Abstract
Genome-wide association studies (GWASs) have yet to be conducted for tuberculosis (TB) susceptibility in China. Two previously identified single nucleotide polymorphisms (SNPs) from tuberculosis GWASs, rs2057178 and rs4331426, were evaluated for TB predisposition. The associations between SNPs and gene expression levels were analyzed using the genomic data and corresponding whole-genome expression of the Han Chinese in Beijing, China. Genotyping was successfully completed for 763 pulmonary TB patients and 763 healthy controls. The T allele of the rare variant rs2057178 was significantly associated with TB predisposition (χ2 = 14.07, P = 0.0002). Meanwhile, the CT genotype of rs2057178 was associated with a decreased risk of TB (adjusted OR = 0.52, 95% CI, 0.34–0.78). The CT genotype of rs2057178 was also associated with decreased expression levels of infection-related gene, suppressor of cytokine signaling 2 (SOCS2) and increased expression levels of v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB). No gene expression levels were found to be associated with the genotype of rs4331426. We found that the rare variant rs2057178 was significantly associated with TB in the Han Chinese population. Moreover, the expression levels of MAFB and SOCS2 correlated with rs2057178 and might be potential candidates for assessing TB susceptibility.
Similar content being viewed by others
Introduction
According to the WHO 2014 global tuberculosis (TB) report1, China has the second largest number of incident TB cases, which accounts for 11% of the global burden. Moreover, the recent TB infection survey conducted in China indicated that one-fifth of the population had been latently infected by Mycobacterium. tuberculosis (M. tb)2. Although a majority of the M. tb-infected population will not progress to clinical TB because of host resistance to the pathogen, it was estimated that approximately 10% of infected individuals will develop TB disease during their lifetime3. The large reservoir of infected people is a crucial threat to China’s TB control program for achieving the END TB goal in 20354. Whether a person who is latently infected with M. tb would become a clinical case may largely depend on the host defense against M. tb. The vast difference between M. tb infection rate and population TB incidence suggests that the human host plays an important role in preventing TB development after infection. Evidence from studies of twins also support the assumption that host genetics contribute to the susceptibility of TB5.
In recent years, TB candidate genes, such as the vitamin D receptor gene6, cytokine genes7 and toll-like receptors genes8, have been studied to explain host genetic susceptibility to TB in various ethnic backgrounds. However, due to limited sample sizes and/or the diversity of ethnicity, evidence of the association between hot spot loci of candidate genes and TB susceptibility is still rare. Genome-wide association studies (GWASs) have provided a large amount of information regarding the genetic contribution of hundreds of thousands of genomic variants to TB susceptibility9,10,11 and some of the variants were highly associated with TB susceptibility. However, no GWAS of TB predisposition has been conducted in China yet.
Common variants of the genome usually have little effect on common disease risk, usually conferring small odds ratios on disease risk effect. Recently, Rivas et al. proposed that rare variants would have a larger effect and higher penetrance in disease occurrence12. In this study, we selected SNPs with allele frequencies of less than 5% in the Han Chinese population and that were found to be significantly associated with TB by previous GWASs. Thus, the single nucleotide polymorphism (SNP) rs2057178 at 11p13 and the SNP rs4331426 at 18q11.2, which were identified in previous GWASs, were selected to verify their association with TB predisposition10,11. More importantly, the suggestion of Wilkinson13 was adopted to test the latent TB infection status of the control group to control for the exposure factor of M. tb infection in this study. Meanwhile, the Gene Expression Omnibus Database was used to explore the relationship between the selected SNPs and whole genome mRNA expression levels14,15. Lymphoblastoid cell lines, which carry the complete set of germline genetic material, have been instrumental, in general, as a source of biomolecules and as a system to carry out various immunological and epidemiological studies16. Dissemination of M. tb in infected persons may be connected with the initiation of adaptive immune responses, which are under strict host genetic control17. The transcriptome level among the lymphoblastoid cell lines may be closely associated with disease-associated genetic variants. Thus, we consider that specific gene expression levels in lymphoblastoid cell lines may be closely regulated by host genomic variants. RNAs extracted from lymphoblastoid cell lines of the 45 unrelated CHB of the HapMap Project were quantified to explore the relationship between selected SNPs and the expression levels of potential TB susceptibility genes.
