Abstract
DNA methylation patterns in plants are dynamically regulated by DNA methylation and active DNA demethylation in response to both environmental changes and development of plant. Beginning with the removal of methylated cytosine by ROS1/DME family of 5-methylcytosine DNA glycosylases, active DNA demethylation in plants occurs through base excision repair. So far, many components involved in active DNA demethylation remain undiscovered. Through a forward genetic screening of Arabidopsis mutants showing DNA hypermethylation at the EPF2 promoter region, we identified the conserved iron-sulfur cluster assembly protein MET18. MET18 dysfunction caused DNA hypermethylation at more than 1000 loci as well as the silencing of reporter genes and some endogenous genes. MET18 can directly interact with ROS1 in vitro and in vivo. ROS1 activity was reduced in the met18 mutant plants and point mutation in the conserved Fe-S cluster binding motif of ROS1 disrupted its biological function. Interestingly, a large number of DNA hypomethylated loci, especially in the CHH context, were identified from the met18 mutants and most of the hypo-DMRs were from TE regions. Our results suggest that MET18 can regulate both active DNA demethylation and DNA methylation pathways in Arabidopsis.
Similar content being viewed by others
Introduction
Epigenetic modifications are important for the silencing of transposable element (TE), chromosome stability, gene expression and many other vital cellular processes1,2,3. DNA methylation is an important epigenetic modification conserved in plants and other higher eukaryotes. Unlike the situation in mammals, wherein DNA methylation predominantly occurs in the CG context4, it occurs in all three cytosine contexts: CG, CHG and CHH (H= A, T, or C) in plants. Whole-genome bisulfite sequencing in Arabidopsis has revealed that gene bodies are mainly associated with CG methylation. While TE- and repeats-enriched heterochromatin regions are the major targets of CHG and CHH methylation, they are also densely CG methylated5. In Arabidopsis, once the DNA methylation pattern is established, symmetric CG and CHG methylation can be maintained by DNA METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) during DNA replication, while asymmetric CHH methylation must be de novo established by DOMAINS REARRANGED METHYLASE 2 (DRM2) through an RNA-directed DNA methylation (RdDM) pathway1,6. In RdDM, two plant-specific RNA polymerases Pol IV and Pol V produce 24-nt siRNAs and long noncoding scaffold transcripts respectively to assist in the recruitment of DRM2 to specific loci1,6. In addition to DRM2, CHROMOMETHYLASE 2 (CMT2) was recently shown to be a methyltransferase that catalyzes CHH methylation7.
DNA demethylation may take place as a passive or an active process. In contrast to the passive demethylation during which DNA methylation is lost due to the inactivation or down-regulation of maintenance DNA methyltransferases (such as DNMT1 and MET1 in mammals and plants, respectively), active DNA demethylation refers to the enzymatic removal of the methylated cytosine in a replication-independent manner8. In plants, the active DNA demethylation pathway is initiated by a subfamily of atypical HhH-GPD enzymes including REPRESSOR OF SILENCING1 (ROS1)9, DEMETER (DME), DEMETER-LIKE2 (DML2) and DEMERTER-LIKE3 (DML3)10,11. All of these enzymes are bifunctional 5-methylcytosine-DNA glycosylases/lyases that not only remove the 5-methylated cytosine, but also cleave phosphodiester backbone at the abasic site10,12,13,14 resulting in a gap with 3′ phosphate or 3′ dRP (3′ α, β-unsaturated aldehyde) termini. A DNA phosphatase (ZDP) and an apurinic/apyrimidinic endonuclease (APE1L) process the 3′ phosphate and 3′ dRP termini, respectively, to generate a 3′ OH group so that downstream polymerases and ligases can fill in the gap with an unmethylated cytosine15,16. Interestingly, DNA demethylation in mammals also utilizes the BER pathway but with a different strategy at the beginning. Instead of being directly removed, in mammals the 5mC is converted into other bases such as 5-hydroxymethylcytosine (5hmC) by the TET family enzymes, which can be further oxidized to produce 5fC and 5caC. The 5fC or 5caC can then be removed by thymine DNA glycosylase (TDG)8,17.
Compared to the well-established RdDM pathway, little is currently known about the locus-specific guidance of active DNA demethylation machinery in plants. Previous studies have shown that the RNA-binding protein ROS3 is required for the recruitment of ROS1 to a subset of its target loci18,19. Recently, an IDM1-IDM2-IDM3-MBD7 complex was identified as another important regulator of DNA demethylation that functions upstream of ROS120,21,22,23,24,25. The IDM1 complex recognizes methylated DNA loci with low H3K4 and H3R2 methylation levels and catalyzes H3K18 and H3K23 acetylation, which facilitates ROS1-mediated DNA demethylation20,21,24,26. Unlike ROS1, which functions in almost all plant tissues, DME is preferentially expressed in the central cell of the female gametophyte. Active DNA demethylation induced by DME in the central cell is essential for the expression of imprinting genes, including FWA and MEA11. In addition to DME, mutations in two cytosolic iron-sulfur cluster assembly (CIA) components NAR1 and DRE2 also influence the expression of FWA27,28.
Many proteins that function in vital processes such as respiration, photosynthesis, sulfur and nitrogen assimilation, amino acid and purine metabolism, plant hormone and coenzyme synthesis, DNA repair and translation have Fe-S clusters as their cofactors29,30. In plants, there are three Fe-S cluster assembly pathways: the SUF (sulfur mobilization) pathway in plastids, the ISC (iron-sulfur cluster) assembly pathway in mitochondria and an emerging CIA pathway in the cytoplasm that is partially dependent on the ISC pathway29,30,31. The CIA pathway, mainly found in eukaryotes, is essential for the maturation of both cytoplasmic and nuclear Fe-S proteins32. In yeast, a mitochondrial cysteine desulfurase complex Nfs1-Isd11 and the mitochondrial ISC machinery initially synthesize a sulfur-containing component, which is then exported to the cytoplasm by the mitochondrial inner membrane ATP-binding cassette transporter Atm130,33,34. In the cytoplasm of yeast and human cells, Fe-S clusters are assembled on the CFD1-NBP35 heterodimer whereas in plants Fe-S clusters are assembled on the NBP35 homodimer35. Fe-S clusters are then transferred to the apoproteins, with the help of the NAR1 and CIA targeting complex comprised of a WD40-repeat protein CIA1, a small acidic protein CIA2 and a HEAT-repeat containing protein MET18/MMS1930,36,37,38. Recent findings suggest that MET18/MMS19 is a late-acting CIA component30,38,39. In yeast, MET18/MMS19 also forms a complex with Dos2, Rik1 and Cdc20 and participates in DNA repair, RNA polymerase II transcription, chromosome segregation and telomere length maintenance40,41,42,43,44. Since the ROS1/DME family members are Fe-S proteins29,45,46, a deficiency in the CIA pathway may have impact on the activities of ROS1/DME family members.
