Abstract
Epithelial differentiation and stratification are essential for normal homeostasis, and disruption of these processes leads to both injury and cancer. The zinc-finger transciption factor KLF4 is a key driver of epithelial differentiation, yet the mechanisms and targets by which KLF4 controls differentiation are not well understood. Here, we define WNT5A, a non-canonical Wnt ligand implicated in epithelial differentiation, repair, and cancer, as a direct transcriptional target that is activated by KLF4 in squamous epithelial cells. Further, we demonstrate functionally that WNT5A mediates KLF4 control of epithelial differentiation and stratification, as treatment of keratinocytes with WNT5A rescues defective epithelial stratification resulting from KLF4 loss. Finally, we show that the small GTPase CDC42 is regulated by KLF4 in a WNT5A dependent manner. As such, we delineate a novel pathway for epithelial differentiation and stratification and define potential therapeutic targets for epithelial diseases.
Similar content being viewed by others
Introduction
Squamous epithelia provide important barriers from the outside world and are the most common sites for human cancer1,2,3. While some common squamous cell cancers are relatively treatable, esophageal squamous cell cancer continues to have an extremely poor prognosis, with a five-year survival of less than 20%4,5. Esophageal cancer is currently the 6th most common cause of cancer death in the world, and more than 90% of these esophageal cancers are squamous cell cancers, arising within the stratified squamous epithelial cells that normally line the esophagus3,6. Moreover, esophagitis and other disorders of the esophageal lining are among the greatest sources of morbidity and healthcare costs in the United States7. The squamous cells that line the esophagus proliferate, differentiate, and stratify to maintain normal homeostasis and for tissue repair while providing protection against damaging luminal substances. As such, perturbation of the pathways of normal esophageal squamous epithelial differentiation contributes to both esophageal injury and cancer8.
Similar to the linings of the skin and several other organs, the esophageal epithelium is organized into several layers with spatial separation of cell proliferation and differentiation9. Esophageal epithelial proliferation occurs in the basal layer, the layer furthest from the luminal surface, and epithelial cells differentiate as they migrate upwards through the overlying suprabasal and superficial cell layers before eventually being extruded into the lumen6,10. The process of stratification involves the stacking and linking of these squamous epithelial cells during differentiation resulting in a permeability barrier. Yet, while much has been learned about the movement of these squamous epithelial cells during differentiation, the transcriptional regulation of squamous epithelial differentiation and stratification is complex11,12, and the signaling pathways that underlie squamous epithelial differentiation are not well understood.
The transcription factor Krüppel-like factor 4 (KLF4) is a key driver of squamous epithelial differentiation, including in the esophagus13,14,15,16. Indicative of this, KLF4 is highly expressed in differentiating esophageal epithelial cells, and genetic ablation of Klf4 results in defective squamous epithelial differentiation. In skin, Klf4 deletion leads to loss of barrier function and defective late-stage differentiation, with early lethality by postnatal day 1 due to these barrier defects14, and in the esophagus, Klf4 ablation results in delayed differentiation, abnormal stratification, and the development of precancerous squamous cell dysplasia16. Moreover, KLF4 loss appears to contribute to human esophageal diseases, as KLF4 is downregulated in human esophageal squamous cell carcinoma17,18, and Klf4 loss also promotes skin carcinogenesis in mice19. Taken together, these data demonstrate a requirement of KLF4 for squamous epithelial differentiation and the relevance of KLF4 to human diseases20,21.
During squamous epithelial differentiation, KLF4 expression is regulated by a number of factors including ZNF750, p63, and PKCδ, as well as several lncRNA15,22,23,24. These factors regulate each other as well, suggesting a coordinated network that controls squamous epithelial differentiation and converges on KLF4. Yet the mechanisms by which KLF4 controls squamous epithelial differentiation and the specific downstream targets of KLF4 during differentiation remain to be delineated. Previously, we identified the non-canonical Wnt ligand Wnt5a25 as a gene that is differentially expressed in esophageal squamous epithelial cells of mice with esophageal specific deletion of Klf4, compared to controls16, suggesting that Wnt5a might be a target of KLF4 in squamous epithelia. WNT5A is particularly intriguing since it is critical for differentiation as well as polarity and migration of multiple cell types26,27,28. In addition, WNT5A is involved in tissue repair following injury29 and is decreased in human esophageal esophageal squamous cell cancer30. In the squamous epidermis, WNT5A induces epithelial differentiation during wound healing31. Taken together, these findings suggest that WNT5A may be important broadly for squamous epithelial differentiation. Yet, to date, KLF4 has not been linked to non-canonical Wnt signaling, and the transcriptional control of WNT5A is not well-defined. Here, employing murine genetic models and primary human and mouse esophageal keratinocytes, we identify WNT5A as a direct transcriptional target of KLF4. Importantly, primary esophageal keratinocytes are a useful model to study epithelial differentiation, as these cells display a predominantly basal cell phenotype in culture but can be induced to terminally differentiate with high concentrations of calcium or by raising cells to the air-liquid interface, allowing the formation of fully stratified epithelia in organotypic culture32,33. Using primary keratinocytes in organotypic culture, we show that loss of KLF4 impairs squamous epithelial differentiation and demonstrate functionally that WNT5A rescues the effects of KLF4 loss on differentiation and stratification. Additionally, we show that KLF4 inhibits activation of the small GTPase CDC42 in a WNT5A dependent manner. As such, we define a novel mechanism for the regulation of squamous epithelial differentiation.
