Abstract
Binary systems based on site-specific recombination have been used for tumor specific transcription targeting of suicide genes in animal models. In these binary systems a site specific recombinase or integrase that is expressed from a tumor specific promoter drives tumor specific expression of a cytotoxic gene. In the present study we developed a new cancer specific binary expression system activated by the Integrase (Int) of the lambdoid phage HK022. We demonstrate the validity of this system by the specific expression of a luciferase (luc) reporter in human embryonic kidney 293T (HEK293T) cells and in a lung cancer mouse model. Due to the absence viral vectors and of cytotoxicity the Int based binary system offers advantages over previously described counterparts and may therefore be developed into a safer cancer cell killing system.
Similar content being viewed by others
Introduction
Site-specific recombinases are widely used for genome manipulations by catalysis of site-specific recombination reactions (SSR) between two defined DNA sequences of ~20–200 base pairs (bp) functioning as recombination sites (RSs)1,2,3,4,5,6. SSR reactions result in integration, excision or inversion, depending on the location and relative orientation of the RSs. A widely used SSR-based technique is the recombinase-mediated-cassette-exchange (RMCE) reaction allowing clean exchange of an existing gene by a gene of interest7. SSR systems that are currently used to manipulate genomes of higher organisms include those of the bacteriophage P1 Cre/lox, the baker yeast Flp/frt and the bacteriophage φC31 Int/att. The recombinases of the former two systems belong to the tyrosine recombinase family and of the latter system to the serine recombinase family8.
SSR systems are also used in binary systems for cell and gene specific expression that are based on two DNA modules. One module carries a recombinase substrate comprising a target gene silenced by a transcription terminator placed between two RSs. The second module carries the recombinase gene controlled by a cell type specific promoter. This promoter targets the expression of the SSR to the desired cells where it excises the terminator and thus activates the expression of the gene of interest9,10,11.
Coliphage HK022 encodes an Integrase recombinase (Int) belonging to the tyrosine integrase family that catalyzes the integration of the phage genome into its RS on the Escherichia coli chromosome as well as its excision from the E. coli chromosome. In these regards it acts very much like the well-documented coliphage λ Integrase12. Int targets a 21 bp long bacterial RS termed attB or BOB′ to indicate its composition of a central 7 bp overlap region (O) and the two flanking incomplete inverted repeats of 7 bp (B and B′). O is the site of DNA exchange while B and B′ are Int binding sites. The phage recombination site termed attP is ~200 bp long. It contains a 21 bp core (COC′) that is similar to attB and is flanked by two long arms (P and P′) that carry additional Int binding sites and others that bind accessory proteins required for the SSR reactions. The phage integration reaction occurs between attP and attB. It yields two hybrid recombination sites flanking the integrated prophage termed attL (BOP′) and attR (POB′). The reverse prophage excision reaction occurs between the attL and attR sites and restores the original attP and attB13. We have previously applied Int for genome manipulation in plants, cyanobacteria and human cells and have shown that the accessory proteins that are required in the E. coli host are dispensable in the eukaryotic milieu14,15,16,17,18. In the present study we developed a cancer specific binary cell expression system that is based on the hTERT promoter. hTERT serves as the catalytic reverse transcriptase subunit of telomerase and is specifically expressed in immortal and cancer cells19,20. In this binary cell expression system hTERT-promoted Int specifically activates a luc reporter in immortalized human embryonic kidney 293 cells (HEK293T) and in mouse lung tumors originating from injected Lewis lung carcinoma line 1 (LLC1). This cancer specific expression system offers certain advantages over those previously described9,10,11 and may be applied to express toxins specifically in cancerous tumors.
Results
Cancer specific luc reporter expression mediated by an Int-based binary system
Two plasmids were constructed to generate a cancer specific binary cell expression system activated by Int. One plasmid expresses the int gene under the control of the cancer specific promoter hTERT19,20 or by the constitutive cytomegalovirus (CMV) promoter as control. The other carries a luc gene separated from a CMV promoter by a transcription terminator (Stop) flanked by tandem attR and attL recombination sites (Fig. 1). Int-catalyzed attR x attL recombination excises Stop allowing expression of the luc reporter.
