submerged batch culture. Microbiology 142, 639–648 (1996).
Liman, R., Facey, P. D., van Keulen, G., Dyson, P. J. & Del Sol, R. A laterally acquired galactose oxidase-like gene is required for aerial development during osmotic stress in Streptomyces coelicolor. PLoS One 8, e54112, 10.1371/journal.pone.0054112 (2013).
Fontaine, T. et al. Cell wall α(1-3) glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Genet Biol 47, 707–712, 10.1016/j.fgb.2010.04.006 (2010).
Metz, B. & Kossen, N. W. F. The growth of molds in the form of pellets – a literature review. Biotechnol Bioeng 19, 781–799 (1977).
Claessen, D. et al. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol 53, 433–443, 10.1111/j.1365-2958.2004.04143.x (2004).
Claessen, D. et al. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol 44, 1483–1492 (2002).
de Jong, W., Wösten, H. A. B., Dijkhuizen, L. & Claessen, D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol 73, 1128–1140, 10.1111/j.1365-2958.2009.06838.x (2009).
Cairns, L. S., Hobley, L. & Stanley-Wall, N. R. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol 93, 587–598, 10.1111/mmi.12697 (2014).
Uhlich, G. A., Chen, C. Y., Cottrell, B. J. & Nguyen, L. H. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett 354, 133–141, 10.1111/1574-6968.12439 (2014).
Speziale, P., Pietrocola, G., Foster, T. J. & Geoghegan, J. A. Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol 4, 171, 10.3389/fcimb.2014.00171 (2014).
Barnes, A. M., Ballering, K. S., Leibman, R. S., Wells, C. L. & Dunny, G. M. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio 3, e00193–00112, 10.1128/mBio.00193-12 (2012).
Hobley, L., Harkins, C., MacPhee, C. E. & Stanley-Wall, N. R. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39, 649–669, 10.1093/femsre/fuv015 (2015).
Martins, A. M., Picioreanu, C., Heijnen, J. J. & Van Loosdrecht, M. C. M. Three-dimensional dual-morphotype species modeling of activated sludge flocs. Environ Sci Technol 38, 5632–5641 (2004).
Kim, Y. M. & Kim, J. H. Formation and dispersion of mycelial pellets of Streptomyces coelicolor A3(2). J Microbiol 42, 64–67 (2004).
Schroeckh, V. et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA 106, 14558–14563,10.1073/pnas.0901870106 (2009).
Wu, C. et al. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci Rep 5, 10868, 10.1038/srep10868 (2015).
Abrudan, M. I. et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA, doi: 10.1073/pnas.1504076112 (2015).
Moody, S. C. Microbial co-culture: harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol 9, 575–578,10.2217/fmb.14.25 (2014).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces genetics (The John Innes Foundation, 2000).
Sun, J., Kelemen, G. H., Fernández-Abalos, J. M. & Bibb, M. J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227 (1999).
Gregory, M. A., Till, R. & Smith, M. C. M. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185, 5320–5323 (2003).
Janssen, G. R. & Bibb, M. J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene 124, 133–134 (1993).
Hindra, Pak, P. & Elliot, M. A. Regulation of a novel gene cluster involved in secondary metabolite production in Streptomyces coelicolor. J Bacteriol 192, 4973–4982, 10.1128/JB.00681-10 (2010).
Petrus, M. L. C., van Veluw, G. J., Wösten, H. A. B. & Claessen, D. Sorting of Streptomyces cell pellets using a complex parametric analyzer and sorter. J Vis Exp 84, e51178 (2014).
Vinck, A. et al. Hyphal differentiation in the exploring mycelium of Aspergillus niger. Mol Microbiol 58, 693–699, 10.1111/j.1365-2958.2005.04869.x (2005).
Acknowledgements
We are grateful to R. Tsien for providing plasmid pRSET-B. VICI (nr. 10379) and VIDI (nr. 12957) grants from the Dutch Applied Research Council (STW) fund work in the van Wezel and Claessen laboratories, respectively.
Author information
Authors and Affiliations
Contributions
B.Z. performed the experiments with the help of J.W., D.v.D., D.C. and B.R. G.P.v.W., H.A.B.W. and D.C. designed the study; B.Z., H.A.B.W. and D.C. wrote the paper with the help of D.v.D. and G.P.v.W. All authors discussed the results and their interpretation and commended on the manuscript at all stages.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zacchetti, B., Willemse, J., Recter, B. et al. Aggregation of germlings is a major contributing factor towards mycelial heterogeneity of Streptomyces. Sci Rep 6, 27045 (2016). https://doi.org/10.1038/srep27045
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27045
This article is cited by
-
Endocytosis-like DNA uptake by cell wall-deficient bacteria
Nature Communications (2022)
-
Transcriptomic Analysis of Morphology Regulatory Mechanisms of Microparticles to Paraisaria dubia in Submerged Fermentation
Applied Biochemistry and Biotechnology (2022)
-
Teichoic acids anchor distinct cell wall lamellae in an apically growing bacterium
Communications Biology (2020)
-
Microencapsulation extends mycelial viability of Streptomyces lividans 66 and increases enzyme production
BMC Biotechnology (2018)
-
Sporulation-specific cell division defects in ylmE mutants of Streptomyces coelicolor are rescued by additional deletion of ylmD
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.