Abstract
Nonhuman primates are valuable for human disease modelling, because rodents poorly recapitulate some human diseases such as Parkinson’s disease and Alzheimer’s disease amongst others. Here, we report for the first time, the generation of green fluorescent protein (GFP) transgenic cynomolgus monkeys by lentivirus infection. Our data show that the use of a human cytomegalovirus immediate-early enhancer and chicken beta actin promoter (CAG) directed the ubiquitous expression of the transgene in cynomolgus monkeys. We also found that injection into mature oocytes before fertilization achieved homogenous expression of GFP in each tissue, including the amnion and fibroblasts, whereas injection into fertilized oocytes generated a transgenic cynomolgus monkey with mosaic GFP expression. Thus, the injection timing was important to create transgenic cynomolgus monkeys that expressed GFP homogenously in each of the various tissues. The strategy established in this work will be useful for the generation of transgenic cynomolgus monkeys for transplantation studies as well as biomedical research.
Similar content being viewed by others
Introduction
To date, many transgenic animals have been developed including mice1,2, rats3,4 and domestic animals5. Although many human disease models have used these transgenic animals, most used mice because the technique of genetic manipulation, such as generation of transgenic animals by DNA microinjection into pronuclear embryos2,6,7 and generation of gene-targeting animals by using homologous recombination in embryonic stem cells8,9, have been established. While rodents are valuable models for biomedical research, the evolutionary distance between rodents and humans is almost 87 million years10 and rodents do not always recapitulate human behavioural and biological responses11. For example, it is difficult to use mice as models for AIDS10, because the host range for human immunodeficiency virus is highly restricted; for influenza11 because the pathogenesis in mice is different from that in humans and influenza virus causes hypothermia12 and severe viral pneumonia13,14; and for lung disorders15 such as asthma, because the original findings of asthma obtained from mouse models had limited success in humans studies16. Additionally, the pathological and behavioural phenotypes of mouse genetic models are often quite different from the human condition. The inactivation of Parkinson’s disease (PD) genes Pink117, Parkin18, Dj-119, Lrrk220 was insufficient to cause PD in mice. Triple knockout mice lacking Parkin, DJ-1 and Pink1 have normal morphology and normal numbers of dopaminergic and noradrenergic neurons in the substantia nigra21. Mouse knockout models of human tumour suppressor genes also often display a tumour spectrum at variance with the human pathology. For example, in humans, germline or somatic RB gene loss is associated with the development of retinoblastomas and osteosarcomas and, later in life, with small cell lung carcinomas, whereas mice with an Rb deletion fail to develop these types of tumours22. Accordingly, it is required to establish transgenic animal models to recapitulate human diseases.
Nonhuman primates (NHPs) are considered one of the most valuable animal models. Several NHPs are used as laboratory animals, including New World monkeys such as common marmosets and Old World monkeys such as rhesus monkeys and cynomolgus monkeys. Common marmosets clearly exhibit anthropoid primate characteristics, are relatively inexpensive to maintain, mature by 1.5–2 years of age, produce next generation offspring by 3 years of age and give birth to 3–5 offspring per year23. However, marmosets exhibit physiological and functional differences relative to humans to a greater degree than do Old World primates. These differences include pituitary gonadotropin secretion and action24, their ability to maintain bone mass without the need for gonadal estrogen25, lack of age-related ovulation decline26 and high fasting glucose and triglyceride levels27. In contrast, Old World monkeys are closer to humans in organ size and structure and therefore have been used for disease models such as stroke28,29, Parkinson’s disease30,31, Huntington’s disease32,33 and transplantation studies34,35. Especially, cynomolgus monkeys are considered a useful animal model because they can be bred throughout the year in contrast to rhesus macaques that have seasonal breeding pattern.
Green fluorescent protein (GFP) is frequently used in biomedical research and GFP-expressing animals are an important source of bone marrow, spermatogonial stem cells and organ transplantation and pre-implantation embryos used to produce chimeric embryos. After the first transgenic NHPs were created by the transduction of GFP retroviral vector in 200136, a transgenic NHP model of Huntington’s disease was developed37. In 2009, the first transgenic NHPs with germline transmission were reported38. Recently, genome editing in monkeys using the CRISPR/Cas9 system39 and TALEN system40 was developed and used to generate human disease models41. Currently, GFP mice42, rats43, rabbits44,45, cats46, pig47, cattle48, common marmosets38 and rhesus monkeys36,37,49 have been produced, yet no GFP cynomolgus monkey has been generated.
We report here for the first time, the generation of a GFP-expressing cynomolgus monkey. Our data show that the use of a human cytomegalovirus immediate-early enhancer and chicken beta actin promoter (CAG) directed the ubiquitous expression of the transgene in cynomolgus monkeys. Furthermore, we show that lentiviral injection into mature oocytes before fertilization is an efficient way to create transgenic NHPs with homogenous expression of GFP in each tissue, including the amnion and fibroblasts. We also clearly demonstrate the fluorescence in the transgenic foetus was due to GFP expression and not autofluorescence, by using wild type tissues as negative controls. This system will contribute to creating human disease models and is also a good tool for transplantation studies.
Result
Production of GFP transgenic cynomolgus monkeys
In rodents, the pronuclear injection of DNA into fertilized eggs is an efficient strategy to generate transgenic animals. An alternative method is the use of genetically engineered embryonic stem cells. However, these strategies have been unsuccessful in NHPs to date. Since a transgenic marmoset with the capability of germ line transmission has been generated by injecting lentivirus into the perivitelline space of a two-cell-stage embryo38, we followed the technique to create a transgenic cynomolgus monkey.