Scheme of the Int-catalyzed binary system to express a luc-reporter in cancer cells.
(A) The recombination substrate carrying a silenced luc reporter separated from its CMV promoter by a transcription terminator (Stop) flanked by attR and attL recombination substrates. (B) The recombination products generated by Int: The activated luc-reporter separated from its CMV promoter by the small bacterial recombination site attB (top) and the excised non-replicative Stop (bottom).
Validation that the luc reporter cancer specific binary cell expression system is activated by Int in HEK293T cells
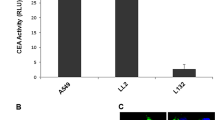
Our first goal was to validate the efficiency of the binary cell expression system activated by Int in cancer cells. To this end we employed the immortalized HEK293T cells that are transformed both by adenovirus and by the SV40 large tumor antigen. This line expresses a high level of hTERT sufficient to maintain their telomeres indefinitely, unlike normal human cells that do not express hTERT21,22. Therefore, placing int under the control of hTERT promoter was expected to express in the HEK293T cells. HEK293T cells were co-transfected with the silent luc substrate carrying the CMV-attR-Stop-attL-luc cassette (pAE1855, Fig. 1) and a plasmid expressing int either under the control of the cancer specific hTERT promoter (pNA1263) or under the constitutive CMV promoter (pNA979) that served as a positive control. The quantitative analysis of the Luc activity assay in HEK293T cells (Fig. 2A) indicated that int driven by the hTERT promoter catalyzed attR x attL excision at ~20% the extent of that observed with the CMV control. In the absence of int Luc activity was negligible. Thus, int expressed from the cancer specific hTERT promoter catalyzed rather efficiently site-specific recombination in the immortalized HEK293T cells.
Luc activity assays of the hTERT-promoted Int-activation of the binary system in HEK293T cells (A) and in LLC-Kat cells (B), each compared with that of a CMV-promoted Int system. The data represent the mean value of three experiments; the error bars indicate standard deviation. RSL represents the attR-Stop-attL cassette (Fig. 1A).
A lung-specific DNA delivery in mice
Efficient in vivo transformation and gene delivery in mammals is a considerable technical challenge. Among different approaches and DNA delivery agents, including lentiviral vectors, we have observed that the linear in vivo-jet PEI (Polyplus, France) is the most effective. This linear polyethylenimine reagent is used to deliver DNA, siRNA and oligonucleotides. It forms stable complexes with the nucleic acid cargo, protecting it from degradation thereby facilitating in vivo delivery. These complexes pass the capillary barrier in the lung, explaining the propensity of in vivo-jet PEI to deliver its cargo specifically into lungs23. As reporter, we found that the Luciferase gene (luc), whose product emits characteristic yellow-green bioluminescence in the presence of its luciferin substrate is most suitable. BALB/c mice were intravenous (IV) tail-injected with a sample of the jet PEI delivery agent complexed with a reporter plasmid (pYM1600) that carries the luc gene under the control of CMV promoter. 24 hours later the mice were injected with the Luciferase substrate luciferin and underwent bioluminescence imaging. The treated mouse (Fig. 3A) was compared with a luciferin-injected luc-transgenic mouse as a positive control (Fig. 3B) and a BALB/c mouse injected with the luciferin substrate alone as a negative control (Fig. 3C). The treated mouse injected with the gene delivery system (Fig. 3A) showed specific bioluminescent activity in the lung area while the transgenic mouse (Fig. 3B) showed abdominal activity. We concluded that the in vivo-jet PEI carried the luc plasmid to the lungs and therefore we set out to develop lung cancer mice as an in vivo model to explore our Int activated cancer specific binary system in vivo.