Results
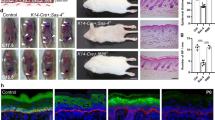
ED-L2-Cre/Klf4loxp/loxp mice have hyperplastic esophageal epithelia with evidence of abnormal differentiation and stratification16. Non-canonical Wnt5a was reduced 3-fold on microarray studies of murine esophagus with Klf4 deletion16, and we postulated that Wnt5a loss might be critical for the effects of Klf4 loss. Initially, we examined the expression and localization of WNT5A in esophageal epithelial of control and ED-L2-Cre/Klf4loxp/loxp mice. In esophageal epithelia of control mice, WNT5A localized to regions of cellular differentiation (Fig. 1A), a pattern of expression that overlapped with the expression domain of KLF4 (Figure S1A)16,34; WNT5A and KLF4 also co-localized in primary esophageal keratinocytes in culture (Figure S1B). In ED-L2-Cre/Klf4loxp/loxp mice, WNT5A was decreased markedly within the suprabasal and superficial layers of esophageal epithelia (Fig. 1B). Wnt5a was also reduced at the mRNA level in esophageal epithelia of mice with Klf4 loss compared to controls (Fig. 1C), suggesting that Wnt5a might be a transcriptional target of KLF4.
(A,B) By immunofluorescence, control mice had extensive WNT5A staining (red) in the suprabasal and superficial layers of their esophageal epithelia (A). In contrast, WNT5A was nearly absent from esophageal epithelia of ED-L2/Cre;Klf4loxP/loxP mice (B). DAPI (blue) was used as a counterstain, and the white dashed line represents the approximate location of the basement membrane. Scale bars: 25 μM. (C) By quantitative real-time PCR, Wnt5a mRNA expression was decreased in the esophageal epithelium of each ED-L2/Cre;Klf4loxP/loxP mouse compared to its littermate control (*p < 0.05). (D) Klf4 knockdown in primary mouse esophageal keratinocytes in culture using either of two shRNA constructs resulted in a 57% decrease in Wnt5a mRNA levels by qPCR. (*p < 0.05) (E) In primary human esophageal keratinocytes, inducible KLF4 knockdown with either of two shRNA constructs led to a 31–39% decrease in WNT5A mRNA expression by qPCR. (*p < 0.05) (F) Right panel: When human primary esophageal keratinocytes were induced to differentiate with CaCl2, KLF4 bound to the region of WNT5A between −945 to −762 upstream of the transcriptional start site. Left panel: No KLF4 binding to WNT5A was observed in actively proliferating keratinocytes. Lack of binding at −1992 to −1796 (not shown) confirmed specificity. (G) Primary mouse esophageal keratinocytes transfected with pCDNA3-Flag-Klf4 to express Klf4 had an 1148-fold increase in luciferase reporter activity compared to cells transfected with pCDNA3.1 control. (*p < 0.05).
To mechanistically dissect the regulation of WNT5A by KLF4, we employed primary esophageal keratinocytes with inducible KLF4 knockdown. Klf4 knockdown decreased Wnt5a mRNA by nearly 60% in primary mouse esophageal keratinocytes (Fig. 1D), and KLF4 knockdown decreased WNT5A mRNA by more than 30% in primary human esophageal keratinocytes (Fig. 1E); of note, the decreases in Wnt5a and WNT5A paralleled the reductions in Klf4 and KLF4. To determine whether WNT5A was a direct transcriptional target for KLF4, we examined the 5′ regulatory region of human WNT5A for putative KLF4-binding sites, using the computational program TESS35 and identified a putative KLF4 binding site between −945 to −762 from the translation start site. Using ChIP assays, we demonstrated binding of KLF4 to WNT5A in the region of the KLF4 site (Fig. 1F). Interestingly, this binding was observed only when cells were induced to differentiate with calcium chloride (Fig. 1F, right panel). To confirm that KLF4 transactivated Wnt5a, we transfected primary mouse esophageal keratinocytes with a Wnt5a luciferase reporter36. Compared to control, Klf4 transfection resulted in a 1148-fold increase in Wnt5a luciferase activity (Fig. 1G). Thus, KLF4 transcriptionally activates WNT5A during keratinocyte differentiation by binding to the 5′ regulatory region of WNT5A.