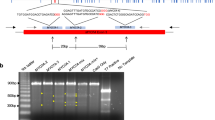
Firstly, we constructed a lentiviral vector that carries GFP cDNA under the control of human cytomegalovirus early enhancer and chicken beta actin (CAG) promoter (Fig. 1A), because CAG promoter is able to achieve ubiquitous expressions in various species50. Transfer of five lentivirus-injected embryos into three recipients resulted in two pregnancies, one of which ended in a miscarriage of twins on day 92 of gestation (Table 1). Detailed organ examination and histology identified no abnormalities (data not shown). Although one of the twins (#1) showed no detectable fluorescence, the other (#2) showed strong fluorescence in the face, skin, placenta and brain (Fig. 1B, data not shown). Strong fluorescence was observed in the organs of #2 foetus including the umbilical cord, placenta and amnion compared with #1 foetus under a fluorescent stereomicroscope (Fig. 1C). PCR analysis was performed to detect GFP transgene and demonstrated that the GFP transgene was integrated ubiquitously in the genome of all tissues in #2 foetus, but not #1 foetus. Consistent with this, RT-PCR analysis indicated that GFP mRNA was expressed ubiquitously in #2 foetus, but not #1 foetus (Fig. 1D). From these results, the #1 and #2 foetuses were determined to be wild type (WT) and transgenic (Tg), respectively. Abundant GFP mRNA expression was detected in the brain, heart and pancreas in #2 foetus, while moderate GFP expression was detected in the kidney, spleen and amnion by RT-quantitative PCR (RT-qPCR) (Fig. 1E). We confirmed GFP protein expression in these organs by immunohistochemistry with an anti-GFP antibody and found that fluorescence was detected in all these organs from the #2 foetus (Fig. 2A). Strong fluorescence was observed in the heart, lung, spleen and stomach from #2 foetus, but not #1 foetus or the miscarried WT foetus (Fig. 2A, Supplementary Fig. 1A), under the same instrumental settings (same laser intensity, please see Materials and Methods). However, we detected moderate fluorescence in the liver and kidney in #1 foetus that was WT as indicated by PCR analysis (Fig. 2A). To investigate carefully the nature of the fluorescence, we examined tissues from the liver and kidney of another miscarried WT foetus and found that these organs also had comparable fluorescence (Fig. 2A). Careful comparison revealed significant differences in the intensity and pattern of fluorescence between WT and #2 foetus, demonstrating that the fluorescence observed in the tissues of #1 and WT foetuses were autofluorescence and that the fluorescence in the #2 foetus was derived from GFP protein. Regarding autofluorescence, as laser power increased, autofluorescence of hepatocytes in the liver and renal tubules in the kidney also increased (Supplementary Fig. 1B). WT monkey face exhibited moderate autofluorescence as if it is GFP transgenic monkey (Supplementary Fig. 1C), showing the importance of negative control. Taken together, we concluded that it was very important to compare fluorescence of WT and Tg at the same time and #2 foetus was Tg with ubiquitous overexpression of the GFP protein.
GFP expression of day 92 aborted twin foetus.
(A) Schematic representation of the lentiviral vector used for the transduction of GFP. LTR: long terminal repeat. CMV IE: human cytomegalovirus immediate early enhancer. WPRE: woodchuck hepatitis posttranscriptional regulatory element. (B) Epifluorescence images of the whole body and brain in day 92 aborted twin foetuses (#1 and #2). Inset in each panel shows bright images. (C) Epifluorescence in umbilical cords, placenta and amnion in day 92 aborted twin (#1 and #2) foetuses. Each organ on the left was obtained from #1 foetus, whereas those on the right were from #2 foetus. Strong epifluorescence was observed under excitation light (489 nm). (Scale bar: 500 μm) (D). Genomic PCR (top panel) and RT-PCR (bottom panel) from #1 and #2 foetus organs. (E) GFP expression in #2 foetus organs evaluated by RT-qPCR. Data are represented as mean ± SD.
GFP protein expression in various tissues.
(A) Immunohistochemistry of tissues from a day 92 aborted twin foetus detected by confocal microscopy. Note that #2 foetus shows overall strong GFP fluorescence in all the tissues examined and that the WT liver and kidney showed moderate fluorescence (autofluorescence). Images were taken under the same instrumental settings (same laser intensity, etc). (Scale bar: 250 μm) (B). Immunohistochemical analysis of #2 brain with anti-GFP, anti-GFAP and anti-NeuN antibodies. Colocalisation of GFP and NeuN or GFAP are shown by white arrows. (Scale bar: 10 μm) (C). Immunohistochemical staining of #2 ovary with anti-OCT4 and VASA antibodies. Colocalisation of GFP and OCT4 or VASA are shown by white arrows. (Scale bar: 10 μm).
GFP expression in the brain tissue
To investigate the detailed expression of GFP in the brain of a day 92 foetus, we examined the colocalisation of GFP, a radial glial and astroglial marker, glial fibrillary acidic protein (GFAP) and a neuronal specific nuclear protein marker, NeuN (Fig. 2B). Widespread GFP expression was observed throughout the cortex, including the forebrain, midbrain and hindbrain. At a higher magnification, neuronal GFP expression was identified by its colocalisation with NeuN in the prefrontal cortex (Fig. 2B). GFP and GFAP were also colocalised in the lateral ventricle (Fig. 2B).
Germline transmission of the transgene
To investigate whether the transgene was transmitted to the germline, we examined the colocalisation of GFP and VASA (DDX4), a marker for primordial germ cell (PGC) of the ovary in the day 92 foetus. GFP and VASA were clearly colocalised in a subset of the ovary cells (Fig. 2C). Expression of OCT4, another marker for PGC was also well overlapped with that of GFP (Fig. 2C).