Methods
Study subjects
This case-control study was carried out in two designated hospitals with TB control programs in Jiangsu Province, eastern China. One was the TB control center of Danyang County and the other was the Nanjing Chest Hospital in the province’s capital city. Incident TB cases of Han ethnicity registered in these two hospitals from July 1st, 2013 to December 31st, 2014 were recruited for inclusion as cases in this study. All enrolled cases were bacteriologically confirmed by Lowenstein-Jensen (LJ) culture and M. tb was identified using the p-nitrobenzoic acid (PNB) method. Meanwhile, the healthy controls were recruited from two communities from Danyang County during the same study period. All control candidates underwent X-ray examination. Sputum culture was provided if the potential controls reported having TB-like clinical symptoms. Only subjects with normal X-ray manifestation, negative LJ culture, if tested and no comorbidity with other infectious diseases (such as HIV/AIDS and hepatitis B virus) were eligible as healthy controls. All of the controls were of Han ethnicity and they were 1:1 matched to the cases by age (±5 years) and gender. In total, 764 TB cases and 764 healthy controls were recruited for the study. All experimental protocols in this study were approved by the Institutional Review Board of the Center for Disease Control and Prevention of Jiangsu Province and written informed consent was obtained from each participant before the study. Additionally, all of the methods in this study were carried out in accordance with the approved guidelines.
Interferon-Gamma Release Assay
The interferon-gamma release assay (QuantiFERON-TB Gold In-Tube [QFT; Qiagen, Valencia, CA, USA]) was used to test the latent TB infection (LTBI) status of the controls. QFT was performed according to the instructions provided by Qiagen18.
Genotyping of rs2057178 and rs4331426
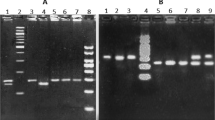
The restriction fragment length polymorphism (RFLP) method was used for the genotyping. The sequences of the primers used to amplify the PCR fragment of rs2057178 were as follows: 5′-TCC ATT GGC CTG AAC TGG AT-3′ (forward); 5′-TGG CCT CCA GTT CTT TAG CA-3′ (reverse). A 186 base pair PCR fragment was amplified by the primers. The restriction endonuclease enzyme StuI (New England BioLabs, inc., Ipswich, MA, USA) was used to digest the PCR fragment. The presence of the C allele results in two fragments: one fragment of 125 base pairs in length and one fragment of 61 base pairs in length. The presence of the T allele results in a single fragment of 186 base pairs in length. The PCR amplification fragment for rs4331426 was 250 base pairs in length and the sequences of the amplification primers were as follows: 5′-AAG GGT GTT GTT CTG TTT CTA GA-3′ (forward), 5′-TGT TGC ACC ACC TCT TGT AGA-3′ (reverse). The restriction endonuclease enzyme HhaI (New England BioLabs, inc., Ipswich, MA, USA) was used to digest the PCR fragment. The presence of the G allele results in two fragments: one fragment of 202 base pairs in length and one fragment of 48 base pairs in length. The presence of the A allele results in a single fragment of 250 base pairs in length.
Genotypic data of rs2057178 and rs4331426 of the 45 Han Chinese in Beijing (CHB) from the HapMap Project and whole-genome expression levels from the Gene Expression Omnibus of PubMed
The genotypic data of rs2057178 and rs4331426 were extracted from the HapMap Genome Browser Release #28 (phases 1, 2 & 3-merged genotypes and frequencies) and the genotypes of each SNP for the 139 CHB individuals were derived from this database. The DNA samples were prepared from blood samples collected from individuals living in the residential community at Beijing Normal University. All of the samples are from unrelated individuals who identified themselves as having at least three out of four Han Chinese grandparents.
Finally, 45 CHB provided validated genotypic data of the two SNPs. Using mRNAs extracted from lymphoblastoid cell lines for the corresponding 45 CHB, the Gene Expression Omnibus (GEO) of PubMed (accession number GSE6536) was used to analyze the relationship between the two SNPs and the whole-genome mRNA expression levels of the 47,293 genes14,15.