Here we report MET18 as a component of the active DNA demethylation pathway in plants. Identified through a forward genetic screening, the met18 mutant plants exhibited DNA hypermethylation at hundreds of genetic loci and conferred transcriptional silencing of reporter genes and some endogenous targets. Most of the hyper-DMRs (differentially methylated regions) identified from the met18 mutants were also hypermethylated in the ros1-4 or rdd (ros1dml1dml2 triple) mutant plants. Interaction between MET18 and ROS1 was observed in vitro as well as in vivo. ROS1 activity was reduced in the met18 mutants. Interestingly, we found that dysfunction of MET18 caused DNA hypomethylation and the activation of some of the TEs. Our results revealed an epigenetic role of MET18 in the regulation of gene expression in Arabidopsis.
Results
Isolation of met18 mutants
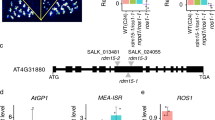
Our previous studies have shown that IDM1 is crucial for the recruitment of ROS1 to only a subset of its targeted loci20, suggesting that there exists an IDM1-independent pathway to regulate ROS1-mediated active DNA demethylation in Arabidopsis. To identify new components in the active DNA demethylation pathway initiated by ROS1, a new chop-PCR marker was designed based on the DNA methylation analysis at the EPF2 promoter (Supplementary Fig. 1a). The chop-PCR analysis of the EPF2 promoter revealed that the DNA methylation level in this region was significantly higher in the ros1-4 and rdd mutants but not in the idm1 and idm2 mutants, which was consistent with the whole-genome bisulfite sequencing results (Supplementary Fig. 1b). The expression level of EPF2 was reduced in ros1-4 but not in idm1-1 (Supplementary Fig. 1c). Screening of the Arabidopsis T-DNA insertion mutants library, which includes mutants of chromatin-related genes, DNA or RNA binding protein-encoding genes and genes co-expressed with ROS1 or ROS3 et al. (Supplementary Table 1), using this chop-PCR marker resulted in the identification of two mutants showing DNA hypermethylation at the EPF2 promoter region. One of them harbors a T-DNA insertion in the At5g48120 gene and is referred to here as met18-1 (Fig. 1a). To confirm that the DNA hypermethylation was due to the T-DNA insertion mutation, we tested another mutant allele (i.e. met18-2). As expected, met18-2 showed a similar DNA hypermethylation phenotype in the EPF2 promoter region (Fig. 1a). Locus-specific bisulfite sequencing of the EPF2 promoter showed that the DNA methylation levels were increased in met18-2 and ros1-4 in all sequence contexts when compared to the levels in the wild-type (WT) control (Fig. 1b). Examination of the methylation status at previously identified marker genes At1g26390 and At1g26400 revealed that both met18 mutants were hypermethylated at these loci in all sequence contexts (Fig. 1a,c,d)20. To further confirm that MET18 is the gene responsible for the DNA hypermethylation phenotype, the met18-1 mutant plants were transformed with an 8.5 kb WT genomic fragment containing the promoter and coding sequence of the At5g48120 gene. The transgene complemented the DNA hypermethylation phenotype of met18-1 (Supplementary Fig. 1d,e). All of these findings suggest that MET18 is a new component of the active DNA demethylation pathway.
Dysfunction of MET18 causes DNA hypermethylation at the EPF2 promoter, At1g26390 and At1g26400 loci.
(a) Analysis of DNA methylation levels at the EPF2 promoter, At1g26390 and At1g26400 loci using chop-PCR. The methylation-sensitive restriction enzymes used were BstUI, HpaII and HhaI. DNA hypermethylation results in no cleavage by the enzymes and increased levels of the PCR product. Undigested controls are shown in the lower panel. (b–d) Bisulfite sequencing data showing the effects of MET18 mutations on DNA methylation in different sequence contexts in the EPF2 promoter (b), At1g26390 (c) and At1g26400 (d). For each sample, at least 20 clones were analyzed and the percentage of DNA methylation was calculated from the indicated number of clones.
MET18 prevents DNA hypermethylation at hundreds of loci
In order to examine the role of MET18 in active DNA demethylation, we compared the genome-wide DNA methylation profiles of met18-2 and WT Col-0 plants by next-generation sequencing after bisulfite conversion4,20. There was no significant difference in the overall genome methylation patterns between met18-2 and WT plants. However, mutation in MET18 caused DNA hypermethylation at 1,228 loci and hypomethylation at 210 loci. The differences between met18-2 and WT plants in DNA methylation at these loci are statistically significant with the Benjamini–Hochberg adjusted p value less than 0.01 (Fig. 2a and Supplementary Table 2). Consistent with the locus-specific bisulfite sequencing results, methylome analysis also showed hypermethylation of At1g26390 and At1g26400 in ros1-4 and met18-2 (Supplementary Fig. 2a,b)20. Analysis of the distribution of the hypermethylated loci on all of the five chromosomes in met18-2, ros1-4 and rdd indicated that the hypermethylated loci in met18-2 were spread evenly over the five chromosomes and that they largely overlapped with the hypermethylated loci in ros1-4 and rdd (Supplementary Fig. 2c). Analysis of the hyper-DMRs in different DNA regions (gene body, intergenic region, TEs out of gene region and TEs overlapping with gene region) showed a similar pattern between met18-2 and ros1-4, with less than 20% of the hyper-DMRs distributed in the gene body (Supplementary Fig. 2d).
Dysfunction of MET18 causes genome-wide DNA hypermethylation.
(a) The number of differentially methylated regions (DMRs) identified in met18-2 and the overlapping DMRs between different mutant plants. (b) Venn diagram showing the numbers of hyper-DMRs that either overlap between or are unique in met18-2, ros1-4 and rdd. (c) Boxplots showing the distribution and the average CG, CHG, or CHH methylation levels calculated from DMRs in the respective subgroups. Dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum percentage of DNA methylation.
About 70% of the 1,228 hyper-DMRs in met18-2 were also hypermethylated in ros1-4 and rdd (Fig. 2a,b). The overlapping hypermethylated loci in these mutants were methylated in all sequence contexts (Fig. 2c and Supplementary Fig. 2e). Notably, the DNA methylation level at the met18-2-specific loci was actually also higher in ros1-4 and rdd (Fig. 2c and Supplementary Fig. 2e), although the increases were not significant enough to be counted as hyper-DMRs as per the parameters defined in this study. These results indicated that MET18 and ROS1 might be functional in the same genetic pathway of active DNA demethylation. However, no increase in the DNA methylation level at the ros1-4- or rdd-specific loci was observed in met18-2 (Fig. 2c), suggesting that MET18 may only control DNA demethylation at a subset of ROS1 targeted loci. To further determine whether MET18 regulates DNA demethylation in an IDM1-independent manner, we compared the hyper-DMRs identified from met18-2 and idm1-1. Our results showed that 181 hyper-DMRs overlapped between them, accounting for only 14.7% of the hyper-DMRs in met18-2 and 16.4% of the hyper-DMRs in idm1-1 (Supplementary Fig. 2f). The low level of hyper-DMR overlap between these two mutants suggests that they may act through different pathways.