Partial GFP expression in the peripheral blood and fibroblasts
Previously, GFP transgenic marmoset showed partial GFP expression ranging from 0 to 19.1% in the whole peripheral blood38. To estimate the percentage of GFP positive cells in the peripheral blood of #2 foetus, immunohistochemistry with an anti-CD45 antibody, a marker for white blood cells, was performed on the spleen and showed that 55% of CD45 positive cells expressed GFP protein (Fig. 3A), indicating that peripheral blood cells only partially expressed GFP.
Partial GFP expression in day 92 aborted twin foetuses.
(A) Immunohistochemistry of spleen with anti-CD45 and GFP antibodies. A subset of CD45-positive white blood cells expressed GFP protein. Colocalisation of GFP and CD45 is shown by white arrows. (Scale bar: 25 μm) (B). Fluorescent image of #2 tail fibroblasts. (Scale bar: 200 μm) (C). Sorting of #2 tail fibroblasts by GFP intensity (upper left panel). Genomic PCR results from #2-①, #2-② and #2-③ tail fibroblasts (lower left panel). GFP expression in #2 foetus fibroblasts evaluated by RT-qPCR (right panel). Data are represented as mean ± SD. (D). Effect of sodium valproate (VPA) on GFP expression in GFP-negative (#2-①) fibroblasts. GFP-expressing fibroblasts are indicated by white arrows. (Scale bar: 300 μm).
To characterize GFP positive and negative cells, we established fibroblasts from the #2 foetus tail and found that about 60% of the cells were GFP-positive by FACS analysis (Fig. 3B,C). Interestingly, we found that GFP-negative cells also carried GFP transgene, suggesting that the transgene was not transcribed (Fig. 3C). Previous studies indicated that lentiviral transgene often undergo silencing through CpG methylation and that treatment with valproic acid, a histone deacetylase inhibitor, was effective to rescue silenced gene transcription51. When GFP-negative fibroblasts were cultured in the presence of valproic acid, a significant number of GFP-positive cells were observed, indicating that GFP silencing may be caused by histone deacetylation (Fig. 3D). Taken together, a GFP cynomolgus monkey foetus was successfully obtained, yet it only showed partial GFP expression and silencing of the transgene.
Optimization of lentivirus infection on GFP expression
To generate a transgenic cynomolgus monkey that expressed GFP homogenously in each of the various tissues, we optimized the lentivirus injection protocol. Since the lentivirus solution had a carryover of GFP protein contaminated in the process of virus production (Fig. 4A), we purified the lentivirus solution further by centrifugation in a sucrose cushion to remove GFP protein and obtained the highly purified lentiviral solution (Fig. 4A).
Optimization of lentivirus injection for homogenous expression of GFP in each tissue.
(A) Fluorescent images of a virus droplet after centrifugation without (−) or with (+) a sucrose cushion. (Scale bar: 500 μm) (B). Optimized transgenesis protocol. PreI: Pre-ICSI. PostI: Post-ICSI. (C). Epifluorescence image of the face in postnatal day 2 PreI Tg #1 offspring. Inset shows a bright image of PreI Tg #1 offspring and postnatal day 1 WT offspring. (D) Epifluorescence images of foetus side of placenta from PreI Tg #1 offspring and a part of WT placenta. Insets show bright images. (E) Fluorescence images of amnion from PreI Tg #1 offspring and WT offspring. Nuclei were stained with Hoechst 33342. (Scale bar: 100 μm) (F). Genomic PCR analysis with DNAs of organs from PreI Tg #1 offspring and WT offspring.
Wongsrikeao et al., reported that when virus was injected into mature oocytes before fertilization, uniform infection was achieved in a cat embryo46. Therefore, matured oocytes were subjected to perivitelline-space injection with lentivirus 4 h before intracytoplasmic sperm injection (ICSI) (PreI) (Fig. 4B). After the injection, the oocytes were cultured for 4 h and subjected to ICSI and cultured until blastocyst stage (day 8).
Full-term development of GFP cynomolgus monkeys
An offspring (named PreI Tg #1) from the PreI embryo was born successfully and showed strong fluorescence in the face and placenta compared with that of WT offspring (Fig. 4C,D). Amnion of PreI Tg #1 monkey showed homogenous GFP expression at the cellular level (Fig. 4E). PCR analysis was performed to detect GFP transgene and demonstrated that the GFP transgene was integrated in the genomic DNA from umbilical cord and placenta (Fig. 4F), demonstrating that PreI Tg #1 monkey is a GFP transgenic.
Although PreI Tg #1 monkey clearly showed strong GFP fluorescence in the skin and homogenous GFP expression in the amnion at the cellular level, GFP expressions in the tissues were not addressed due to consideration for animal welfare. Another offspring (named PreI Tg #2) was born, but died 3 days after birth (Table 2). Hypoplasia of the pituitary grand and pancreas were observed at the necropsy (data not shown). The whole body of the transgenic cynomolgus monkey showed strong fluorescence under an excitation light (Fig. 5A). The fluorescence was also detected in the placenta (Fig. 5A). PCR analysis indicated that GFP lentiviral transgene was integrated into the genome of various tissues (Fig. 5B). RT-PCR analysis indicated that GFP was expressed abundantly at the mRNA level in the brain, lung, kidney and stomach, while moderate GFP expression was observed in the heart, liver, pancreas, spleen, intestine and testis (Fig. 5B,C). Immunohistochemistry revealed abundant GFP expression in all tissues tested including the heart, lung, liver, kidney, spleen, stomach and intestine (Fig. 6A, Supplementary Fig. 2A,B). This was in sharp contrast with the autofluorescence observed in WT tissues (Fig. 6A). GFP protein expressions were also evident in lateral ventricle and grey matter layer (Fig. 6B). To investigate the contribution of the transgene to the germ cell lineage, we examined GFP expression in PGCs and found that almost all VASA-positive PGCs expressed GFP (Fig. 6C). To examine the ratio of GFP positive cells in offspring, high magnification images of sections were analysed and showed that >95% of cells were GFP positive (data not shown). Furthermore, CD45 positive peripheral blood was analysed and 99% of CD45 positive cells expressed GFP (Fig. 6D). To demonstrate this more clearly, fibroblasts were established from the tail and ear of offspring and almost all cells were GFP-positive under microscopic observation (Fig. 6E). Consistent with this, FACS analysis of fibroblasts showed that nearly 100% of cells were GFP-positive (Fig. 6E). Collectively, these results demonstrated that the GFP cynomolgus monkey created by PreI technique expressed GFP homogenously in each of the various tissues.