Statistics
An unpaired Student t test was applied to numerical variables, whereas the differences in categorical variables were tested using the χ2 test. The Cochran-Armitage trend test was used to compare the genotype dosage among the TB cases and controls. Hardy-Weinberg equilibrium (HWE) was assessed by Pearson χ2 test. The strength of associations between genotypes and TB were estimated by odds ratio (OR) and its 95% confidence interval (95% CI) through univariate and multivariate logistic regression analyses adjusted for age and gender. A P value of less than 0.05 was considered statistically significant. The relationships between the genotypes of the SNPs and the gene expression levels of the 45 CHB were analyzed by the online software GEO2R based on the moderate t test. Meanwhile, the usual t test was also applied to analyze the relationship between the genotypes and the gene expressions. The Benjamini & Hochberg (False discovery rate) was adopted for the multiple comparison correction19. The significance level for gene expression among different genotype groups was P < 0.2020. The value of fold change (FC) was calculated for showing the fluctuation of the gene expression among different genotype groups. The software Quanto (Version: 1.2.3, South California, USA) was employed to calculate the power of the OR on TB risk. All of the analyses were performed using Stata software (Version 13.0, StataCorp, Texas, USA).
Results
A total of 763 TB cases and 763 controls were included in this analysis, with one case and one control failing the genotyping. The mean ages for TB cases and controls were 49.17 ± 17.48 years and 52.03 ± 17.33 years, respectively. The QFT results showed that the positive rate of LTBI for the controls was 23.1% (176/763). As shown in Table 1, the minor allele (T allele) frequency of rs2057178 was 0.048 in the TB cases and 0.027 in the healthy controls (χ2 = 14.07, P = 0.0002). As the minor allele of rs2057178 was less than 0.05 and the TT genotype was only found in six subjects, the TT genotype among TB cases and controls showed a decreased risk of TB without reaching significance (adjusted OR = 0.56, 95% CI, 0.10–3.10). However, the CT genotype was significantly associated with a decreased risk of TB (adjusted OR = 0.52, 95% CI, 0.34–0.78). The dominant model (CT + TT vs. CC) demonstrated a protective effect on TB (adjusted OR = 0.52, 95% CI, 0.35–0.78). Based on the T allele frequency (0.048) of rs2057178 and the estimated TB prevalence (51/100000) in this region21, the corresponding power for the dominant OR of rs2057178 was 87.77%. For SNP rs4331426, the minor allele (G allele) frequencies between TB cases and controls showed no statistically significant difference (χ2 = 0.04, P = 0.8390). The Hardy-Weinberg equilibrium test demonstrated that genotypes of each locus in the controls were all in Hardy-Weinberg equilibrium (χ2 = 2.79, P = 0.095 for rs2057178 and χ2 = 0.142, P = 0.704 for rs4331426).
Then, the controls were classified as QFT-positive and QFT-negative to compare the genotype distributions of the two variants among the TB cases, non-infected controls and LTBI controls. The data in Table 2 show that the CT genotype of rs2057178 was significantly associated with a 0.60-fold (adjusted OR = 0.40, 95% CI, 0.22–0.70) decreased risk of TB in QFT-positive controls. The protective effect of the CT genotype was also observed in the QFT-negative controls (adjusted OR = 0.57, 95% CI, 0.36–0.90). The Cochran-Armitage trend test showed that the proportion of the CT genotype was increasing from the QFT-negative controls (8.0%) to the QFT-positive controls (11.4%, Ptrend = 0.0006). No associations of rs4331426 with TB were found among the TB cases and the subgroups of the controls.
The liner regression analysis was conducted between the genotypes of the two SNPs and the whole genome mRNA expression levels in the lymphoblastoid cell lines. The 45 CHB were classified into two groups based on the genotypic data of rs2057178 and rs4331426. For rs2057178, 42 CHB had the CC genotype, two CHB had the CT genotype and one CHB failed the genotyping (no TT genotype was found). For rs4331426, 39 CHB had the AA genotype, four CHB had the AG genotype and two CHB failed the genotyping (no GG genotype was found). The GEO2R analyzed the expression levels of 47293 genes in each CHB subject and 28 genes revealed significantly different expression levels between the two genotype groups of rs2057178 by moderate t test after multiple comparison (Table 3). However, the usual t test only found the first 20 genes were in relationship with SNP rs2057178. Thus, we included the first 20 genes for both reaching significance. No gene expression levels were found to be associated with the genotypes of rs4331426 after multiple comparison adjustment (data not shown).