Int activates the luc reporter binary system in the lungs of BALB/c mice
We next tested if our binary system is also active in vivo in mice as it does in cell culture. Two mice were IV-tail injected with jet PEI complexed with different doses of the constitutive CMV-int expressing plasmid and the silent attR-Stop-attL-luc substrate plasmid (20 μg + 30 μg and 10 μg + 40 μg respectively). As a positive control two mice were each similarly treated with two doses of a CMV-luc expressing plasmid (pYM1600, 50 μg and 10 μg). 24 hours later bioluminescence imaging of these mice (Fig. 4A) has shown that in mouse No. 4 injected with the higher dose of the silent substrate along with the lower dose of the Int expressing plasmid Int has catalyzed the recombination reaction that lead to the activation of the silent luc reporter in the lungs’ area (Fig. 1). Quantification of these assays (Fig. 4B) shows that the efficiency of the binary reaction is comparable to that acquired by control mouse No. 2 injected with higher dose of a CMV-luc expressing plasmid. These results show that under a constitutive CMV-int expression the binary system is also active in vivo in the mouse lungs. The sensitivity of Luc detection strongly depends on the concentration ratios of the injected plasmid, explained by the virtual absence of a positive signal in mice Nos 1 and 3.
Int activity in the lungs of BALB/c mice.
(A) Bioluminescence analysis. BALB/c mice were IV-injected with the in vivo-jet PEI complexed with indicated amounts of a CMV-luc plasmid as control (mice Nos 1 and 2) or with the complexed binary system (Nos 3 and 4). (B) Quantitative analysis of the bioluminescence data in the lung areas. The bars show the mean value of three experiments; the error bars indicate standard deviation.
An experimental lung cancer mice model
Having demonstrated that the jet PEI DNA delivery reagent has delivered the luc plasmid exclusively to the lungs, our next step was to develop a mouse lung cancer model. To this end we used the Lewis lung carcinoma line 1 (LLC1)24 labelled with a far-red mutant of the red fluorescent gene of the sea anemone Entacmaea quadricolor known as Katushka cells (LLC-Kat)25,26. LLC1 cells remain highly tumorigenic in C57BL/6 mice and produce primary tumors and lung metastases. LLC1 cells are widely used as a model for the development of lung-metastatic tumors in mice and are useful in studying the mechanisms of cancer chemotherapeutic agents27. The presence of the Katushka label in LLC1 cells facilitates the detection of the lung metastases. A preliminary experiment with these cells shows that the binary system in these cells works as efficiently as in HEK293T cells (Fig. 2B).
Lung tumor metastases development was examined nine days following the injection of the LLC-Kat cells using in vivo micro-CT scanning. No tumors were detected in the lungs of the non-injected control [Fig. 5A(a)] whereas in the injected mice metastases were detected and localized in the lungs [arrows in Fig. 5B(a)]. The presence of metastases in LLC-Kat lungs were verified by ex-vivo fluorescence detection using the Maestro imaging system [Fig. 5B(b)] as compared to non-injected control [Fig. 5A(b). Further validation that the injected mice have acquired lung cancer were performed by H&E staining [Fig. 5B(c)] and by immunostaining using proliferating cancer nuclear antigen (PCNA) [Fig. 5B(d)] compared to control healthy lungs [Fig. 5A(c,d)]. In both cases the cancer metastases were evident. These data demonstrate the generation of a mouse lung cancer model using injection of LLC-Kat cells in C57BL/6 mice. Moreover the mice died after approximately 14 days post injections.
In vivo imaging and immunohistochemistry analysis of lungs from healthy (A) and LLC-Kat injected C57BL/6 mice (B). Panels (a), in vivo imaging using a micro-CT device. Arrows mark metastases location; panels (b), ex vivo fluorescence imaging of the lungs using a Maestro system; panels (c), H&E staining; panels (d), immunostaining for PCNA.