MET18 prevents the silencing of reporter genes and some endogenous genes
Previous studies have reported that ROS1 was required for the anti-silencing of reporter genes and endogenous genes9. We introduced 35S::LUC and 35S::NPT II transgenes into the met18-2 background via crossing and we found that both LUC and NPT II were silenced in met18-2 (Fig. 3a,b). We also examined the expression levels of twelve endogenous genes by using real-time PCR to determine whether DNA hypermethylation affects their expression in the met18 mutants. Nine of the tested genes showed a significant reduction in their transcript levels in the met18 mutants (Fig. 3c and Supplementary Fig. 3). Unexpectedly, the expression level of EPF2 was not reduced in the met18 mutants in spite of the fact that we did identify the met18 mutants via chop-PCR using the EPF2 marker (Supplementary Fig. 3a). Bisulfite sequencing results indicated that the increase in DNA methylation in the EPF2 promoter in met18-2 was not as high as that observed in ros1-4, suggesting that the increase in DNA methylation in the met18 mutants may not be sufficient to silence the EPF2 gene (Supplementary Fig. 3a). To further explore the anti-silencing role of MET18 genome-wide, we analyzed previously published RNA-seq data generated using met18-247. However, we only identified 4 genes (AT1G48660, AT1G77960, AT3G26742 and AT3G27940), out of 132 down-regulated genes in met18-2, that are hypermethylated (Supplementary Table 2). Our analysis suggests that most of the down-regulated genes are not controlled by MET18 directly through DNA methylation.
MET18 prevents the silencing of reporter genes and endogenous genes.
(a) The MET18 mutation causes the silencing of 35S-LUC and 35S-NPTII reporter genes. The reporter genes were introduced into met18-2 by crossing. Seedlings grown in MS plates were imaged after being sprayed with luciferase substrate. (b) Real-time PCR analysis of the expression levels of the LUC and NPTII reporter genes in the different genotypes. (c) Effect of MET18 mutation on the DNA methylation levels in WT control, ros1-4 and met18-2 and the expression levels of endogenous genes At1g77780 and At1g77790. (d) Effect of MET18 mutation on the DNA methylation levels in WT control, ros1-4 and met18-2 and the expression levels of endogenous genes At5g38540 and At5g38550 in the mutants. Snapshots in the Integrated Genome Browser show the DNA methylation levels, while the bar diagrams show the real-time PCR analysis of the expression levels of the hypermethylated genes or nearby genes in different genotypes. TUB8 was used as an internal control. Standard errors were calculated from three biological replications, *P < 0.05.
MET18 is a component of the CIA complex
To investigate the role of MET18 in the regulation of active DNA demethylation, the protein complex associated with MET18 was purified from floral tissues of the met18-1 plants harboring MET18-3Flag transgene driven by native MET18 promoter, which could complement the DNA methylation phenotype (Supplementary Fig. 1d,e). As expected, the most abundant peptides identified by the mass spectrometric analysis corresponded to MET18. Meanwhile, several other components of the CIA complex, including CIA1, CIA2/AE7, NAR1 and AtDRE2 were also identified (Supplementary Tables 4 and 5). The peptides from CIA1 and CIA2/AE7 were much more abundant than those from NAR1 and AtDRE2, suggesting that MET18, CIA2/AE7 and CIA1 are major components of the CIA targeting complex and that the ratio for these three proteins should be 1:1:1 in the complex (Supplementary Table 5). Moreover, many Fe-S cluster proteins including ROS1, DME and ACO1 were co-purified with MET18, suggesting the interaction between MET18 and these proteins (Supplementary Table 5). Taken together, our results suggest that as in mammals and yeast38,39, MET18 is also a component of the CIA complex in plants. Our results are consistent with the findings of two previous studies done by Luo et al.45 and Han et al.47.
MET18 interacts with ROS1 in vitro and in vivo
ROS1 and DME were co-purified with MET18, suggesting that MET18 interacts with these proteins (Supplementary Table 5). Since MET18 functions as a platform to facilitate Fe-S cluster transfer to targeted proteins in the CIA pathway in Arabidopsis45 and as ROS1 contains a Fe-S cluster binding motif (Supplementary Fig. 4), we speculated that MET18 physically interacts with ROS1. To detect such a physical interaction, we performed a yeast two-hybrid (Y-2-H) assay. The results showed that ROS1 can directly interact with MET18 but not with AE7, another component of the CIA complex (Fig. 4a). The in vivo interaction of MET18 and ROS1 was validated by a luciferase complementation imaging assay wherein the co-infiltration of tobacco leaves with MET18-cLUC and AE7-nLUC or MET18-cLUC and ROS1-nLUC activated the expression of the luciferase reporter gene and transmitted fluorescence (Fig. 4b).
MET18 can interact with ROS1 in vivo.
(a) Interaction of MET18 with ROS1 as determined by Y-2-H assay. Yeast cells harboring different fusion protein combinations (listed on the right) in pGBK-T7 (BD) and pGAD-T7 (AD) vectors were plated on the medium lacking Leu, Trp (SD-TL) or the medium lacking Leu, Trp and His (SD-TLH). Interaction between BD-MET18 and AD-AE7 serves as a positive control. (b) Interaction of MET18 with ROS1 as demonstrated by firefly luciferase complementation imaging assay in Nicotiana benthamiana leaves. Interaction between MET18-cLUC and AE7-nLUC serves as a positive control. nLUC, C-terminal region of firefly luciferase; cLUC, N-terminal region of firefly luciferase. (c) BiFC assay showing ROS1 and MET18 interaction in the cytoplasm. YFPC, C-terminal region of YFP; YFPN, N-terminal region of YFP. The experiments were repeated three times.
ROS1 has been reported to be localized in the nucleus9, while MET18 was proposed to be a component of the cytosolic CIA complex in eukaryotes38,39,45, although it was also detected in the nucleus40,45. To examine where ROS1 and MET18 interact in vivo, we first examined the subcellular localization of MET18 using stably expressing MET18-3Flag transgenic plants. Nuclear and cytoplasmic proteins extracted from the transgenic plants were subjected to Western blot analysis using HSP90 and histone H3 as markers for the cytoplasmic proteins and nuclear proteins, respectively. The results indicated that MET18 was specifically localized in the cytoplasm but not in the nucleus (Supplementary Fig. 5). Secondly, to check for interaction between ROS1 and MET18, we performed a bimolecular fluorescence complementation (BiFC) assay. It primarily detected a YFP signal in the cytoplasm (Fig. 4c), suggesting that ROS1 and MET18 interact mainly in the cytoplasm.
MET18 is required for ROS1 activity
ROS1 function and RdDM pathway have recently been found to be tightly interconnected48,49. In the RdDM pathway mutants, the ROS1 promoter is hypomethylated and the transcript level of ROS1 is dramatically reduced48,49. By contrast, in the ros1 and rdd mutants, the ROS1 promoter is hypermethylated and the transcript level of ROS1 is increased48,49. Our whole-genome bisulfite sequencing results and real-time PCR analysis showed that met18-2 was similar to ros1-4 in their methylation levels at ROS1 promoter and their ROS1 transcript levels (Fig. 5a,b), suggesting that the DNA hypermethylation in met18 was not due to decreased ROS1 transcription. As a key component of the CIA pathway, MET18 is essential for the stability of the targeted proteins38. Hence, we investigated whether the loss of MET18 led to DNA hypermethylation through affecting the localization or stability of ROS1 in the nucleus. To this end, we transiently expressed ROS1-GFP fusion protein in ros1-4 or met18-2 protoplasts. We observed that ROS1 was localized exclusively in the nucleus both in met18-2 and ros1-4 (Fig. 5c). Meanwhile, we conducted Western blot using the GFP antibody and our results confirmed that MET18 mutation did not affect the stability of ROS1 protein (Fig. 5d). These results suggest that MET18 mutation did not affect the localization or the stability of ROS1 protein. We then compared the nicking activity of ROS1 protein on methylated DNA using immunoprecipitated ROS1-GFP fusion protein from met18-2 and ros1-4. Our results showed that the nicking activity of ROS1-GFP protein purified from met18-2 was reduced compared with that from ros1-4 (Fig. 5e,f), suggesting that MET18 mutation mainly affected active DNA demethylation by compromising the enzymatic activity of ROS1 and ROS1 family proteins.