GFP expression in transgenic cynomolgus monkey offspring at postnatal day 3.
(A) Whole body and placenta epifluorescence imaging of Pre-ICSI transgenic (PreI Tg #2) offspring. Inset shows a bright image of 3 month old WT and PreI Tg #2 offspring. (Scale bar: 5 μm) (B). Genomic PCR (upper panel) and RT-PCR (lower panel) results from PreI Tg #2 organs. (C). GFP expression of PreI Tg #2 organs evaluated by RT-qPCR. Data are represented as mean ± SD.
Whole body analyses of PreI Tg #2 offspring.
(A) Expression of GFP protein revealed by immunohistochemistry with anti-GFP antibodies of PreI Tg #2 offspring by confocal microscopy. Upper and lower panel show WT and PreI Tg #2 tissues, respectively. Images were taken under the same instrumental settings (same laser intensity, etc). (Scale bar: 100 μm) (B). Immunohistochemical analysis of PreI Tg #2 brain with anti-GFP antibodies. Inset a: white arrows show astrocyte and GFP colocalisation. Inset b: white arrows show neuron nuclei and GFP colocalisation. (Scale bar: upper panel 100 μm, lower panel 25 μm) (C). Immunohistochemical staining of PreI Tg #2 testis with anti-VASA antibodies. Colocalisation of GFP and VASA is shown by white arrows. (Scale bar: upper panel 50 μm, lower panel 25 μm) (D). Immunohistochemistry of PreI Tg #2 spleen with anti-CD45 and GFP antibodies. Homogenous GFP protein expression in CD45-positive white blood cells was observed. Colocalisation of GFP and CD45 are shown by white arrows. (Scale bar: 25 μm) (E). Merged (Epifluorescence and Bright) images of fibroblasts from WT tail and PreI Tg #2 tail and ear. FACS analysis using fibroblasts from WT tail and PreI Tg #2 tail and ear. (Scale bar: 100 μm).
Comparison of expression levels of GFP protein in tissues from Tg monkeys and the GFP mouse
The green mice that ubiquitously express GFP have been created and been used for many biomedical researches such as transplantation studies52. Although the generation of GFP rhesus monkey and GFP marmoset have been reported36,37,38,49 previously, there was no study about the comparison of the level of GFP expression with that of the GFP mice. To evaluate the usefulness of GFP cynomolgus monkey established in this study, we compared GFP expression levels in tissues from GFP cynomolgus monkey with those from GFP mice42 by western blot analysis. Strong GFP expressions were detected in lung, kidney and spleen of PreI Tg cynomolgus monkey compared to those of GFP mouse (Fig. 7). #2 (Tg) foetus also expressed moderate level of GFP protein (Fig. 7). Thus, GFP cynomolgus monkeys generated in this study expressed even higher level of GFP protein in various tissues compared with those of GFP mice and the technique established in this study will be very useful for the creation of human disease models.
Comparison of expression levels of GFP protein from tissues in Tg monkeys and the GFP mice.
Western blot was performed with 2.5 μg of proteins extracted from tissues of Tg monkeys and the GFP mouse that carries one copy of the CAG promoter-GFP expression unit. Anti-GFP and β-actin antibodies were used to detect GFP and β-actin, respectively. Full-length blots are presented in Supplementary Fig. 3.
Discussion
Transgenic NHPs will be valuable for human disease models, such as Parkinson’s disease and Alzheimer diseases. So far, transgenic NHP models of Huntington’s disease37 and Parkinson’s disease53 have been created and reported. GFP is frequently used in biomedical research and GFP-expressing animals are an important source of transplantation studies. Previously, generation of GFP rhesus monkeys and GFP marmosets have been reported36,37,38,49, yet no GFP cynomolgus monkeys. To the best of our knowledge, this is the first study reporting the generation of GFP expressing cynomolgus monkeys. The strategy established in this study will be valuable for generating cynomolgus monkeys that homogenously overexpress a causative gene for human disease in each tissue.
During the generation of GFP cynomolgus monkeys, we were aware that moderate fluorescence expression is detectable even in WT tissues. It was critically important to use WT tissues as a negative control to confirm the fluorescence derived from GFP protein in transgenic tissues. It is important to note that some previous reports of GFP transgenic monkeys showed only GFP transgenic tissues but not corresponding WT tissues36,37,49.
Our analysis indicated that when lentivirus was injected into a two-cell-stage embryo, partial GFP expression was achieved. Although lentivirus can transduce non-dividing cells, integration is significantly slowed by the nuclear membrane48,54. This has previously been observed in other species including marmosets38.This mosaicism may be caused by the delay of lentiviral integration into the genome. Interestingly, a bovine study showed that the lentiviral genome could not efficiently enter the pronucleus48. Wongsrikeao et al. examined the effect of infection timing and found mosaic expression was able to be avoided by injecting lentivirus into cat oocytes before IVF46. Consistent with this, when lentivirus was injected into cynomolgus monkey oocytes before fertilization, rather than the two-cell embryo stage, we obtained offspring with homogenous GFP expression in each tissue, showing that lentiviral injection into mature oocytes before fertilization is an effective way to create transgenic NHPs with homogenous expression of GFP in each of the various tissues, including the amnion and fibroblasts.