Cancer specificity of the Int-based binary cell expression system in mice
Finally, to test the cancer specificity of the binary Int-activated cell expression system in mice we performed a comparative analysis of this system between healthy C57BL/6 mice (Fig. 6A) and LLC-Kat-infected mice (Fig. 6B). In each of these two types of C57BL/6 mice, there were four groups (4 mice/group) that were IV tail-injected with the in vivo-jet PEI carrier complexed with different combinations of plasmids [panels (b–e)] while groups (a) remained untreated. In groups (b) the jet PEI carrier was complexed with the CMV-luc reported as positive control; in groups (c) the carrier was complexed with the silent recombination substrate (CMV-RSL-luc) alone to validate the absence of luc leakage; in groups (d,e) the carrier was complexed with the silenced substrate and the CMV-promoted Int (d) or with the hTERT-promoted Int (e). 24 hours later all mice were injected with luciferin and underwent bioluminescence imaging. As expected, in both healthy and cancerous mice the negative controls (a) and the ones treated with the silent substrate (c) showed no bioluminescence while the positive controls (b) did. So did the two groups (d) treated with the silent substrate along with the constitutive CMV-Int. However, in groups (e) only the cancerous mice treated with the silent substrate along with the hTERT-Int showed bioluminescence [Fig. 6B(e)] while in the healthy mice [Fig. 6A(e)] no bioluminescence was evident. Quantitative data of the bioluminescence mice are shown in Fig. 6C. Though in the cancerous mice the bioluminescence that was specifically catalyzed by the activity of hTERT-Int was lower compared to the constitutive CMV-Int, it was consistent with the expected specificity of our binary cancer cell expression system. To reinforce this cancer specific effect, the treated healthy and cancer mice underwent an immunohistochemical analysis of Luc expression (Fig. 7). The expected negative controls [panels (a,c)] did not show any immune response, the positive treatments [panels (b,d)] showed an immune response in both types of mice, while among the mice treated with the hTERT-Int binary system [panels (e)] only the cancerous ones showed an immune response. These results unambiguously confirmed the cancer specificity of our binary system.
Cancer specificity of the binary hTERT-int based expression system using a cancerous mouse model.
(A) Healthy C57BL/6 mice, (B) LLC-Kat injected counterparts. Panels (a), untreated mice; panels (b–e), IV-tail injected mice with the following plasmids complexed with the in vivo-jetPEI (b) CMV-luc, (pYM1600 - 50 μg) (c) CMV-RSL-luc, (pAE1855 - 40 μg) (d) CMV-RSL-luc, (pAE1855 - 40 μg) + CMV-int, (pNA979 - 10 μg) (e) CMV-RSL-luc, (pAE1855 - 40 μg) + hTERT-int, (pNA1263 -10 μg). The mice were injected 24 hr later with luciferin and their bioluminescence visualized as detailed in the Methods section. Each figure shows a typical mouse from a cohort of at least four mice. (C) Quantitative bioluminescence data. The bars show the mean value of four experiments; the error bars indicate standard deviation. RSL represents the attR-Stop-attL cassette.
Immunohistochemical analyses of Luc expression in the lungs of the healthy mice (A) and of the LLC-Kat injected mice (B). The mice were treated in the same order as in Fig. 6. Each figure shows a typical mouse from a cohort of at least four mice.
Int does not show a toxic effect
Cre is one of the most widely SSRs used in mammalian gene manipulations, however, Cre-dependent cellular toxicity has been reported in eukaryotic cells and organisms some even under transient conditions. These include reduced cell growth, chromosome aberrations, brain development, cardiac fibrosis, heart failure, thymic atrophy and severe hematological toxicity28,29,30,31,32. To examine if, under the conditions of the above treatments, Int has any toxic effect we performed a TUNEL assay33 on the lungs of the same treated and untreated mice, thereby evaluating the apoptotic effect of the binary system. As evident from Fig. 8A all healthy lungs showed only negligible or no levels of apoptosis while the cancer lungs (Fig. 8B) were apoptotic as expected in those types of cells34. However, neither the lungs of the healthy mice treated with Int [Fig. 8A(d,e)] showed any apoptosis nor did their parallel cancer lungs [Fig. 8B(d,e)] show any increase in apoptosis relative to the ones in the absence of Int [panels (a,b)]. Quantitative data of the TUNEL assay confirm these observation (Fig. 8C), concluding that under our experimental conditions the amounts of supplied Int had no toxic effect.