Dysfunction of MET18 affects ROS1 activity.
(a) Snapshots in the Integrated Genome Browser showing the DNA methylation levels at the ROS1 promoter. The specific region important for ROS1 regulation is highlighted with red box. (b) Real-time PCR analysis of ROS1 transcripts in different mutants. Standard errors were calculated from three biological repeats, *P < 0.05. (c) Localization of ROS1-GFP in the protoplasts of met18-2. NLS-RFP was used as a control for nuclear localized protein. ROS1-GFP and NLS-RFP constructs were co-introduced into the Col-0 or met18-2 mutant protoplasts. GFP signal (green) and overlay of the signals from GFP and RFP (red) are shown. (d) Detection of ROS1-GFP protein by GFP antibody. Anti-H3 serves as a loading control. (e,f) Nicking activity of ROS1-GFP fusion protein from met18-2 and ros1-4 transgenic plants. OC, open circular form plasmid DNA; CC, closed circular plasmid DNA. Standard errors were calculated from three biological replications, *P < 0.05.
The iron-sulfur cluster binding motif is essential for ROS1 function
ROS1/DME family members possess four conserved cysteine residues adjacent to the DNA glycosylase domain that may function to bind the [4Fe-4S] cluster (Supplementary Fig. 4). Any cysteine mutation in DME protein has been reported to cause the loss of 5mC excision activity in vitro46, suggesting that the iron-sulfur binding motifs are essential for the enzymatic activity of the ROS1/DME family proteins. To investigate whether the binding of the Fe-S cluster has any impact on the activity of ROS1, we introduced C1038S and C1045S mutations in the conserved iron-sulfur cluster binding motif of ROS1 via site-directed mutagenesis (Supplementary Fig. 4). Our Split-LUC assay showed that the mutations didn’t affect the interaction between ROS1 and MET18 (Supplementary Fig. 6). We further tested whether these mutations affect ROS1 activity by measuring the viability of E.coli cells expressing WT or mutated ROS1. Since a previous study demonstrated that the expression of active DME induces lethality in methylation-proficient E.coli46, we speculated that the expression of other members of the demethylating DNA glycosylase family, including ROS1, may also cause lethality in E.coli and measuring E.coli viability could be a way to determine whether ROS1 is active. Indeed we found that the expression of WT ROS1 caused lethality in E.coli, but the expression of C1038S or C1045S mutant form of ROS1 had no effect on the viability of E.coli (Fig. 6a), suggesting that C1038S or C1045S mutation abolished the enzymatic activity of ROS1 protein. Moreover, we found that the expression of WT, but not the C1045S mutant version of ROS1, under a constitutive promoter could complement the luciferase and DNA hypermethylation phenotypes of the ros1-1 mutant (Fig. 6b,c), despite equal amounts of WT and mutated ROS1 being expressed (Fig. 6d). These results suggest that the Fe-S cluster binding motif is crucial for ROS1 function in vivo.
Iron-Sulfur cluster binding site is essential for ROS1 function.
(a) Enzymatic activity of WT and mutated ROS1 in E. coli indicated by colony formation of WT and mutated (C1038S and C1045S) ROS1 transformants at 0 and 20 μM IPTG. (b) Complementation assay showing that WT, but not the mutant form of ROS1-3Flag, can restore LUC expression in ros1-1. Leaves were collected for luminescence imaging after the treatment with 3% NaCl for 6 h. (c) DNA hypermethylation phenotype of ros1-1 plant transformed with the WT or mutant form (C1045S) of ROS1. Methylation-sensitive restriction enzymes (HhaI, HpaII and BsmAI) were used for the chop-PCR analysis and undigested DNA was amplified as a control. (d) Detection of WT ROS1 protein and site-directed mutated (C1045S) ROS1 protein in transgenic Arabidopsis plants by Flag antibody. About 10 μg protein extracts were loaded in each well. The gel for loading control was stained with Coomassie blue.
Mutation in MET18 causes DNA hypomethylation in the CHH context at TE regions
Although the overall DNA methylation level in the genic regions of met18-2 and WT control was similar, methylation in the CHH context in TEs was slightly reduced in met18-2 while it was slightly increased in ros1-4 or rdd (Supplementary Fig. 7a–c). CHH hypomethylation of 2038 loci was observed in met18-2 compared to a significantly lower number of hypomethylated loci in ros1-4 and rdd (Fig. 7a and Supplementary Table 3). Examination of the effect of MET18 mutation on DNA methylation in the CHH context in TEs of different lengths revealed that MET18 mutation had a larger impact on shorter TEs (Supplementary Fig. 7d). Distribution analysis of the CHH hypo-DMRs on all of the five chromosomes in met18-2 indicated that the CHH hypo-DMRs were enriched at the pericentromeric regions (Supplementary Fig. 8a). Most of the CHH hypo-DMRs were from TE regions and more than half of them were overlapped with CHH hypo-DMRs in the Pol IV mutant (nrpd1-3) and Pol V mutant (nrpe1-11) (Fig. 7a,b). We also examined the methylation level in nrpd1-3 and nrpe1-11 at those regions that were hypomethylated in met18-2 in the CHH context and found that most of them were hypomethylated, albeit to different degrees (Supplementary Fig. 8b). The overlapping loci in these mutants were hypomethylated in the CHH context (Fig. 7c). Methylation in the CHG context was also reduced in nrpd1-3 and nrpe1-11, but not in met18-2 for the three overlapped loci (Fig. 7c). DNA methylation at the met18-2-specific loci was also decreased in nrpd1-3 and nrpe1-11 (Fig. 7c) although the DNA methylation levels at the nrpd1- and nrpe1-specific loci were not decreased in met18-2. Taken together, our results indicated that MET18 may also regulate DNA methylation pathway.
Comparison of CHH hypo-DMRs among met18-2, nrpd1-3 and nrpe1-11 mutants.
(a) Number and location of CHH hypo-DMRs identified in met18-2 and other mutant plants. (b) Venn diagram showing the numbers of CHH hypo-DMRs that either overlap between or are unique in met18-2, nrpd1-3 and nrpe1-11. (c) Boxplots showing the distribution and the average CG, CHG and CHH methylation levels calculated from hypo-DMRs in the respective subgroups. Dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum percentage of DNA methylation.