NHPs are invaluable models for high order brain research into disorders such as Parkinson’s disease, Autism and Alzheimer’s disease. Alzheimer pathology is correlated strongly with the density of activated astrocytes55. In these cells, the expression of GFAP is strongly upregulated56,57. In Parkinson’s disease patients, the phosphorylation level of GFAP at serine 13 was significantly lower compared with control subjects58. As the present study indicated that the GFP was expressed ubiquitously in various organs and cells including neuronal cells of transgenic cynomolgus monkeys, this transgenic technique will be useful to create human disease models for Parkinson’s disease and Alzheimer’s disease amongst others. The GFP cynomolgus monkey will also provide a source of green pre-implantation stage embryos, which can be used for the production of chimeric cynomolgus monkeys by the injection or aggregation with non-green embryonic stem cells.
Materials and Methods
Animals
Experimental procedures were approved by the Animal Care and Use Committee of Shiga University of Medical Science and methods were carried out in accordance with the approved guideline (Approval number: 26–42). Oocytes were collected from seven sexually mature female cynomolgus monkeys, aged 4–8 years and weighing 2.1–3.9 kg. Twenty sexually mature females aged 4 years old and weighing 2.0–3.8 kg, were used as recipients. Sperm were collected from one sexually mature male cynomolgus monkey, aged 12 years and weighing 6.2 kg. Temperature and humidity in the animal rooms were maintained at 25 ± 2 °C and 50 ± 5%, respectively. Monkeys were housed individually in cages (800 × 500 × 800 mm). The light cycle was 12 h of artificial light from 8 am. to 8 pm., Each animal was fed 20 g/kg of body weight of commercial pellet monkey chow (CMK-1; CLEA Japan, Inc., Tokyo, Japan) in the morning, supplemented with 20–50 g of sweet potato in the afternoon. Water was available ad libitum.
Oocyte collection
Ovarian stimulation and oocyte collection were carried out as previously described by Yamasaki et al.59 with some modifications. In brief, beginning at menses, the level of sex steroid hormones was reduced by subcutaneous injection of 0.9 mg of a gonadotropin-releasing hormone antagonist (Leuplin; Takeda Chemical Industries, Ltd., Osaka, Japan). Two weeks later, Micro infusion pump (iPRECIO SMP-200, ALZET Osmotic Pumps Co., Cupertino, CA, USA) with 15 IU/kg human follicle-stimulating hormone (hFSH; Asuka Pharmaceutical Co., Tokyo, Japan) was embedded subcutaneously under anesthesia and injected 7 μl/h for 10 days. On the day after the last hFSH injection, 400 IU/kg human chorionic gonadotropin (hCG; Puberogen, Nippon Zenyaku Kogyo Co., Ltd., Fukushima, Japan) was injected intramuscularly. Oocytes were collected by follicular aspiration 40 h after hCG treatment, using a laparoscope (LA-6500, Machida Endoscope Co., Ltd., Chiba, Japan). Cumulus-oocyte complexes were recovered in Alpha modification of Eagle’s medium (MP Biomedicals LLC, Solon, OH, USA), containing 10% Serum Substitute Supplement (SSS; Irvine Scientific, Santa Ana, CA, USA) at 38 °C, in an atmosphere of humidified 5% CO2 and 95% in air for 1–2 h. Oocytes were stripped off cumulus cells by mechanical pipetting after brief exposure (<1 min) to 0.5 mg/mL hyaluronidase (Sigma Chemical Co., St. Louis, MO, USA), adjusted with m-TALP (pH 7.4), a modified Tyrode solution, with, lactate, pyruvate, 0.3% bovine serum albumin (Sigma Chemical Co.) and HEPES. Then, oocytes were transferred to m-TALP without hyaluronidase, at 38 °C in 5% CO2 until further use. Oocytes were classified by stages as germinal vesicle, metaphase I, metaphase II or degenerate.
Intracytoplasmic sperm injection
Intracytoplasmic sperm injection was carried out on metaphase II-stage oocytes, as previously described59, in m-TALP containing HEPES (mTALP-HEPES). A glass needle (Humagen Fertility Diagnostics, Charlottesville, VA, USA), connected to an injector and an inverted microscope (Olympus Tokyo, Tokyo, Japan) with a micromanipulator, was used for sperm injection. Intracytoplasmic sperm injection was performed with fresh sperm collected by electric stimulation of the penis with no anaesthesia. Following ICSI, embryos were cultured in GIBCO CMRL Medium-1066 (Invitrogen, Carlsbad, CA, USA) supplemented with 20% bovine serum (Invitrogen) at 38 °C in 5% CO2 and 5% O2 then blastocyst stage embryos were generated in vitro.
Lentiviral vector construction
pCSII-CAG-EGFP was constructed by introducing CAG promoter from pCAGGS and GFP cDNA into pCSII-EF-MCS-IRES2-Venus plasmid. pCAGGS was provided by Dr. Hitoshi Niwa (Kumamoto Univ.).
Lentiviral vector package and transduction
Viral particles were obtained through Lipofectamine 2000 (Life Technologies, Calsbad, CA, USA) transfection in 293FT cells and with packaging plasmids VSVG, RSV-Rev, HIVgp plasmids. Viral supernatants were harvested after 48 and 72 h of transfection. The supernatant then was clarified by centrifugation (1,000 × g for 5 min at room temperature), passed through a PVDF filter (pore size, 0.22 μm) and concentrated by ultracentrifugation (50,000 × g for 2 h at 4 °C). The pellet was suspended in PBS and centrifuged on a 20% (w/v) sucrose cushion. After the viral pellet was resuspended in CMRL, the infectious unit (IU) was determined by Lenti-X™ p24 Rapid Titer Kit (Takara bio, Shiga, Japan).