Discussion
In this case-control study, the T allele of SNP rs2057178 was significantly associated with a decreased risk of TB in the Han Chinese population and 28 mRNA levels were found to be associated with the genotypes of rs2057178 in the 42 CHB, which indicated a potential functional role of rs2057178 in modulating those gene expression levels. However, no genotype of rs4331426 was found to be associated with TB susceptibility and no gene expression levels were associated with any genotype of rs4331426.
Thy et al. first reported that rs4331426 was associated with TB susceptibility in a GWAS of the African population in 201011. The HapMap data showed that the G allele frequency of rs4331426 in the Han Chinese population was 0.044, whereas the G allele frequency in the African population was 0.51. Because of the vast difference in the G allele frequencies between the African and Asian populations, the results of repeated association studies would be different. Another replicated association study conducted in the Chinese population by Wang et al. found that the G allele of rs4331426 had an opposite effect on TB susceptibility22 compared with the results of Thy et al. The G allele frequency was 0.0338 in the control group of Wang’s study, while the G allele frequency in our control group was 0.0301, all were close to the G allele frequency of 0.044 of the CHB of the HapMap data. Based on our data, we did not find any association between the genotypes of rs4331426 and TB predisposition. Another two association studies conducted in the Chinese population also did not find a relationship between rs4331426 and TB risk23,24. As SNP rs4331426 was located in a gene desert region, it was difficult to determine the function of the locus. Generally, it was postulated that other functional loci, in linkage with rs4331426, would be the target loci that were involved in the mechanism of predisposition to TB. In this study, the relationship between the genotypes of rs4331426 and the whole-genome expression levels indicated that no gene expression levels were correlated with the genotypes of rs4331426 after multiple comparison adjustment.
A subsequent GWAS by Thy et al. revealed that rs2057178 was associated with TB susceptibility10. In Thy’s study, the effect of the T allele on TB susceptibility was further verified in the Gambian and Russian populations. However, the association between rs2057178 and the predisposition to TB failed to replicate in the Indonesian population. Another study conducted in the Asian population also failed to replicate the protective effect of the T allele of rs2057178 on TB susceptibility23. It is interesting that a recent GWAS conducted in the African population also revealed the relationship between rs2057178 and TB susceptibility9. For the populations discussed above, the HapMap data showed that the T allele frequency of rs2057178 varied with a broad spectrum; it was highest in the African population (0.33) and lowest in the Asian population (0.02). Inter-population heterogeneity cannot be ignored for genetic susceptibility for TB. However, in this study, SNP rs2057178 was found to be significantly associated with TB susceptibility in the Han Chinese population. When the control group was stratified into QFT-positive and QFT-negative groups, the proportion of the CT genotype in the QFT-positive group (11.4%) was higher than that of the QFT-negative group (8%). The Cochran-Armitage trend test revealed that the QFT-positive group with the CT genotype would be more resistant to TB, which suggested that the T allele of rs2057178 might protect people latently infected with TB from developing TB disease. Although the locus was associated with TB susceptibility, the functional role of the locus was not clearly determined as the SNP was located in an intergenic region. SNP rs2057178 was in the 45 Kb downstream of Wilms’ tumor 1 (WT1) gene, which had been shown to be associated with the occurrence of Wilms’ tumor25. It was reported that WT1 variants might play a role in altering the effects of interferon-beta on vitamin D26, which had been shown to be beneficial in the treatment of TB27. Meanwhile, the WT1 gene was involved in the activation of the vitamin D receptor28, which was critically important for binding with 1,25-dihydroxyvitamin D3 to modulate the immune system in fighting M. tb infection29.
Although the HapMap genotypic data of rs2057178 and the whole-genome expression levels of the 42 CHB did not reveal a significant association between the genotypes of rs2057178 and WT1 gene expression, another 20 gene expression levels were found to be significantly associated with rs2057178. More importantly, v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB, Fig. 1), suppressor of cytokine signaling 2 (SOCS2, Fig. 2) were found to be associated with SNP rs2057178 in infectious diseases. According to the fold change (FC) of the gene expressions, MAFB was up expressed by 31% while SOCS2 was down expressed by 57%. MAFB was first reported to be a candidate gene for TB susceptibility in a GWAS by Mahasirimongkol et al.30 and the expression level of MAFB was found to be higher in patients with active TB compared with the healthy controls and previous TB cases31. Our study provided evidence for rs2057178 in modulating the TB susceptible gene MAFB in trans effect and the mechanism of modulation needs to be further explored.