To determine whether KLF4 regulates esophageal epithelial differentiation and stratification via WNT5A, we examined the effects of KLF4 and WNT5A on primary esophageal keratinocytes in three-dimensional organotypic culture. In organotypic culture, control EPC2-hTERT cells formed mature, stratified epithelia featuring rounded, proliferative cells with high nuclear-cytoplasmic ratios in the basal layer that gave rise to flattened cells with compacted nuclei in the suprabasal and superficial layers (Fig. 2A). In contrast, KLF4 knockdown in EPC2-hTERT cells in organotypic culture yielded hyperplastic epithelia and rounded, immature-appearing cells with discernible nuclei in the suprabasal and superficial layers (Fig. 2B,C). Thus, KLF4 knockdown in esophageal keratinocytes in organotypic culture recapitulated the esophageal phenotype of the ED-L2-Cre/Klf4loxp/loxp mice (Figure S2). To test the requirement of WNT5A for KLF4 effects on esophageal epithelial stratification, we treated EPC2-hTERT cells in organotypic culture with recombinant WNT5A. Interestingly, addition of WNT5A had no effect on control EPC2-hTERT cells (Fig. 2D) while WNT5A treatment restored normal stratification of EPC2-hTERT cells with KLF4 knockdown (Fig. 2E,F). Thus, the abnormalities of epithelial stratification resulting from KLF4 loss are mediated by WNT5A.
(A) In three-dimensional organotypic culture, primary human esophageal keratinocytes form stratified epithelia. Cells in the basal layer were rounded with large nuclear-cytoplasmic ratios, and cells in the suprabasal and superficial layers were flattened with compacted nuclei. (B,C) In contrast, KLF4 knockdown in primary human esophageal keratinocytes in organotypic culture yielded epithelia that were hyperplastic, and cells appeared less mature, with cells outside of the basal layer maintaining a rounded appearance and large nuclei. (D–F) Treatment of the cultures with recombinant WNT5A had little effect on control epithelia (D) but restored normal epithelial stratification of primary esophageal keratinocytes with inducible KLF4 knockdown (E,F). Scale bars, 50 μm.
Normal cellular differentiation is essential for proper epithelial stratification6,37. As such, we sought to determine whether WNT5A corrected defective keratinocyte differentiation resulting from KLF4 knockdown by examining the expression patterns of keratin 14, which marks immature, proliferative keratinocytes typically located in the basal layer, and keratin 4, a marker of keratinocyte differentiation6,38. Compared to controls (Fig. 3A), esophageal epithelia from organotypic cultures with KLF4 knockdown had marked expansion of keratin 14 expression indicative of more immature keratinocytes (Fig. 3B,C). When control esophageal keratinocytes were treated with recombinant WNT5A (Fig. 3D), the localization of keratin 14 positive cells was similar to untreated cultures; WNT5A treatment of organotypic cultures with KLF4 knockdown resulted in a reduction of keratin 14 expressing cells and a more normal pattern of keratin 14 expression (Fig. 3E,F). The differentiation marker keratin 4, which was expressed in suprabasal and superficial layers of control cultures (Fig. 3G), was nearly absent from organotypic cultures with KLF4 knockdown (Fig. 3H,I), consistent with the consequences of Klf4 loss in vivo16. Again, WNT5A showed little effect on differentiation of control keratinocytes (Fig. 3J). However, WNT5A treatment was sufficient to rescue the effects of KLF4 knockdown on keratin 4 expression and therefore esophageal epithelial differentiation (Fig. 3K,L). Thus KLF4 controls esophageal epithelial differentiation and stratification through WNT5A.
(A) Keratin 14 (red), which marks immature keratinocytes, was restricted to the basal layer in organotypic cultures of control keratinocytes. (B,C) When KLF4 was knocked down in primary human esophageal keratinocytes, keratin 14 staining was more extensive, including in cells of the suprabasal layer. (D–F) Treatment of primary human esophageal keratinocytes with recombinant WNT5A had little effect in control cells (D), but restored the normal pattern of keratin 14 expression in cells with KLF4 knockdown, with staining again restricted to the basal layer in these cells (E,F). (G–I) Keratin 4 was expressed in the suprabasal and superficial layers of organotypic cultures of control human esophageal keratinocytes (G) while expression was nearly absent from cells with KLF4 knockdown (B,C). (J–L) Treatment of control primary human esophageal keratinocytes with recombinant WNT5A had minimal effect (J), but WNT5A treatment of organotypic cultures of cells with KLF4 knockdown normalized keratin 4 expression, with keratin 4 staining again seen in the suprabasal and superficial layers (K,L). DAPI (blue) was used as a counterstain. Scale bars, 25 μm.