Immunohistochemical analysis of the TUNEL assays in the lungs of the healthy mice (A) and of the LLC-Kat injected mice (B) shown in the same order as in Fig. 6. Each figure shows a typical mouse from a cohort of at least four mice. (C) Quantitative data of the TUNEL assays in the lungs of the healthy mice (a) and of the LLC-Kat injected mice (b). The bars show the mean value of four experiments; the error bars indicate standard deviation.
Discussion
In the present study a new cancer specific binary cell expression system was developed that is based on the cancer-specific hTERT promoter showing that when the expression of coliphage HK022 Int is promoted by hTERT it activates, by a site-specific recombination reaction, the expression of the luc reporter gene exclusively in lung cancer cells without affecting normal tissues. In previous two reports that employed the Cre/lox system in such binary systems one used an adenovirus vector as the gene delivery carrier10 and the other combined the two binary elements (recombinase and RS) by crossing two transgenic mice, each carrying one of them9. The advantages of our system are that it is not involved with a risky mammalian viral DNA carrier, that it can be delivered transiently and that under the experimental conditions Int does not show any toxicity. However, to test if Int may have some toxic effect a more extensive study that uses dose calibration and more than one toxicity assay are still missing.
When using a toxin gene to be specifically expressed in cancer cells it is essential that in the binary system the recombinase, being an enzyme, be promoted by a weak and specific promoter and that the toxic gene of the substrate be completely blocked by a tight transcription terminator. Because more substrate than enzyme needs to be delivered (Fig. 4A) the direct use of an hTERT promoted substrate, though being cancer specific, may be somewhat leaky on its voyage from its delivery spot to the cancer tissue thereby loosing specificity by damaging healthy tissues. Admittedly, a successful direct adenoviral carrier delivery of an hTERT promoted DTA toxin that cured ovary cancer in mice was reported35, nevertheless extra caution is always an advantage when it goes with cancer therapy.
With regards to possible off-target chromosomal integration events catalyzed by Int, we have previously shown that for Int to promote integration into secondary attB sites, such sites need to to carry a perfect wild type 7 bp overlap with two flanking palindromic sequences18. Moreover, the attL and attR RS sites in the binary system are incomplete attPs. These and the fact that Int is weakly promoted by hTERT reduce the likelihood of off target Int-catalyzed integration events into human secondary attB sites.
In the Int-catalyzed binary system we employ the excisive attL and attR as recombination sites because they proved more efficient in mammalian recombination activity than does the attB + attP pair. Another reason is because their recombination leads to the short 21 bp attB site located between the promoter and the Luc gene (Fig. 1) that does not interfere with the expression of Int.
Preliminary experiments in cell culture with the binary system that carry subunit A of the diphtheria toxin (DTA) have already shown that with the DTA toxicity is clearly specific in the LLC-Kat as compared to normal human foreskin BJ cells and experiments with live mice using DTA and other toxins to eradicate the lung cancer are in progress. Success may render the system useful with other cancer cases using relevant cell specific promoters and cell specific DNA carries.
Methods
Cells, growth conditions, mice, plasmids and oligomers
The bacterial host used was E. coli K12 strain TAP114 (lacZ)M1536. It was grown and plated on Luria-Bertani (LB) rich medium with the appropriate antibiotics. Plasmid transformations were performed by electroporation37. Plasmids and oligomers are listed in Tables 1 and 2, respectively.
Human embryonic kidney cells HEK293T (ATCC CRL-3216) were cultured in Dulbecco’s modified Eagle’s medium. Mouse Lewis lung carcinoma LLC1 (ATCC CRL-1642) cells labelled by Katushka fluorescent gene reporter26 were cultured in RPMI medium.
For transient transfection of HEK293T and LLC-Kat, the cells (~6×105) were plated in a 6 well plate and 24 hours later treated with 2 μg circular plasmid DNA using Puro-Fection transfection reagent (System Biosciences, USA) or TransIT-XT2 reagent (Mirus, Madison, WI, USA), respectively.