Simultaneously, the SOCS2 expression level was significantly decreased in the CT genotype of rs2057178 for the CHB compared with the CC genotype. A previous study showed that SOCS2 was required to mediate the effects of lipoxin32, which was thought to negatively regulate protective Th1 responses against mycobacterial infection in vivo33.
Even though the interferon regulatory factor 5 (IRF5, Fig. 3) was not found in association with SNP rs2057178 by the usual t test and the FC showed that IRF5 was only slightly decreased (14%) among the CT genotype of rs2057178, IRF5 has an important role in the type 1 interferon response to M. tb34. Thus, there is a contradiction between the protective effect of the CT genotype of rs2057178 and the decreased expression level of IRF5 and the level of IRF5 needs to be further validated in TB cases and controls.
Several limitations need to be noted in this study. First, the QFT method is an indirect method for detecting latent TB infection and it may not accurately represent the existence of M. tb in vivo because we do not know how long the immunological reaction to M. tb will last. However, the differentiation ability of the QFT is more convincing in detecting M. tb-induced infection rather than other Mycobacteria when compared with the tuberculin skin test. Second, the transcriptome varies considerably across different cell populations and developmental stages. A previous study revealed the different cell-type associated gene expression profiles of tuberculosis35. Even some researchers found that the interferon-inducible genes were predominantly expressed in neutrophils and, to some extent, in monocytes, but not in T cells36. Gene expression levels in other cell types should be evaluated to comprehensively reveal the potentially distinct gene expression profiles of different cell populations. Third, the limited sample size for revealing the associations between SNPs and gene expression levels may be confounded by other factors and it is worthwhile to compare the actual mRNA expression levels among the TB cases and the healthy controls with larger samples in future studies.
In conclusion, we replicated the loci of TB GWAS in the Han Chinese population. We found that rs2057178 was significantly associated with TB predisposition and that the expression levels of MAFB and SOCS2 were significantly associated with the genotypes of rs2057178. We assume that MAFB and SOCS2 could be potential candidate genes for TB susceptibility in the Han Chinese population. Further functional studies are required to reveal the mechanism of host genetics on TB susceptibility. Additionally, the liner regression analysis for the association between SNP genotypes and gene expression levels could be a choice for exploring the potential functional role of disease predisposition loci.
Additional Information
How to cite this article: Chen, C. et al. A rare variant at 11p13 is associated with tuberculosis susceptibility in the Han Chinese population. Sci. Rep. 6, 24016; doi: 10.1038/srep24016 (2016).
References
WHO. Global Tuberculosis Report 2014. World Health Organization Genova (2014).
Gao, L. et al. Latent tuberculosis infection in rural China: baseline results of a population-based, multicentre, prospective cohort study. Lancet Infect Dis 15, 310–9 (2015).
Parrish, N. M., Dick, J. D. D. & Bishai, W. R. Mechanisms of latency in Mycobacterium tuberculosis. Trends in Microbiology 6, 107–12 (1998).
WHO. The END TB strategy. WHO: Genova,, Switzerland (2015).
Comstock, G. W. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis 117, 621–4 (1978).
Wilkinson, R. J. et al. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet 355, 618–21 (2000).
Lindenau, J. D. et al. Cytokine gene polymorphisms are associated with susceptibility to tuberculosis in an Amerindian population. Int J Tuberc Lung Dis 18, 952–7 (2014).
Velez, D. R. et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans and West Africans. Hum Genet 127, 65–73 (2010).
Chimusa, E. R. et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet 23, 796–809 (2014).
Thye, T. et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet 44, 257–9 (2012).
Thye, T. et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 42, 739–41 (2010).
Rivas, M. A. et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43, 1066–73 (2011).
Wilkinson, R. J. Human genetic susceptibility to tuberculosis: time for a bottom-up approach? J Infect Dis 205, 525–7 (2012).
Stranger, B. E. et al. Population genomics of human gene expression. Nat Genet 39, 1217–24 (2007).