WNT5A typically signals via the receptor tyrosine kinase ROR2 to activate β-catenin-independent Wnt pathways39,40,41. Members of the Rho family of GTPases, including CDC42 and RHOA, are important downstream targets of WNT5A and are critical for cellular processes such as differentiation, migration, and polarity42,43,44. To identify whether CDC42 and RHOA were downstream targets of KLF4-WNT5A signaling, we examined the consequences of KLF4 knockdown on CDC42 and RHOA activation in primary human esophageal keratinocytes. Interestingly, while RHOA activation was not altered by KLF4 knockdown (Figure S3A), KLF4 knockdown activated CDC42, and this activation was blocked by treatment with recombinant WNT5A (Fig. 4A). Additionally, CDC42 mRNA levels were not affected by KLF4 knockdown (Figure S3B), indicating that KLF4 did not regulate CDC42 transcription. Thus, in esophageal keratinocytes, KLF4 upregulates WNT5A to inhibit CDC42 activity.
(A) Quantification of GTPase activation (n = 3) demonstrated increased CDC42 activity with KLF4 knockdown in primary human esophageal keratinocytes induced to differentiate with CaCl2; activation of CDC42 in cells with KLF4 knockdown was abolished by treatment with recombinant WNT5A. (B) Proposed model for the regulation of squamous epithelial cell differentiation and stratification via KLF4 and WNT5A. Previously described regulators of KLF4 are indicated in gray.
Discussion
The mucosal barrier of the esophagus is an essential line of defense against external damaging agents, and proper keratinocyte differentiation and stratification are required to maintain the integrity of the epithelial barrier6,8. The Krüppel-like factor family member KLF4 is critical for the regulation of epithelial homeostasis and disease, including in the squamous esophagus and skin, and mice with Klf4 deletion in esophageal keratinocytes develop altered cell morphology, delayed differentiation, and abnormal stratification, leading to precancerous esophageal squamous cell dysplasia14,16,20. Yet, the molecular mechanisms by which KLF4 controls squamous epithelial differentiation and stratification have not been clear.
Non-canonical WNT5A signaling has been extensively studied in development where it regulates cell polarity and directional cell movement, but the importance of WNT5A for adult epithelial stratification is less clear43. Recently, WNT5A was shown to restore stratification of the apical ectodermal ridge in the developing limb45. Wnt5a deletion in mice compromises differentiation of the hair follicle36, and in the interfollicular epidermis, WNT5A activation induces keratinocyte differentiation during wound healing31, consistent with a role for WNT5A in tissue repair seen in other contexts29. Our findings here highlight the importance of non-canonical WNT5A for epithelial squamous differentiation and stratification, and since WNT5A treatment of normal epithelia has no overt effects on differentiation and stratification, WNT5A might be effective to therapeutically target defective differentiation and/or stratification and to promote esophageal wound healing following injury. Moreover, Wnt5a is decreased in a murine model of esophageal squamous cell carcinogenesis46 suggesting that WNT5A may have tumor suppressive function in esophageal squamous cell carcinogenesis. Nonetheless, while organotypic cultures are useful models of carcinogenesis32,47,48,49, overexpression of Klf4 results in esophageal squamous cell cancer via activation of inflammatory pathways in vivo50 and thus further study is required to exclude that higher levels of WNT5A, which is downstream of KLF4, promote esophageal inflammation and carcinogenesis in vivo.
In squamous epithelia, KLF4 is regulated directly and/or indirectly by the transcription factors p63, ZNF750, MAF, and MAFB, by PKCδ, and by the lncRNA ANCR and TINCR15,22,23,24. Integrating our data with the published literature, we propose a broad network that controls squamous epithelial differentiation and stratification, converging on KLF4 and WNT5A (Fig. 4B). Interestingly, CDC42 is inhibited by WNT5A in esophageal keratinocytes, while WNT5A may activate CDC42 or the WNT5A and CDC42 pathways may cooperate in other contexts51,52,53. CDC42 itself can either promote or inhibit differentiation54,55, and in fact, both upregulation and downregulation of CDC42 activity can inhibit cell growth in the same cell type, suggesting that tight regulation of CDC42 may be essential for normal differentiation54,56. In addition, both WNT5A and CDC42 can regulate β-catenin-dependent Wnt signaling, which may play a role in esophageal squamous cell carcinogenesis57,58,59,60. In the stomach, WNT5A from gastric innate lymphoid cells activates epithelial RHOA, in contrast to our findings, suggesting that the pathways downstream of WNT5A vary by context61. Thus the contextual and coordinate functions of KLF4, WNT5A, and CDC42 in epithelial differentiation, stratification, and carcinogenesis require further study.