All mice procedures were performed in compliance with Tel Aviv University guidelines and protocols approved by the Institutional Animal Care and Use Committee. BALB/c and C57BL/6 strains were used for the mice experiments. In experiments involving injected LLC-Kat cells we used only the C57BL/6 strain.
All experiments described below were at least triplicated.
Plasmid construction
The pNA1263 plasmid (Table 1) carrying the int gene of HK022 under the control of hTERT promoter (hTERT) was constructed as follows. A BglII-HindIII hTERT PCR fragment, was generated using primers oEY638 + oEY639 (Table 2) and an hTERT-plasmid as template provided by Dr. S. C. Teng at the Department of Microbiology, National Taiwan University. and this PCR product was ligated into the matching sites of plasmid pNA97938. The pYM1600 plasmid, carrying the luc gene under the control of CMV promoter was constructed by the RF cloning procedure39. The CMV fragment that was obtained by PCR using plasmid pCDNA3 as a template and primers oEY843 + 850 was introduced into the luciferase reporter vector pGL2 vector (Promega, Madison, WI, USA). The pAE1855 plasmid, used as a substrate for the CMV-Luc assays, was constructed by ligation of a AgeI-NotI luc PCR fragment, obtained using pYM1600 as template and primers oEY807 + 808, into the same sites of the intermediate plasmid construct pAE1855inter carrying CMV- attR-Stop-attL cassette on pCDNA3. All plasmid constructs were verified by DNA sequencing.
Luc activity assay in cell culture
HEK293T or LLC-Kat cells transfected by the appropriate luc plasmids were collected 48 hours later by trypsin treatment and centrifugation, resuspension in 100 μl of x1 Passive Lysis Buffer (Promega, Madison, WI, USA) followed by sonication and centrifugation. Quantitative Luc analysis of the supernatant was performed using Dual-luciferase reporter assay kit (Promega, Madison, WI, USA) and Synergy2 plate reader (Biotek, USA) according to the manufacturer’s instructions.
DNA delivery and Luc activity imaging in mice
BALB/c or C57BL/6 mice were IV tail-injected with 250 μl of the in vivo-jet PEI delivery reagent (Polyplus transfection, France) complexed with an appropriate luc reporter plasmid. 24 hours later the mice were injected with 250 μl of 3 mg/ml luciferin solution (Gold Biotechnology, USA) and underwent bioluminescence imaging by Biospace Photon Imager (Biospace Lab, France).
Lung cancer development and fluorescence imaging in mice
0.8 × 106 LLC-Kat cells were IV tail-injected into 8 week old (17–21 gr) female C57BL/6 mice. Lung tumor metastases development was examined nine and twelve days following the injection using in vivo micro-CT scanner (TomoScope® In-vivo CT, Germany). The Katushka fluorescent signal in the lung metastases was analyzed on day 12 by an in vivo fluorescence imaging system (CRi MaestroTM, USA).
Preparation of Sections, Immunohistochemistry, Confocal Microscopy, apoptosis TUNEL assay and Image analysis
At the end of the experiments, biopsy specimens from lungs were collected, fixed in 4% paraformaldehyde (PFA), embedded in paraplast and 5 μm sections were prepared. Healthy and cancer lungs carrying metastases were treated in parallel with H&E stained and immune-stained anti-Luciferase antibody (Abcam, Cambridge, UK) at 1:100 dilution, with a PCNA kit (Life Technologies, Carlsbad, CA, USA) and with a TUNEL kit (MBL international, Woburn, MA, USA) according to manufacturer’s instructions. Donkey anti-Goat IgG (H + L) secondary antibodies conjugated with Alexa Fluor 488 dye (Life Technologies, Carlsbad, CA, USA) were used for Luciferase detection. Nuclei were detected with DAPI (Biolegend, San Diego, CA, USA). At least 20 images of five tissue sections from 3 different mice per group were obtained for each analysis. Microscopic observation and image acquisition were performed using a Zeiss 510 Meta – NLO confocal laser-scanning microscope (Zeiss, Oberkochen, Germany). TUNEL assay analysis was performed using Image J software.