Virus injection to embryos
For virus injection to the two-cell-stage embryos, two-cell-stage embryos were prepared after 24 h of ICSI and lentivirus was injected into the perivitelline space in 0.25 M sucrose/ mTALP-HEPES. For injection to oocytes, metaphase II stage oocytes were selected and lentivirus was injected into the perivitelline space in 0.25 M sucrose/ mTALP-HEPES. After 4 h of virus injection, ICSI was performed.
Embryo transfer and pregnancy detection
When embryos developed to expanded blastocysts, one or two embryos were transferred into each female recipient. Embryo transfer was performed as previously described59, with some modifications. Laparoscopy was carried out on alternate days from days 10–14 of the menstrual cycle after menstruation (ovulation day = day 0) until ovulation occurred. Recipients were selected for embryo transfer 1–6 day after ovulation. Neither the stage of the embryos nor the number of days after the recipient’s ovulation were matched. Embryos were aspirated into a catheter (ETC3040SM5-17; Kitazato Medical Service Co., Ltd., Tokyo, Japan) under a stereomicroscope. The catheter was inserted into the oviduct of the recipient via the fimbria under the laparoscope and the cultured embryo was transplanted with a small amount of medium. Pregnancy was determined by ultrasonography 30 days after ICSI.
RT- quantitative PCR
Total RNA was extracted from cells or tissues using RNeasy Mini kits (Qiagen, Hilden, Germany). For reverse transcription, ReverTra Ace (Toyobo Co., Ltd, Osaka, Japan) and oligo (dT) 20 primer were used. For real-time PCR, THUNDERBIRD SYBR qPCR Mix (Toyobo) was used. Transcript levels were determined in triplicate reactions and normalized against the corresponding levels of GAPDH. Primer sequences are shown in Supplementary Table 1.
Immunohistochemical analysis
Tissues were fixed by 4% PFA at 4 °C overnight, embedded in OCT compound, frozen in liquid nitrogen, sliced into 10-μm sections and placed onto glass slides that were treated with Blocking One for 30 min at room temperature. Primary antibodies and dilutions used were rabbit anti-GFP AlexaFluor 488 conjugate (1:400, Life Technologies A21311), mouse anti-Oct4 (1:400, Santa Cruz sc-5279), mouse anti-NeuN (1:500, Millipore MAB377), rabbit anti-GFAP (1:500, Biomedical Technologies Inc. BT-575), rabbit anti-CD45 (1:100, Dako M0701), rabbit anti-DDX4 (VASA) (1:400, Abcam ab13840), which were then detected with appropriate secondary AlexaFluor 488, 568 or 647 antibodies. Cells were counterstained with Hoechst 33342 and observed using a Leica TCS SP8 confocal microscope.
FACS analysis
Tail fibroblasts were established from day 92 foetuses (#1 and #2) and day 3 offspring (PreI Tg #2). After removal of tail skin, the remaining tails were washed in PBS and maintained in Dulbecco’s modified eagle medium (DMEM, Invitrogen) containing 10% foetal bovine serum (FBS, JRH Biosciences, Corston Bath, UK), penicillin, streptomycin (Invitrogen) and primocin (Invivogen). Ear fibroblasts were established from PreI Tg #2 by cutting ears into 5 mm3 fragments, washing in PBS and then cultured in DMEM containing 10% FBS, penicillin, streptomycin and primocin. The fibroblasts were cultured until confluent and then collected by trypsin treatment. Cells were resuspended in a final volume of 500 μl of PBS/2% FCS and analysed with a Becton Dickinson FACSAria cell sorter.
Observation of green fluorescence in transgenic offspring
Images were captured under 470 nm excitation light with 520-nm wavelength filters using a Nikon D3300 digital camera. No image intensifying procedures were applied to any of the images. A 1-day-old and 3-month-old cynomolgus monkeys were used as a negative control for PreI Tg #1 and #2 respectively.
Western blot analysis
One mm2 of tissues were incubated in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP40 and 0.1% SDS) with protease and phosphatase inhibitors for 10 min at 4 °C. After centrifugation (15,000 × g for 5 min), protein samples were obtained from supernatant. Samples were diluted in sample buffer (Wako, Osaka, Japan) at a final concentration (10 μg/μl) and stored at −20 °C just before assessment. After denaturing by boiling at 95 °C for 5 min, 2.5 μl of sample was separated by SDS-PAGE on 10% polyacrylamide gel at 250 V for 80 min and then transferred onto a polyvinylidene fluoride membrane (Merck Millipore, Darmstadt, Germany). The membrane was blocked using Blocking One and then incubated with rabbit anti-GFP AlexaFluor 488 conjugate (1:2,000) or rabbit anti-β-actin horseradish peroxidase (HRP) conjugate (1:4,000) antibody overnight at 4 °C in Blocking One. After three washes in Tween20-PBS (T-PBS), the membrane incubated with rabbit anti-GFP was treated with HRP-labeled anti-rabbit immunoglobulin G (1:2,000; Invitrogen) in Blocking One for 1 h at room temperature. After one wash of 15 min and five washes of 5 min each with T-PBS, peroxidase activity was visualized using the Chemi-Lumi One Super (Nacalai Tesque) according to the manufacturer’s instructions. Tissues from a GFP mouse (C57BL/6-Tg(CAG-EGFP)10sb/J)42 that carries one copy of the CAG promoter-GFP expression unit were provided by Drs. H. Kojima and M. Katagi (Shiga University of Medical Science) and used as a positive control.