In sum, we delineate a novel pathway for epithelial differentiation and stratification acting via the key differentiation-promoting transcription factor KLF4 and the non-canonical Wnt ligand WNT5A. A number of key differentiation factors converge on KLF415,22,23,24, and we demonstrate that loss of KLF4 leads to defects in epithelial differentiation and stratification that are rescued by WNT5A. Thus, in outlining the mechanisms underlying squamous epithelial differentiation and stratification, we define potential therapeutic targets for diseases and disorders of the esophagus and other stratified squamous epithelia, sources of significant human morbidity and mortality1,2,3,7. Moreover, as KLF4 is important for cellular differentiation and carcinogenesis more broadly20,21, these targets may also be relevant to other tissues and cell types.
Methods
ED-L2-Cre/Klf4loxP/loxP mice
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania and carried out in accordance with the approved guidelines. Mice homozygous for the floxed Klf4 gene and hemizygous for the ED-L2/Cre transgene have been previously described16. For analyses, esophagi from 3 month-old mice were removed and processed as described16. For experiments with ED-L2-Cre/Klf4loxP/loxP mice, sex-matched littermate Klf4loxP/loxP mice lacking the Cre transgene served as controls. All mice used for experiments were on a mixed genetic background.
Cell Culture and Treatment
The isolation and culture of primary mouse esophageal keratinocytes were described elsewhere62. Primary human esophageal keratinocytes (EPC2) retrovirally transduced with hTERT to generate EPC2-hTERT cells63 were cultured as previously described47. HEK 293T cells used for lentivirus production were purchased from ATCC. For WNT5A treatment, recombinant mouse/human WNT5A (R&D Systems) was added at 100 ng/ml into growth media.
Viral constructs and infections
The lentiviral vector pLKO.1 puro64, a gift from Bob Weinberg (Addgene plasmid # 8453) was used to express 2 distinct short hairpin RNAs (shRNA) against mouse Klf4, and the lentiviral vector TET-pLKO-neo65, a gift from Dmitri Wiederschain (Addgene plasmid # 21916), was used to express 2 distinct shRNA against human KLF4. For TET-pLKO-neo, shRNA was induced in cells in two-dimensional culture with 4 μg/ml doxycycline for 7 days. Additional methods are available in Supplemental Information.
Immunohistochemistry/Immunofluorescence/Western Blotting
Immunohistochemistry, immunofluorescence, and Western blots were performed using standard protocols. For descriptions of the protocols and antibodies used, see the Supplemental Information.
RNA analyses
RNA was extracted from primary esophageal keratinocytes using the GeneJet RNA Purification Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Reverse transcription was performed with the Maxima First-Strand cDNA Synthesis kit (Thermo Fisher Scientific). Quantitative real-time PCR (qPCR) was performed in triplicate using an ABI Step-One Plus sequence detection system (Thermo Fisher Scientific) and SYBR Green PCR master mix (Thermo Fisher Scientific). TATA box binding protein gene (TBP) and GAPDH were used as internal controls. Primer sequences are available in Supplemental Information.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed in triplicate with the ChIP assay kit (Millipore) as described previously47. Cells were treated with 1% formaldehyde for 10 minutes to cross-link associated protein to DNA, lysed, and sonicated. After a 10-fold dilution, samples were pre-cleared with protein A-agarose/salmon sperm DNA for 30 minutes at 4 °C and incubated overnight at 4 °C with 1:500 anti-KLF4 antibody66 or 1:500 anti-mouse IgG (Sigma) as a negative control. Additional methods are available in Supplemental Information.
Reporter assays
The Wnt5a-luc reporter plasmid, containing the region from −1.66 to + 2.29 kb relative to the mouse Wnt5a transcription start site in the pGL4 Luciferase Reporter Vector (Promega), was a gift of G. Paolo Dotto36. To express Klf4, a Flag-tagged full-length mouse Klf4 cDNA was subcloned into the pCDNA3.1 vector (Life Technologies). Mouse primary esophageal keratinocytes were transfected with either pCDNA3.1 or pCDNA3-Flag-Klf4 and with either pGL4 or Wnt5a-luc at 70% confluence in triplicate on 24-well plates using Turbofect transfection reagent (Thermo Fisher Scientific). Cells were lysed after 48 hours with Cell Lysis Buffer (Pharmingen), and luciferase reporter activity was analyzed using luciferase assay reagent (Promega) with a GLOMAX multi detection system (Promega). Luciferase activity was normalized to Renilla and expressed as relative luciferase activity.
Organotypic culture
EPC2-hTERT cells containing TET-pLKO-neo constructs were grown in three-dimensional organotypic culture as described previously32. KLF4 knockdown was induced with 4 μg/ml of doxycycline from days 7–15, and recombinant WNT5A was added from days 11–15. Cultures were fixed overnight in 10% buffered formalin phosphate (Fisher Scientific) before paraffin embedding and sectioning.