Dysfunction of MET18 causes impaired production of 24-nt siRNAs and long noncoding scaffold RNAs as well as abolished transcriptional silencing at some loci
In parallel with the whole-genome bisulfite sequencing, we characterized 24-nt siRNAs in met18-2, nrpd1-3 and nrpe1-11. The total amount of 24-nt siRNAs in met18-2 was reduced but the reduction was less compared to that in nrpd1-3 and nrpe1-11 (Supplementary Fig. 9a). Consistently, we identified 501, 4498 and 2963 siRNA-generating regions where 24-nt siRNAs were strongly decreased in met18-2, nrpd1 and nrpe1 respectively (Supplementary Table 6). Almost all (99.8%) MET18-regulated siRNAs are Pol IV-dependent (Supplementary Fig. 9b), in agreement with the fact that Pol IV is required for the production of the majority of 24-nt siRNAs. Pol V is also required for the production of most MET18-dependent siRNAs, although the proportion of siRNAs affected by Pol V is smaller (80.6%) (Supplementary Fig. 9b). We further examined whether the generation of Pol V transcripts was affected by MET18 dysfunction. Mutation of MET18 reduced the levels of Pol V-dependent transcripts at multiple tested loci (Fig. 8a), including IGN22, P2 and P650,51. The changes in 24-nt siRNA and Pol V transcript levels may lead to altered CHH DNA methylation level. To examine this, we classified siRNA-generating loci into four groups based on the dependence of siRNA generation on specific proteins and made detailed analysis. As expected, the slight reduction of CHH DNA methylation level in all groups coincided with the changes in 24-nt siRNA levels (Supplementary Fig. 9c). Altogether, our results suggest that MET18 was required for the accumulation of 24-nt siRNAs, Pol V transcripts and CHH methylation at some loci in Arabidopsis.
MET18 is required for the generation of Pol V-dependent noncoding RNAs and transcriptional gene silencing.
(a) Detection of Pol V-dependent noncoding RNAs by real-time PCR, NP is a none NRPE1 enrichment region. (b,c) Snapshots in the Integrated Genome Browser showing the DNA methylation levels and bar diagrams showing the real-time PCR-based expression analysis of the hypomethylated TEs in different genotypes. TUB8 serves as an internal control. Standard errors were calculated from three biological replications, *P < 0.05.
CHH methylation has been suggested to be critical for TE silencing. To determine whether CHH hypomethylation affects the expression of targeted TEs in met18, we examined the expression levels of three TEs which showed CHH DNA hypomethylation in met18-2 (Fig. 8b). All of the tested TEs showed a substantial increase in their transcript levels (Fig. 8c). Recently, Han et al. reported that MMS19/MET18 associates with ABO4 and ICU2 to confer transcriptional gene silencing via a DNA methylation-independent mechanism47. To explore the genome-wide correlation between DNA hypo-methylation and expression of TEs or genes in met18-2, we analyzed our whole-genome bisulfite sequencing results and their RNAseq data and we identified 16 TEs showing increased expression from the CHH hypo-DMR list (Supplemental Table 7). Those 3 TEs showing increased expression as determined by our q-PCR experiments were not detected by the RNAseq assay possibly due to the low sensitivity of RNAseq (Fig. 8b,c). Thus, there might be more TEs whose expression is controlled by DNA methylation and MET18 may regulate TE expression through promoting DNA methylation. In contrast to TEs, none of the genes showing increased expression in met18-2 as determined by RNAseq is hypomethylated, suggesting that MET18 regulates gene expression mainly through DNA methylation-independent mechanisms. Taken together, our results suggest that MET18 regulates transcriptional silencing through both DNA methylation-dependent and -independent mechanisms and the choice of mechanism is locus-specific in Arabidopsis.
Discussion
In plants, active DNA demethylation is initiated by the ROS1/DME family of 5mC DNA glycosylases/lyases and it is presumably continued through a base excision repair (BER) pathway3,17. Currently, little is known about how the enzymatic activity of ROS1 is regulated. To identify new components in the active DNA demethylation pathway initiated by ROS1, we designed a new chop-PCR marker based on the whole-genome DNA methylation analysis of ros1-4. We performed forward genetic screening for DNA hypermethylation mutants at the EPF2 promoter region and identified MET18 as a new factor modulating DNA methylation level through regulating ROS1 enzymatic activity.
The effects of MET18 mutations on DNA methylation and gene expression resembled those of the ROS1 mutations, suggesting that MET18 and ROS1 may function in the same genetic pathway to prevent DNA hypermethylation. ROS1 and DME were co-purified with MET18 (Supplementary Table 5) and a Y-2-H assay as well as split-LUC assay demonstrated interaction between MET18 and ROS1 (Fig. 4). Western blot analysis results showed that MET18 was largely distributed in the cytoplasm, while ROS1 was mainly localized in the nucleus (Fig. 5c). It thus seems that MET18 and ROS1 are not present in the same cellular compartment. It is likely that MET18 regulates the incorporation of the Fe-S cluster into the ROS1 protein in the cytoplasm before ROS1 enters the nucleus. Our BiFC results confirmed their interaction in the cytoplasm and implicated the cytoplasmic regulation of ROS1 by MET18 (Fig. 4c). In Arabidopsis, the CIA pathway components NBP35, NAR1, CIA1 and CIA2/AE7 were reported to be essential for embryo development and the null mutants for these components were lethal45,52. MET18 is not an essential component of the CIA complex in Arabidopsis as the met18 knock-out mutants are viable in Arabidopsis and they did not show severe developmental phenotype under normal growth conditions47. It may act to enhance Fe-S cluster assembly for a small subset of Fe-S proteins, including the ROS1 family, but it does not mediate the assembly of the majority of the Fe-S proteins essential for plant development. Even for the ROS1 family, MET18 mutation may only affect the efficiency of Fe-S cluster assembly, rather than totally abolishing the Fe-S cluster assembly, since the ROS1 activity was reduced but not totally abolished in met18 (Fig. 5e,f). Moreover, not all of the ROS1 targets were affected in the met18-2 mutant (Fig. 2). Another CIA pathway component AE7 has also been reported to mediate active DNA demethylation and the DNA methylation levels in the ae7-1 mutant were reported to be elevated at two loci (XBAT34 and MRD1)45. However, in our study, the interaction between AE7 and ROS1 was not detected by either the Y-2-H assay or the split-LUC assay (Fig. 4), which was not consistent with the result got by Duan et al.53. Our whole-genome bisulfite sequencing results showed that the DNA methylation levels at the XBAT34 and MRD1 loci were also increased in met18-2 (Supplementary Fig. 10). These results suggest that, although AE7 may not directly bind ROS1, both AE7 and MET18 are required for Fe-S cluster assembly into ROS1 protein.
As a key component of the CIA pathway, MET18/MMS19 was reported to be essential for the stability of the targeted proteins in human cells38. In contrast, our results showed that MET18 dysfunction in Arabidopsis had no effect on ROS1-GFP transgene stability and sub-localization although we could not detect the endogenous ROS1 protein due to the unavailability of an appropriate antibody. However, the mRNA level for ROS1 was found to be higher in met18, which is consistent with the findings in the ros3 mutant18. Based on the ROS1 site-directed mutagenesis results and the previous finding regarding DME46, we propose that MET18 facilitates interaction between the Fe-S cluster assembly complex and the ROS1/DME family and that it acts late in the Fe-S cluster assembly pathway (Supplementary Fig. 11). Dysfunction of MET18 therefore affects the enzymatic activity of ROS1.