Statistical analysis
Statistical analyses of all data comparisons were carried out using the t-test using Excel software. P < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Seita, Y. et al. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci. Rep. 6, 24868; doi: 10.1038/srep24868 (2016).
References
Jaenisch, R. & Mintz, B. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc. Natl. Acad. Sci. USA 71, 1250–1254 (1974).
Gordon, J. W., Scangos, G. A., Plotkin, D. J. & Barbosa, J. A. & Ruddle, F. H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 77, 7380–7384 (1980).
Bachmann, S., Peters, J., Engler, E., Ganten, D. & Mullins, J. Transgenic rats carrying the mouse renin gene–morphological characterization of a low-renin hypertension model. Kidney Int. 41, 24–36 (1992).
Mullins, J. J., Peters, J. & Ganten, D. Fulminant hypertension in transgenic rats harbouring the mouse Ren-2 gene. Nature 344, 541–544 (1990).
Hammer, R. E. et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315, 680–683 (1985).
Brinster, R. L. et al. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27, 223–231 (1981).
Wagner, T. E. et al. Microinjection of a rabbit beta-globin gene into zygotes and its subsequent expression in adult mice and their offspring. Proc. Natl. Acad. Sci. USA 78, 6376–6380 (1981).
Doetschman, T. et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330, 576–578 (1987).
Kuehn, M. R., Bradley, A., Robertson, E. J. & Evans, M. J. A potential animal model for Lesch-Nyhan syndrome through introduction of HPRT mutations into mice. Nature 326, 295–298 (1987).
Springer, M. S., Murphy, W. J., Eizirik, E. & O’Brien, S. J. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100, 1056–1061 (2003).
Kalter, S. S. & Heberling, R. L. Serologic response of primates to influenza viruses. Proc. Soc. Exp. Biol. Med. 159, 414–417 (1978).
Bouvier, N. M. & Lowen, A. C. Animal Models for Influenza Virus Pathogenesis and Transmission. Viruses 2, 1530–1563 (2010).
Tripp, R. A. & Tompkins, S. M. Animal models for evaluation of influenza vaccines. Curr. Top. Microbiol. Immunol. 333, 397–412 (2009).
Fukushi, M. et al. Serial histopathological examination of the lungs of mice infected with influenza A virus PR8 strain. PLos One 6, e21207 (2011).
Seok, J. et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 110, 3507–3512 (2013).
Wenzel, S. & Holgate, S. T. The mouse trap: It still yields few answers in asthma. American Journal of Respiratory and Critical Care Medicine 174, 1173–1176 (2006).
Gautier, C. A., Kitada, T. & Shen, J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. USA 105, 11364–11369 (2008).
Goldberg, M. S. et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278, 43628–43635 (2003).
Andres-Mateos, E. et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. USA 104, 14807–14812 (2007).
Hinkle, K. M. et al. LRRK2 knockout mice have an intact dopaminergic system but display alterations in exploratory and motor co-ordination behaviors. Mol. Neurodegener. 7, 25 (2012).
Hennis, M. R., Marvin, M. A., Taylor, C. M. & Goldberg, M. S. Surprising behavioral and neurochemical enhancements in mice with combined mutations linked to Parkinson’s disease. Neurobiol. Dis. 62, 113–123 (2014).
Rangarajan, A. & Weinberg, R. A. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat. Rev. Cancer 3, 952–959 (2003).
Abbott, D. H. & Hearn, J. P. Physical, hormonal and behavioural aspects of sexual development in the marmoset monkey, Callithrix jacchus. J. Reprod. Fertil. 53, 155–166 (1978).
Gromoll, J. et al. A new subclass of the luteinizing hormone/chorionic gonadotropin receptor lacking exon 10 messenger RNA in the New World monkey (Platyrrhini) lineage. Biol. Reprod. 69, 75–80 (2003).
Binkley, N. et al. Zoledronate prevents the development of absolute osteopenia following ovariectomy in adult rhesus monkeys. J. Bone Miner. Res. 13, 1775–1782 (1998).
Tardif, S. D. & Jaquish, C. E. Number of ovulations in the marmoset monkey (Callithrix jacchus): relation to body weight, age and repeatability. Am. J. Primatol. 42, 323–329 (1997).
Tardif, S. D. et al. Characterization of obese phenotypes in a small nonhuman primate, the common marmoset (Callithrix jacchus). Obesity (Silver Spring). 17, 1499–1505 (2009).
Monsein, L. H. et al. Irreversible regional cerebral ischemia: serial MR imaging and proton MR spectroscopy in a nonhuman primate model. Ajnr Am. J. Neuroradiol. 14, 963–970 (1993).
Marshall, J. W. B. et al. Serial MRI, functional recovery and long-term infarct maturation in a non-human primate model of stroke. Brain Res. Bull. 61, 577–585 (2003).
Barraud, Q. et al. Sleep disorders in Parkinson’s disease: the contribution of the MPTP non-human primate model. Exp. Neurol. 219, 574–582 (2009).
Soderstrom, K., O’Malley, J., Steece-Collier, K. & Kordower, J. H. Neural repair strategies for Parkinson’s disease: insights from primate models. Cell Transplant. 15, 251–265 (2006).
DeLong, M. R. Primate models of movement disorders of basal ganglia origin. Trends Neurosci. 13, 281–285 (1990).
Isacson, O., Riche, D., Hantraye, P., Sofroniew, M. V. & Maziere, M. A primate model of Huntington’s disease: cross-species implantation of striatal precursor cells to the excitotoxically lesioned baboon caudate-putamen. Exp. brain Res. 75, 213–220 (1989).
Kisu, I. et al. Uterus allotransplantation in cynomolgus macaque: a preliminary experience with non-human primate models. J. Obstet. Gynaecol. Res. 40, 907–918 (2014).