CDC42 and RHOA activation assays
G-LISA CDC42 and RHOA activation assays (Cytoskeleton Inc.) were performed according to the manufacturer’s instructions. Briefly, 50 μg of protein lysates were incubated at 4 °C for 30 minutes under agitation. After washes and incubation with the antigen presenting buffer, the plates were incubated with primary and secondary antibodies at room temperature for 45 minutes. Absorbance at 490 nm was measured using a Tecan Infinite 200 PRO microplate reader (Tecan) following a 15 minute incubation with the HRP detection reagent.
Additional Information
How to cite this article: Tetreault, M.-P. et al. KLF4 transcriptionally activates non-canonical WNT5A to control epithelial stratification. Sci. Rep. 6, 26130; doi: 10.1038/srep26130 (2016).
References
Albert, M. R. & Weinstock, M. A. Keratinocyte Carcinoma. CA Cancer J Clin 53, 292–302, doi: 10.3322/canjclin.53.5.292 (2003).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2015. CA Cancer J Clin 65, 5–29, doi: 10.3322/caac.21254 (2015).
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108, doi: 10.3322/caac.21262 (2015).
Stoner, G. D. & Gupta, A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis 22, 1737–1746 (2001).
Kim, E., Koroukian, S. & Thomas, C. R., Jr. Conditional Survival of Esophageal Cancer: An Analysis from the SEER Registry (1988–2011). J Thorac Oncol 10, 1490–1497, doi: 10.1097/jto.0000000000000649 (2015).
Squier, C. A. & Kremer, M. J. Biology of oral mucosa and esophagus. J Natl Cancer Inst Monogr 29, 7–15 (2001).
Peery, A. F. et al. Burden of Gastrointestinal, Liver, and Pancreatic Diseases in the United States. Gastroenterology 149, 1731–1741.e1733, doi: 10.1053/j.gastro.2015.08.045 (2015).
Marchiando, A. M., Graham, W. V. & Turner, J. R. Epithelial barriers in homeostasis and disease. Annu Rev Pathol 5, 119–144 (2010).
Rosekrans, S. L., Baan, B., Muncan, V. & van den Brink, G. R. Esophageal development and epithelial homeostasis. Am J Physiol Gastrointest Liver Physiol 309, G216–G228, doi: 10.1152/ajpgi.00088.2015 (2015).
Karam, S. M. Lineage commitment and maturation of epithelial cells in the gut. Front Biosci 4, D286–298 (1999).
Dai, X. & Segre, J. A. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev 14, 485–491, doi: 10.1016/j.gde.2004.07.002 (2004).
Nagarajan, P., Romano, R. A. & Sinha, S. Transcriptional control of the differentiation program of interfollicular epidermal keratinocytes. Crit Rev Eukaryot Gene Expr 18, 57–79 (2008).
McConnell, B. B. & Yang, V. W. Mammalian Kruppel-like factors in health and diseases. Physiol Rev 90, 1337–1381, doi: 10.1152/physrev.00058.2009 (2010).
Segre, J. A., Bauer, C. & Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22, 356–360 (1999).
Sen, G. L. et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 22, 669–677, doi: 10.1016/j.devcel.2011.12.001 (2012).
Tetreault, M. P. et al. Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel-like factor 4 in murine esophagus. Gastroenterology 139, 171-181 e179, doi: 10.1053/j.gastro.2010.03.048 (2010).
Luo, A. et al. Discovery of Ca2 + -relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNA microarray. Oncogene 23, 1291–1299 (2004).
Wang, N. et al. Down-regulation of gut-enriched Krüppel-like factor expression in esophageal cancer. World J Gastroenterol 8, 966–970 (2002).
Li, J. et al. Deficiency of the Kruppel-like factor KLF4 correlates with increased cell proliferation and enhanced skin tumorigenesis. Carcinogenesis 33, 1239–1246, doi: 10.1093/carcin/bgs143 (2012).
Tetreault, M. P., Yang, Y. & Katz, J. P. Kruppel-like factors in cancer. Nat Rev Cancer 13, 701–713, doi: 10.1038/nrc3582 (2013).
McConnell, B. B., Ghaleb, A. M., Nandan, M. O. & Yang, V. W. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 29, 549–557, doi: 10.1002/bies.20581 (2007).
Chew, Y. C., Adhikary, G., Xu, W., Wilson, G. M. & Eckert, R. L. Protein kinase C delta increases Kruppel-like factor 4 protein, which drives involucrin gene transcription in differentiating keratinocytes. J Biol Chem 288, 17759–17768, doi: 10.1074/jbc.M113.477133 (2013).