MET18 may also regulate active DNA demethylation through facilitating Fe-S cluster assembly on other enzyme(s) involved in the BER pathway. In fission yeast, the replicative DNA polymerases have been demonstrated to be Fe-S proteins54. In Arabidopsis, Han et al. identified the catalytic subunits of DNA polymerases α, δ and ε (ICU2, AT5G63960 and ABO4) co-purified with MET1847. Consistently, we identified DNA polymerases subunits (AT5G63960 and ICU2) associated with MET18 (Supplementary Table 5). These results suggest that DNA polymerases in plants are very likely to be Fe-S proteins. Dysfunction of MET18 may thus compromise DNA polymerase activities and result in defective DNA demethylation. However, the identities of DNA polymerases that participate in the DNA demethylation pathway and whether their full activities require MET18 need further investigation. Besides DNA polymerases, other proteins involved in active DNA demethylation may also be Fe-S proteins and regulated by MET18 even though they do not have conserved Fe-S binding motifs.
When preparing our manuscript, Duan et al. reported that MET18 was required for preventing transgene silencing as well as active DNA demethylation through a different forward genetic screening53. The identification of the same protein by two independent studies further underlines the importance of MET18 in active DNA demethylation. More importantly, in our study, we investigated the precise role of MET18 in active DNA demethylation and found that the ROS1 enzymatic activity was compromised in met18-2 (Fig. 5e,f). In addition to the DNA hypermethylation phenotype, we also identified lots of hypo-DMRs, especially in the CHH context in met18-2. Most of the CHH hypo-DMRs were also identified in nrpd1-3 and nrpe1-11. Dysfunction of MET18 resulted in reduced production of Pol IV-dependent 24-nt siRNAs and Pol V-dependent scaffold transcripts and eventually led to abolished transcriptional gene silencing (Fig. 8), suggested that MET18 plays a role in modulating the RdDM pathway. In fission yeast, during DNA replication, Mms19 interacts with Cdc20 and Dos2 to recruit Pol II to produce siRNAs, which mediate heterochromatin silencing40. In Arabidopsis, however, whether MET18 modulates the RdDM pathway through similar mechanisms or through the CIA pathway remains unknown.
In summary, our study reveals that MET18 mutation caused DNA hypermethylation at more than 1000 loci, of which about 70% were overlapped with the ROS1 targeted loci. This indicates that MET18 and ROS1 belong to the same genetic pathway. Our results clearly indicated that MET18 regulates active DNA demethylation by affecting the enzymatic activity of the ROS1/DME family proteins. Meanwhile, MET18 dysfunction gave rise to a DNA hypomethylation phenotype partially similar to that in RdDM mutants. Thus, MET18 can regulate both active DNA demethylation and DNA methylation pathways in Arabidopsis.
Methods
Plant materials and growth conditions
The met18-1 (SALK_121963) and met18-2 (SALK_147068) seeds were kindly provided by Dr. Xiaofeng Cui. Arabidopsis seedlings (Columbia background) were grown on Murashige-Skoog (MS) nutrient agar plates at 23 °C with 16 h of light and 8 h of darkness. After two weeks, the seedlings were transplanted into soil and grown at 23 °C with a 16 h photoperiod. Nicotiana benthamiana were also grown under the similar conditions.
Chop-PCR analysis
A number of loci with higher DNA methylation levels than those of the WT Col-0 plants were identified on the basis of the genome-wide DNA methylation profiling of ros1-4. Based on the DNA methylation data, PCR-based markers were developed for detecting DNA methylation status20. For the chop-PCR analysis (detection of DNA methylation levels by PCR after DNA digestion by methylation-sensitive restriction enzymes), 1 μg of genomic DNA was digested with a methylation-sensitive enzyme (HhaI, BstUI or HpaII) in a 20 μl reaction mixture. PCR was performed using 1 μl of the digested DNA as template in a 10 μl reaction mixture using the loci-specific primers (Supplementary Table 8).
Cloning strategy
For the complementation studies, an 8.5 kb genomic DNA fragment of the MET18 gene was amplified from Col-0 by PCR and then cloned into the pCAMBIA1305 vector using an In-Fusion® HD Cloning Kit (Clontech). Full-length ROS1 CDS was amplified by PCR and introduced into sp1300:GFP (or Flag tag) with SalI and SpeI digestion. A ROS1 mutation site was introduced into Psuper::ROS1:3Flag (super1300 vector) through site-directed mutagenesis using a MutanBEST Kit according to the manufacturer’s instructions (TaKaRa). Agrobacterium tumefacines strains (GV3101) harboring different constructs were used for Arabidopsis transformation following the standard floral dipping method. Primary transformants were selected on an MS medium containing 50 mg L−1 hygromycin.
Bisulfite sequencing
About 100 ng of genomic DNA was modified using the BisulFlash DNA Modification Kit (Epigentek) according to the manufacturer’s instructions. An aliquot (1 μl) of bisulfite-treated DNA was used for PCR in a reaction volume of 20 μl using ExTaq DNA polymerase (Takara) and gene-specific primers. The PCR amplified products were cloned into a pGEM-T Easy vector (Promega) and at least 20 independent clones of each sample were sequenced.
Whole-genome bisulfite sequencing and data analysis
Genomic DNA was extracted from 2 g leaf tissues of 14-day-old seedlings using DNeasy Plant Mini Kit (Qiagen) and then sent for bisulfite treatment, library preparation and sequencing by the High-throughput Sequencing Center (Biodynamic Optical Imaging Center, Peking University). The raw paired end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. All clean reads were mapped to the Arabidopsis thaliana TAIR 10 (10th release of the Arabidopsis thaliana genome sequence from the Arabidopsis Information Resource) genome with the BSMAP aligner allowing up to 2 mismatches. Uniquely mapped reads were used to determine the cytosine methylation levels as previously stated4. To identify DMRs, only cytosines covered with at least four reads in a library were considered. DMRs were searched by using a 200 bp sliding-window with 50 bp as step-size. DNA methylation levels of different plants were compared pairwise through Fisher’s exact test and the p values were adjusted for multiple comparisons using the Benjamini-Hochberg method. Windows with an adjusted p value less than 0.01 and an over 2-fold change in the methylation level were retained for further analysis. Moreover, the p value of each cytosine in the selected regions was calculated by Fisher’s exact test and it was considered as differentially methylated cytosine (DMC) if its p value was <0.01. The regions were retained only if it contained at least 7 DMCs. Finally neighboring DMRs were combined if the gap was <100 bp. The length of the DMR was adjusted to start from the first 5mC and end at the last 5mC.
Small RNA Sequencing and data analysis
Total RNAs were extracted from 12-day-old seedlings using TRIzol reagent. Small RNA (sRNA) data were analyzed as described by Zhang et al.55.
Yeast two-hybrid assay
For the yeast two-hybrid assays, coding sequences of MET18, ROS1 and AE7 were amplified by PCR. After verification of the sequences, the genes were cloned into pGBK-T7 or pGAD-T7 vector (Clontech) to generate DNA binding or activation domain fusion constructs, respectively. NdeI and BamHI sites were used for cloning MET18 and AE7 in pGBKT7 and pGAD-T7, while NdeI and EcoRI sites were used for cloning ROS1 in pGBKT7 and pGAD-T7. For the protein interaction analysis, two combinatory constructs were simultaneously transformed into yeast strain AH109 (Clontech) and then tested for Leu-, Trp- and His- auxotrophy according to the manufacturer’s protocols.