DNA manipulations
Plasmid DNA from E. coli was prepared using a DNA Spin Plasmid DNA purification Kit (Intron Biotechnology, Korea) or a NucleoBondTM Xtra Maxi Plus EF kit (Macherey-Nagel, Germany). General genetic engineering experiments were performed as described by Sambrook and Russell37.
Additional Information
How to cite this article: Elias, A. et al. Cancer-specific binary expression system activated in mice by bacteriophage HK022 Integrase. Sci. Rep. 6, 24971; doi: 10.1038/srep24971 (2016).
References
Branda, C. S. & Dymecki, S. M. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6, 7–28 (2004).
Glaser, S., Anastassiadis, K. & Stewart, A. F. Current issues in mouse genome engineering. Nature Genetics 37, 1187–1193 (2005).
Wirth, D. et al. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biothecnol. 18, 411–419 (2007).
Nafissi, N. & Slavcev, R. Bacteriophage recombination systems and biotechnical applications. Appl. Microbiol. Biotechnol. 98, 2841–2851 (2014).
Krappmann, S. Genetic surgery in fungi: employing site-specific recombinases for genome manipulation. Appl. Microbiol. Biotechnol. 98, 1971–1982 (2014).
Gaj, T., Sirk, S. J. & Barbas, C. F. III Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol. Bioeng. 111, 1–15 (2014).
Schlake, T. & Bode, J. Use of Mutated Flp Recognition Target (Frt) Sites for the Exchange of Expression Cassettes at Defined Chromosomal Loci. Biochemistry 33, 12746–12751 (1994).
Grindley, N., Whitestone, K. & Rice, P. Mechanism of site-specific recombination. Annu. Rev. Biochem. 75, 567–605 (2006).
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93, 5860–5865 (1996).
Lee, E. J. & Jameson, J. L. Cell-specific Cre-mediated activation of the diphtheria toxin gene in pituitary tumor cells: potential for cytotoxic gene therapy. Hum. Gene Ther. 13, 533–542 (2002).
Jia, H., Pang, Y., Chen, X. & Fang, R. Removal of the selectable marker gene from transgenic tobacco plants by expression of Cre recombinase from a tobacco mosaic virus vector through agroinfection. Transgenic Res. 15, 375–384 (2006).
Azaro, M. A. & Landy, A. Integrase and the Lambda int family. In Mobile DNAII (eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. ) 118–148 (ASM Press, 2002).
Weisberg, R. A., Gottesmann, M. E., Hendrix, R. W. & Little, J. W. Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet. 33, 565–602 (1999).
Harel-Levy, G., Goltsman, J., Tuby, C. N. J. H., Yagil, E. & Kolot, M. Human genomic site-specific recombination catalyzed by coliphge HK022 integrase. J. Biotechnol. 134, 45–54 (2008).
Voziyanova, E. et al. Efficient Flp-Int HK022 dual RMCE in mammalian cells. Nucleic Acids Res. 41, e125 (2013).
Melnikov, O. et al. Site-specific recombination in the cyanobacterium Anabaena sp. strain PCC 7120 catalyzed by the integrase of coliphage HK022. J. Bacteriol. 191, 4458–4464 (2009).
Gottfried, P. et al. Site-specific recombination in Arabidopsis plants promoted by the Integrase protein of coliphage HK022. Plant Molecular Biology 57, 435–444 (2005).
Kolot, M., Malchin, N., Elias, A., Gritsenko, N. & Yagil, E. Site promiscuity of coliphage HK022 integrase as tool for gene therapy. Gene Ther. 22, 602 (2015).
Ito, H. et al. Expression of human telomerase subunits and correlation with telomerase activity in urothelial cancer. Clin. Cancer Res. 4, 1603–1608 (1998).
Takakura, M., Kyo, S., Kanaya, T., Tanaka, M. & Inoue, M. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 58, 1558–1561 (1998).
Counter, C. M. et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11, 1921–1929 (1992).