Enskog, A. et al. Uterus transplantation in the baboon: methodology and long-term function after auto-transplantation. Hum. Reprod. 25, 1980–1987 (2010).
Chan, A. W., Chong, K. Y., Martinovich, C., Simerly, C. & Schatten, G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science 291, 309–312 (2001).
Yang, S.-H. et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature 453, 921–924 (2008).
Sasaki, E. et al. Generation of transgenic non-human primates with germline transmission. Nature 459, 523–527 (2009).
Niu, Y. et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843 (2014).
Liu, H. et al. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14, 323–328 (2014).
Chen, Y. et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 24, 3764–3774 (2015).
Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
Hakamata, Y. et al. Green fluorescent protein-transgenic rat: a tool for organ transplantation research. Biochem. Biophys. Res. Commun. 286, 779–785 (2001).
Wang, H. J., Lin, A. X., Zhang, Z. C. & Chen, Y. F. Expression of porcine growth hormone gene in transgenic rabbits as reported by green fluorescent protein. Anim. Biotechnol. 12, 101–110 (2001).
Takahashi, R. et al. Establishment and characterization of CAG/EGFP transgenic rabbit line. Transgenic Res. 16, 115–120 (2007).
Wongsrikeao, P., Saenz, D., Rinkoski, T., Otoi, T. & Poeschla, E. Antiviral restriction factor transgenesis in the domestic cat. Nat. Methods 8, 853–859 (2011).
Naruse, K. et al. Production of a transgenic pig expressing human albumin and enhanced green fluorescent protein. J. Reprod. Dev. 51, 539–546 (2005).
Reichenbach, M. et al. Germ-line transmission of lentiviral PGK-EGFP integrants in transgenic cattle: new perspectives for experimental embryology. Transgenic Res. 19, 549–556 (2010).
Niu, Y. et al. Transgenic rhesus monkeys produced by gene transfer into early-cleavage-stage embryos using a simian immunodeficiency virus-based vector. Proc. Natl. Acad. Sci. USA 107, 17663–17667 (2010).
Niwa, H., Yamamura, K. & Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991).
Pikaart, M. J. & Felsenfeld, G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes & Dev. 12, 2852–2862 (1998).
Herbst, F. et al. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol. Ther. 20, 1014–1021 (2012).
Niu, Y. et al. Early Parkinson ’ s disease symptoms in α -synuclein transgenic monkeys. Hum. Mol. Genet. 24, 2308–2317 (2015).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Muramori, F., Kobayashi, K. & Nakamura, I. A quantitative study of neurofibrillary tangles, senile plaques and astrocytes in the hippocampal subdivisions and entorhinal cortex in Alzheimer’s disease, normal controls and non-Alzheimer neuropsychiatric diseases. Psychiatry Clin. Neurosci. 52, 593–599 (1998).
Hanzel, D. K., Trojanowski, J. Q., Johnston, R. F. & Loring, J. F. High-throughput quantitative histological analysis of Alzheimer’s disease pathology using a confocal digital microscanner. Nat. Biotechnol. 17, 53–57 (1999).
Beach, T. G., Walker, R. & McGeer, E. G. Patterns of gliosis in Alzheimer’s disease and aging cerebrum. Glia 2, 420–436 (1989).
Clairembault, T. et al. Enteric GFAP expression and phosphorylation in Parkinson’s disease. Journal of Neurochemistry 130, 805–815 (2014).
Yamasaki, J. et al. Vitrification and transfer of cynomolgus monkey (Macaca fascicularis) embryos fertilized by intracytoplasmic sperm injection. Theriogenology 76, 33–38 (2011).
Acknowledgements
We would like to thank Research Center for Animal Life Science research support team for animal care, Shiga University of Medical Science. This work was supported in part by a grant from PRESTO, Japan Science and Technology Agency (JST) (to M. E.), JSPS KAKENHI Grant number 15K21080 (to T. T.), in-house grant from Shiga University of Medical Science (to M. E., Y. S., T. T.) and a Grant-in-Aid for JSPS Fellows (to T. A.) and a grant from the Network Program for Realization of Regenerative Medicine from the Japan Agency for Medical Research and Development (AMED) (to K. O., T. I., M. E.).
Author information
Authors and Affiliations
Contributions
Y.S. performed the biochemical and cell biological experiments and co-wrote the paper. Y.S., T.T., M.S., E.S. and M.E. designed the experiments. Y.S., H.M., E.S. and T.T. designed GFP expression vectors and prepared lentivirus particle. Y.S. and T.T. established fibroblasts from tails and ears. C.I., H.T., J.M., J.O., S.N., E.S. and Y.S. assisted in embryological technique development. T.A. and Y.S. contributed to immunohistochemistry and Y.I., T.I., M.N., I.T. and K.O. analysed the data. Y.H. and S.H. developed high-resolution immunohistochemistry of brain. T.T. edited the paper. M.E. conducted the project and contributed to development of the hypothesis. M.E. supervised the whole project and co-wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Seita, Y., Tsukiyama, T., Iwatani, C. et al. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci Rep 6, 24868 (2016). https://doi.org/10.1038/srep24868
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24868
This article is cited by
-
Generation of a familial hypercholesterolemia model in non-human primate
Scientific Reports (2023)
-
Screening for gene doping transgenes in horses via the use of massively parallel sequencing
Gene Therapy (2022)
-
Monkeys mutant for PKD1 recapitulate human autosomal dominant polycystic kidney disease
Nature Communications (2019)
-
MHC-identical and transgenic cynomolgus macaques for preclinical studies
Inflammation and Regeneration (2018)
-
Genetic engineering in nonhuman primates for human disease modeling
Journal of Human Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.