Split-LUC assay
For the split-LUC assays, the coding sequences of MET18 and AE7 were amplified by PCR. After verification of the sequences, the genes were cloned into pCAMBIA1300-NLUC or pCAMBIA1300-CLUC to generate N terminal or C terminal luciferase protein fusion constructs, respectively. The KpnI and BamHI sites were used for cloning MET18 and AE7 in pCAMBIA1300-nLUC or pCAMBIA1300-cLUC. WT or mutant forms of ROS1 were introduced to nLUC using In-Fusion® HD Cloning Kit (Clontech). For the protein interaction analysis, two combinatory constructs were simultaneously transformed into Nicotiana benthamiana leaves. To prevent the silencing of the genes, a construct expressing viral p19 protein was co-transformed56 .
Bimolecular fluorescence complementation (BiFC) assay
The BiFC assay was performed as described by Walter et al.57.
Scaffold RNA and mRNA analyses
Total RNA was extracted from 14-day-old Arabidopsis seedlings using an RNeasy Plant Mini Kit (Qiagen) and any contaminating DNA was removed by RNase-free DNase (Qiagen) treatment. mRNA (2 μg) was utilized for the first-strand cDNA synthesis using the SuperScriptTM III First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. Oligo dT primers were used during RT reaction to analyze mRNA expression, while gene-specific primers were used for detecting Pol V-dependent noncoding RNAs. The cDNA was then diluted five times and 2 μl was used as a template in a 25 μl reaction mixture using iQ SYBR Green Supermix (Bio-Rad). The primers used for real-time PCR are listed in Supplementary Table 8.
Nuclear-cytoplasmic fractionation and Western blot analysis
Nuclear-cytoplasmic fractionation was performed as described by Wang et al.58. Briefly, 14-day-old MET18-3Flag transgenic plants (about 1 g) were ground in liquid N2 using a pestle and mortar. The resultant fine powder was suspended in 2 ml of lysis buffer (20 mM Tris-HCl, pH 7.5, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM sucrose, 5 mM DTT and 1 protease inhibitor cocktail tablet (Roche)) and then centrifuged for 10 min at 1500 g at 4 °C after filtering through a double layer of Miracloth. The supernatant was centrifuged for 10 min at 10,000 g at 4 °C and the cytoplasmic fraction (supernatant) was collected. The pellet (nuclear fraction) was washed four times with nuclear resuspension buffer NRBT (20 mM Tris-HCl, pH 7.4, 25% glycerol, 2.5 mM MgCl2 and 0.2% Triton X-100) and then resuspended in NRB2 buffer (20 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 10 mM MgCl2, 0.5% Triton X-100 and 5 mM DTT) before being overlaid on top of NRB3 buffer (20 mM Tris-HCl, pH 7.5, 1.7 M sucrose, 10 mM MgCl2, 0.5% Triton X-100 and 5 mM DTT) and centrifuged for 45 min at 16,000 g at 4 °C. HSP90 was detected using a rabbit anti-HSP90 polyclonal antibody (1:5,000, at-115; Santa Cruz Biotechnology) as the cytoplasmic marker and histone H3 (05-499 Millipore) was used as the nuclear marker. MET18-3Flag was detected by anti-Flag (Sigma, F1804).
Fluorescence microscopy
ROS1-GFP fluorescence was detected with a confocal microscope (Zeiss, LSM710).
ROS1 in vitro activity assay
The ROS1 in vitro activity assay was performed as described by Mok et al.46. Briefly, WT control and two mutant forms (C1038S and C1045S) of full-length ROS1 CDS were cloned into pMal-c2X vector (New England Biolabs). The pMal-c2X, pMal-WT-ROS1, pMal-C1038S and pMal-C1045S were transformed into the E. coli strain DH5α. Overnight cultures of the four above-mentioned four transformed bacteria were diluted to 1.0 O.D. and then grown on LB agar medium containing 100 mg L−1 ampicillin with or without 20 μM IPTG at 37 °C for 16 h.
To compare the ROS1 activity in WT and met18 mutant plants, approximately 5 g of floral tissues from pSP1300::ROS1-GFP transgenic plants in ros1-4 or met18-2 background were grounded into a fine powder using liquid N2 and then suspended in 25 ml lysis buffer (50 mM Tris pH8.0, 230 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1 mM PMSF and 1 protease inhibitor cocktail tablet (Roche) and then centrifuged for 15 min at 18300 g 4 °C. The supernatants was incubated with 12.5 μl anti-GFP tag rabbit polyclonal antibody (EAZYBIO) at 4 °C for 2 h. About 50 μl of Protein A agarose (Chromo Tek) beads were added to trap the anti-GFP antibody and incubated at 4 °C for another 2 h. The beads were washed twice with 25 ml lysis buffer and twice with 1 ml lysis buffer.
The nicking activity assay was performed as described by Agius et al.13. Plasmid pBluescript KS+ (Stratagene) was purified from E. coli Trans110 (TRANSGEN BIOTECH), a dam and dcm strain. About 50 μg of the plasmid was methylated in vitro in a 50 μl reaction containing 5 U of SssI methylase (New England Biolabs) under the conditions recommended by the manufacturer. The non-methylated plasmid was subjected to the same procedure, but without the addition of the methyltransferase. For the nicking activity assay, a 20 μl reaction mixture containing 400 ng of each of the plasmid and 10 mM Tris-HCl, pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.2 mg ml−1 BSA was incubated overnight at 37 °C with an equal amount of ROS1-GFP protein from either ros1-4 or met18-2 background.
Affinity purification and mass spectrometry
About 6 g of flower tissue collected from MET18-3Falg transgenic plants was ground into fine powder in liquid N2 using a pestle and mortar and then suspended in 25 ml of lysis buffer (50 mM Tris, pH8.0, 230 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1 mM PMSF and 1 protease inhibitor cocktail tablet (Roche)) and centrifuged for 20 min at 18,300 g at 4 °C. The supernatant was incubated with 50 μl of anti-Flag M2 Magnetic beads (Sigma) for 3 h at 4 °C. The beads were washed twice with 25 ml of lysis buffer and then four times with 1 ml of lysis buffer. Proteins bound to the Flag beads were eluted with Flag peptides (Sigma). After SDS-PAGE, the entire gel lane was excised into six equal portions and digested in-gel with sequencing grade trypsin, followed by the mass spectrometric analysis as previously described by Li et al.25.
Additional Information
Accession codes: Whole-genome bisulfite sequencing was used to analyze the genome-wide methylation status in WT (Col-0) and met18-2 mutant plants. The dataset was deposited at NCBI (Col-0: SRX747290, met18-2: SRX747291). The ros1-4 and rdd whole-genome bisulfite sequencing data used were from GEO accession GSE3307120 and nrpd1-3 and nrpe1-11 methylation data were from GSE4420955. The small RNA dataset was deposited at NCBI (PRJNA299084).
How to cite this article: Wang, X. et al. The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Sci. Rep. 6, 26443; doi: 10.1038/srep26443 (2016).
References
Law, J. A. & Jacobsen, S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11, 204–220 (2010).