Cordani, N. et al. Mutant p53 subverts p63 control over KLF4 expression in keratinocytes. Oncogene 30, 922–932, doi: 10.1038/onc.2010.474 (2011).
Lopez-Pajares, V. et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell 32, 693–706, doi: 10.1016/j.devcel.2015.01.028 (2015).
Veeman, M. T., Axelrod, J. D. & Moon, R. T. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell 5, 367–377, doi: 10.1016/S1534-5807(03)00266-1 (2003).
Witze, E. S., Litman, E. S., Argast, G. M., Moon, R. T. & Ahn, N. G. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science 320, 365–369, doi: 10.1126/science.1151250 (2008).
Lai, S. L., Chien, A. J. & Moon, R. T. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res 19, 532–545, doi: 10.1038/cr.2009.41 (2009).
Chien, A. J., Conrad, W. H. & Moon, R. T. A Wnt survival guide: from flies to human disease. J Invest Dermatol 129, 1614–1627, doi: 10.1038/jid.2008.445 (2009).
Miyoshi, H., Ajima, R., Luo, C. T., Yamaguchi, T. P. & Stappenbeck, T. S. Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113, doi: 10.1126/science.1223821 (2012).
Li, J. et al. WNT5A antagonizes WNT/beta-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol Ther 10, 617–624 (2010).
Fathke, C. et al. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol 7, 4, doi: 10.1186/1471-2121-7-4 (2006).
Kalabis, J. et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 7, 235–246, doi: 10.1038/nprot.2011.437 (2012).
Compton, C. C., Warland, G., Nakagawa, H., Opitz, O. G. & Rustgi, A. K. Cellular characterization and successful transfection of serially subcultured normal human esophageal keratinocytes. J Cell Physiol 177, 274–281, doi: 10.1002/(sici)1097-4652(199811)177:2 <274::aid-jcp9> 3.0.co;2-k (1998).
Ali, I. et al. Intramucosal distribution of WNT signaling components in human esophagus. J Clin Gastroenterol 43, 327–337 (2009).
Schug, J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics Chapter 2, Unit 2.6, doi: 10.1002/0471250953.bi0206s21 (2008).
Hu, B. et al. Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev 24, 1519–1532, doi: 10.1101/gad.1886910 (2010).
Watt, F. M. Involucrin and other markers of keratinocyte terminal differentiation. J Invest Dermatol 81, 100s–103s (1983).
Moll, R., Divo, M. & Langbein, L. The human keratins: biology and pathology. Histochem Cell Biol 129, 705–733, doi: 10.1007/s00418-008-0435-6 (2008).
Nishita, M., Enomoto, M., Yamagata, K. & Minami, Y. Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol 20, 346–354, doi: 10.1016/j.tcb.2010.03.001 (2010).
O’Connell, M. P. et al. The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene 29, 34–44, doi: 10.1038/onc.2009.305 (2010).
Angers, S. & Moon, R. T. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 10, 468–477, doi: 10.1038/nrm2717 (2009).
Melendez, J., Grogg, M. & Zheng, Y. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem 286, 2375–2381, doi: 10.1074/jbc.R110.200329 (2011).
Kikuchi, A., Yamamoto, H., Sato, A. & Matsumoto, S. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol (Oxf) 204, 17–33, doi: 10.1111/j.1748-1716.2011.02294.x (2012).
McCormack, J., Welsh, N. J. & Braga, V. M. Cycling around cell-cell adhesion with Rho GTPase regulators. J Cell Sci 126, 379–391, doi: 10.1242/jcs.097923 (2013).
Conte, D. et al. The apical ectodermal ridge of the mouse model of ectrodactyly Dlx5;Dlx6-/- shows altered stratification and cell polarity, which are restored by exogenous Wnt5a ligand. Hum Mol Genet 25, 740–754, doi: 10.1093/hmg/ddv514 (2016).
Osei-Sarfo, K., Urvalek, A. M., Tang, X. H., Scognamiglio, T. & Gudas, L. J. Initiation of esophageal squamous cell carcinoma (ESCC) in a murine 4-nitroquinoline-1-oxide and alcohol carcinogenesis model. Oncotarget 6, 6040–6052 (2015).
Yang, Y. et al. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res 71, 6475–6484, doi: 10.1158/0008-5472.CAN-11-1702 (2011).
Rhim, A. D. & Rustgi, A. K. Three-Dimensional Organotypic Culture of Stratified Epithelia. Cold Spring Harbor Protocols 2015, 349–353, doi: 10.1101/pdb.prot078311 (2015).
Kim, S. H. et al. Tumorigenic conversion of primary human esophageal epithelial cells using oncogene combinations in the absence of exogenous Ras. Cancer Res 66, 10415–10424, doi: 10.1158/0008-5472.CAN-06-2104 (2006).