Xi, L., Schmidt, J. C., Zaug, A. J., Ascarrunz, D. R. & Cech, T. R. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 16, 231 (2015).
Goula, D. et al. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 5, 1291–1295 (1998).
Bertram, J. S. & Janik, P. Establishment of a cloned line of Lewis Lung Carcinoma cells adapted to cell culture. Cancer Lett. 11, 63–73 (1980).
Shcherbo, D. et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 4, 741–746 (2007).
Rask, L., Fregil, M., Hogdall, E., Mitchelmore, C. & Eriksen, J. Development of a metastatic fluorescent Lewis Lung carcinoma mouse model: identification of mRNAs and microRNAs involved in tumor invasion. Gene 517, 72–81 (2013).
Zarogoulidis, P. et al. Management of malignant pleural effusion by suicide gene therapy in advanced stage lung cancer: a case series and literature review. Cancer Gene Ther. 19, 593–600 (2012).
Loonstra, A. et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 98, 9209–9214 (2001).
Baba, Y., Nakano, M., Yamada, Y., Saito, I. & Kanegae, Y. Practical range of effective dose for Cre recombinase-expressing recombinant adenovirus without cell toxicity in mammalian cells. Microbiol. Immunol. 49, 559–570 (2005).
Forni, P. E. et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J. Neurosci. 26, 9593–9602 (2006).
Bersell, K. et al. Moderate and high amounts of tamoxifen in alphaMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis. Model. Mech. 6, 1459–1469 (2013).
Higashi, A. Y. et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J. Immunol. 182, 5633–5640 (2009).
Weigel, M. T. et al. Nilotinib in combination with carboplatin and paclitaxel is a candidate for ovarian cancer treatment. Oncology 87, 232–245 (2014).
Wouters, B. G. & Chiu, R. K. Evaluating the importance of apoptosis and other determinatns of cell death and suvival. In Apoptosis, Senescence and Cancer ( Gewirtz, D. A., Holt, S. E., Grant, S. eds.) 57–72 (Humana press, 2007).
Xie, X. et al. A novel hTERT promoter-driven E1A therapeutic for ovarian cancer. Mol. Cancer Ther. 8, 2375–2382 (2009).
Dorgai, L., Yagil, E. & Weisberg, R. Identifying determinants of recombination specificity: construction and characterization of mutant bacteriophage integrases. J. Mol. Biol. 252, 178–188 (1995).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual. (Cold Spring Harbor Laboratory, 1989).
Malchin, N. et al. Optimization of coliphage HK022 Integrase activity in human cells. Gene 437, 9–13 (2009).
Peleg, Y. & Unger, T. Application of the Restriction-Free (RF) cloning for multicomponents assembly. Methods Mol. Biol. 1116, 73–87 (2014).
Acknowledgements
We thank Dr. N. Malchin for pNA1263 plasmid construction, Dr. Teng S.C. for providing the hTERT-carrying plasmid, Dr. O. Ernst for help with the Luc activity assay in cell cultures and Prof. Gabi Kaufmann for valuable comments and corrections. This work was supported by the Kamin fund of the Israel Ministry of Economy and Industry and by G.I.F., the German Israeli Foundation for Scientific Research and development [1062/2008 to E. Y. and M. K].
Author information
Authors and Affiliations
Contributions
A.E., I.S., I.S.-B., N.G., Y.M., Y.Z., E.Y. and M.K. designed research. A.E., I.S., I.S.-B., N.G., Y.M. and Y.Z. performed experiments. A.E., I.S., I.S.-B., N.G., L.R., Y.M., Y.Z. and M.K. analyzed data. L.R. contributed LLC-Kat cell line and advised on their use. M.K. and E.Y. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Elias, A., Spector, I., Sogolovsky-Bard, I. et al. Cancer-specific binary expression system activated in mice by bacteriophage HK022 Integrase. Sci Rep 6, 24971 (2016). https://doi.org/10.1038/srep24971
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24971
This article is cited by
-
Bacteriophages: cancer diagnosis, treatment, and future prospects
Journal of Pharmaceutical Investigation (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.