Zilberman, D. The evolving functions of DNA methylation. Curr Opin Plant Biol 11, 554–559 (2008).
Zhu, J. K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43, 143–166 (2009).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008).
Zhang, X. et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201 (2006).
Zhang, H. & Zhu, J. K. Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol 77, 161–173 (2012).
Zemach, A. et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205 (2013).
Wu, S. C. & Zhang, Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11, 607–620 (2010).
Gong, Z. et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 (2002).
Ortega-Galisteo, A. P., Morales-Ruiz, T., Ariza, R. R. & Roldán-Arjona, T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 67, 671–681 (2008).
Choi, Y. et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42 (2002).
Morales-Ruiz, T. et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA 103, 6853–6858 (2006).
Agius, F., Kapoor, A. & Zhu, J. K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA 103, 11796–11801 (2006).
Gehring, M. et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506 (2006).
Martínez-Macías, M. I. et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell 45, 357–370 (2012).
Li, Y. et al. An AP endonuclease functions in active DNA dimethylation and gene imprinting in Arabidopsis. PLoS Genet 11, e1004905 (2015).
Gong, Z. & Zhu, J. K. Active DNA demethylation by oxidation and repair. Cell Res 21, 1649–1651 (2011).
Zheng, X. et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 455, 1259–1262 (2008).
Stroud, H., Greenberg, M. V., Feng, S., Bernatavichute, Y. V. & Jacobsen, S. E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152, 352–364 (2013).
Qian, W. et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336, 1445–1448 (2012).
Qian, W. et al. Regulation of active DNA demethylation by an α-crystallin domain protein in Arabidopsis. Mol Cell 55, 361–371 (2014).
Lang, Z. et al. The Methyl-CpG-Binding Protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol Cell 57, 971–983 (2015).
Wang, C. et al. Methyl-CpG-Binding Domain Protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol 167, 905–914 (2015).
Li, X. et al. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci USA 109, 11425–11430 (2012).
Li, Q. et al. Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in Arabidopsis thaliana. PLoS Genet 11, e1005210 (2015).
Zhao, Y. et al. REPRESSOR OF SILENCING5 encodes a member of the small heat shock protein family and is required for DNA demethylation in Arabidopsis. Plant Cell 26, 2660–2675 (2014).
Buzas, D. M., Nakamura, M. & Kinoshita, T. Epigenetic role for the conserved Fe-S cluster biogenesis protein AtDRE2 in Arabidopsis thaliana. Proc Natl Acad Sci USA 111, 13565–13570 (2014).
Nakamura, M. et al. The role of Arabidopsis thaliana NAR1, a cytosolic iron-sulfur cluster assembly component, in gametophytic gene expression and oxidative stress responses in vegetative tissue. New Phytol 199, 925–935 (2013).
Balk, J. & Pilon, M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16, 218–226 (2011).
Netz, D. J., Mascarenhas, J., Stehling, O., Pierik, A. J. & Lill, R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol 24, 303–312 (2014).
Lill, R. & Mühlenhoff, U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol 22, 457–486 (2006).
Lill, R. & Mühlenhoff, U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes and diseases. Annu Rev Biochem 77, 669–700 (2008).
Kispal, G., Csere, P., Prohl, C. & Lill, R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18, 3981–3989 (1999).
Adam, A. C., Bornhövd, C., Prokisch, H., Neupert, W. & Hell, K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J 25, 174–183 (2006).
Bych, K. et al. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J Biol Chem 283, 35797–35804 (2008).
Balk, J. & Lobréaux, S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci 10, 324–331 (2005).
Tsaousis, A. D., Gentekaki, E., Eme, L., Gaston, D. & Roger, A. J. Evolution of the cytosolic iron-sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes. Eukaryot Cell 13, 143–153 (2014).
Stehling, O. et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199 (2012).
Gari, K. et al. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337, 243–245 (2012).
Li, F., Martienssen, R. & Cande, W. Z. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475, 244–248 (2011).
Ito, S. et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell 39, 632–640 (2010).
Kou, H., Zhou, Y., Gorospe, R. M. & Wang, Z. Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc Natl Acad Sci USA 105, 15714–15719 (2008).
Askree, S. H. et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA 101, 8658–8663 (2004).
Lauder, S. et al. Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol Cell Biol 16, 6783–6793 (1996).
Luo, D., Bernard, D. G., Balk, J., Hai, H. & Cui, X. The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24, 4135–4148 (2012).
Mok, Y. G. et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci USA 107, 19225–19230 (2010).
Han, Y. F. et al. The cytosolic iron-sulfur cluster assembly protein MMS19 regulates transcriptional gene silencing, DNA repair and flowering time in Arabidopsis. PLoS One 10, e0129137 (2015).
Williams, B. P., Pignatta, D., Henikoff, S. & Gehring, M. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet 11, e1005142 (2015).
Lei, M. et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA 112, 3553–3557 (2015).
Zhong, X. et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19, 870–875 (2012).
Wierzbicki, A. T., Haag, J. R. & Pikaard, C. S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008).
Kohbushi, H. et al. Arabidopsis cytosolic Nbp35 homodimer can assemble both [2Fe-2S] and [4Fe-4S] clusters in two distinct domains. Biochem Biophys Res Commun 378, 810–815 (2009).
Duan, C. G. et al. MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in Arabidopsis. PLoS Genet 11, e1005559 (2015).
Netz, D. J. et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol 8, 125–132 (2012).
Zhang, H. et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA 110, 8290–8295 (2013).
Shamloul, M., Trusa, J., Mett, V. & Yusibov, V. Optimization and utilization of Agrobacterium-mediated transient protein production in Nicotiana. J Vis Exp 86, doi: 10.3791/51204 (2014).
Walter, M. et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40, 428–438 (2004).
Wang, W. et al. An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23, 3565–3576 (2011).
Acknowledgements
We thank Dr. Xiaofeng Cui for providing met18 seeds, Dr. Xuemei Chen for the WT-LUC and ros1-5 seeds, the Mass Spectrometry Facility of the National Center for Protein Sciences at Peking University for mass spectrometry and Dr. Meng Rong for the help in data analyses, Liang Zeng for the DNA methylation data analyses. This work was supported by the National Natural Science Foundation of China (31571326 and 31522005) and Thousand Youth Talents Program to W.Q.
Author information
Authors and Affiliations
Contributions
X.W. and W.Q. designed the research. X.W., Q.L., W.Y., B.Q. and W.Q. performed the experiments. Q.L., X.W., Z.C. and W.Q. analyzed the data. Y.L., S.K. and W.Q. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, X., Li, Q., Yuan, W. et al. The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Sci Rep 6, 26443 (2016). https://doi.org/10.1038/srep26443
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26443
This article is cited by
-
Transcriptome analysis of a near-isogenic line and its recurrent parent reveals the role of Pup1 QTL in phosphorus deficiency tolerance of rice at tillering stage
Plant Molecular Biology (2022)
-
Insights into Salt Stress-Induced Biochemical, Molecular and Epigenetic Regulation of Spatial Responses in Pigeonpea (Cajanus cajan L.)
Journal of Plant Growth Regulation (2019)
-
Active DNA demethylation: mechanism and role in plant development
Plant Cell Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.