Stranger, B. E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–53 (2007).
Hussain, T. & Mulherkar, R. Lymphoblastoid Cell lines: a Continuous in vitro Source of Cells to Study Carcinogen Sensitivity and DNA Repair. Int J Mol Cell Med 1, 75–87 (2012).
Chackerian, A. A., Alt, J. M., Perera, T. V., Dascher, C. C. & Behar, S. M. Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun 70, 4501–9 (2002).
Whitworth, H. S., Scott, M., Connell, D. W., Donges, B. & Lalvani, A. IGRAs–the gateway to T cell based TB diagnosis. Methods 61, 52–62 (2013).
Benjamini, Y. & Yekutieli, D. Quantitative trait Loci analysis using the false discovery rate. Genetics 171, 783–90 (2005).
Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journa of the Royal Statistical Society 57, 289–300 (1995).
Wang, Y. National Fifth Epidemiological Survey of Tuberculosis. Military Medical Science Press, Beijing (2011).
Wang, X. et al. Association of polymorphisms in the Chr18q11.2 locus with tuberculosis in Chinese population. Hum Genet 132, 691–5 (2013).
Ji, L. D. et al. Lack of association between polymorphisms from genome-wide association studies and tuberculosis in the Chinese population. Scand J Infect Dis 45, 310–4 (2013).
Dai, Y. et al. Fine mapping of genetic polymorphisms of pulmonary tuberculosis within chromosome 18q11.2 in the Chinese population: a case-control study. BMC Infect Dis 11, 282 (2011).
Huff, V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 11, 111–21 (2011).
Lin, R. et al. Modulating effects of WT1 on interferon-beta-vitamin D association in MS. Acta Neurol Scand 131, 231–9 (2015).
Coussens, A. K. et al. Vitamin D accelerates resolution of inflammatory responses during tuberculosis treatment. Proc Natl Acad Sci USA 109, 15449–54 (2012).
Wagner, K. D., Wagner, N., Sukhatme, V. P. & Scholz, H. Activation of vitamin D receptor by the Wilms’ tumor gene product mediates apoptosis of renal cells. J Am Soc Nephrol 12, 1188–96 (2001).
Liu, P. T. et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311, 1770–3 (2006).
Mahasirimongkol, S. et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet 57, 363–7 (2012).
Satproedprai, N. et al. Diagnostic value of blood gene expression signatures in active tuberculosis in Thais: a pilot study. Genes Immun 16, 253–60 (2015).
Russell, C. D. & Schwarze, J. The role of pro-resolution lipid mediators in infectious disease. Immunology 141, 166–73 (2014).
Bafica, A. et al. Host control of Mycobacterium tuberculosis is regulated by 5-lipoxygenase-dependent lipoxin production. J Clin Invest 115, 1601–6 (2005).
Pandey, A. K. et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLos Pathog 5, e1000500 (2009).
Maertzdorf, J. et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 12, 15–22 (2011).
O’Garra, A. et al. The immune response in tuberculosis. Annu Rev Immunol 31, 475–527 (2013).
Acknowledgements
We are especially thankful to Dr. Jianling Bai (Department of Epidemiology and Biostatistics, Nanjing Medical University, Nanjing, China) for his contributions on the statistics. This study was supported by the National Natural Science Foundation of China [grant number 81302480, 81202256] and Key Laboratory of Public Health Safety of Fudan University [grant number GW2015-4].
Author information
Authors and Affiliations
Contributions
C.C. wrote the main manuscript text, B.X. and W.L. designed the study and scientifically edited the manuscript, C.C., Y.S. and G.L. designed and conducted the experiments. Q.Z. and Y.H. collected the field data, L.Z. conducted the quality control. All the authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, C., Zhao, Q., Hu, Y. et al. A rare variant at 11p13 is associated with tuberculosis susceptibility in the Han Chinese population. Sci Rep 6, 24016 (2016). https://doi.org/10.1038/srep24016
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24016
This article is cited by
-
Gene expression pattern analysis using dual-color RT-MLPA and integrative genome-wide association studies of eQTL for tuberculosis suscepitibility
Respiratory Research (2021)
-
2′-5′-Oligoadenylate synthetase 1 polymorphisms are associated with tuberculosis: a case-control study
BMC Pulmonary Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.