Tetreault, M. P. et al. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology 139, 2124–2134, doi: 10.1053/j.gastro.2010.08.048 (2010).
Florian, M. C. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396, doi: 10.1038/nature12631 (2013).
Lee, J. G. & Heur, M. Interleukin-1beta-induced Wnt5a enhances human corneal endothelial cell migration through regulation of Cdc42 and RhoA. Mol Cell Biol 34, 3535–3545, doi: 10.1128/mcb.01572-13 (2014).
Schlessinger, K., McManus, E. J. & Hall, A. Cdc42 and noncanonical Wnt signal transduction pathways cooperate to promote cell polarity. J Cell Biol 178, 355–361, doi: 10.1083/jcb.200701083 (2007).
Kesavan, G. et al. Cdc42/N-WASP signaling links actin dynamics to pancreatic beta cell delamination and differentiation. Development 141, 685–696, doi: 10.1242/dev.100297 (2014).
Sakamori, R. et al. Cdc42 and Rab8a are critical for intestinal stem cell division, survival, and differentiation in mice. J Clin Invest 122, 1052–1065, doi: 10.1172/jci60282 (2012).
Yang, L., Wang, L. & Zheng, Y. Gene Targeting of Cdc42 and Cdc42GAP Affirms the Critical Involvement of Cdc42 in Filopodia Induction, Directed Migration, and Proliferation in Primary Mouse Embryonic Fibroblasts. Mol Biol Cell 17, 4675–4685, doi: 10.1091/mbc.E06-05-0466 (2006).
Tarapore, R. & Katz, J. In Molecular Pathology of Neoplastic Gastrointestinal Diseases Vol. 7 Molecular Pathology Library (eds Antonia R. Sepulveda & John P. Lynch ) Ch. 4, 53–66 (Springer: US,, 2013).
Wu, X. et al. Cdc42 controls progenitor cell differentiation and beta-catenin turnover in skin. Genes Dev 20, 571–585, doi: 10.1101/gad.361406 (2006).
Mikels, A. J. & Nusse, R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 4, e115, doi: 10.1371/journal.pbio.0040115 (2006).
Webster, M. R. & Weeraratna, A. T. A Wnt-er Migration: The Confusing Role of β-Catenin in Melanoma Metastasis. Science Signaling 6, pe11–pe11, doi: 10.1126/scisignal.2004114 (2013).
Hayakawa, Y. et al. Mist1 Expressing Gastric Stem Cells Maintain the Normal and Neoplastic Gastric Epithelium and Are Supported by a Perivascular Stem Cell Niche. Cancer Cell 28, 800–814, doi: 10.1016/j.ccell.2015.10.003 (2015).
Yang, Y., Goldstein, B. G., Nakagawa, H. & Katz, J. P. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J 21, 543–550, doi: 10.1096/fj.06-6694com (2007).
Harada, H. et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res 1, 729–738 (2003).
Stewart, S. A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003).
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009).
Yang, Y., Goldstein, B. G., Chao, H. H. & Katz, J. P. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther 4, 1216–1221 (2005).
Acknowledgements
This work was supported by NIH K99 DK094977 to M.-P.T. and NIH NIDDK R01 DK069984 to J.P.K. and by the University of Pennsylvania Center for Molecular Studies in Digestive and Liver Diseases (NIH NIDDK P30 DK050306) through the Molecular Pathology and Imaging Core, the Molecular Biology/Gene Expression Core, the Cell Culture Core, and the Transgenic and Chimeric Mouse Core and by NIH NCI P01 CA098101 (“Mechanisms of Esophageal Carcinogenesis”).
Author information
Authors and Affiliations
Contributions
M.-P.T. designed and performed experiments, analyzed and interpreted data, obtained funding and wrote the manuscript; D.W. designed and performed experiments, analyzed and interpreted data; K.S. designed and performed experiments, analyzed and interpreted data; Y.Y. designed and performed experiments, analyzed and interpreted data; and J.P.K. designed experiments, analyzed and interpreted data, obtained funding, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tetreault, MP., Weinblatt, D., Shaverdashvili, K. et al. KLF4 transcriptionally activates non-canonical WNT5A to control epithelial stratification. Sci Rep 6, 26130 (2016). https://doi.org/10.1038/srep26130
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26130
This article is cited by
-
Effect of overexpression of KLF4 on the growth and development of hair follicles in mice
Development Genes and Evolution (2023)
-
KLF4 inhibition promotes the expansion of keratinocyte precursors from adult human skin and of embryonic-stem-cell-derived keratinocytes
Nature Biomedical Engineering (2019)
-
A novel DLX3–PKC integrated signaling network drives keratinocyte differentiation
Cell Death & Differentiation (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.