Abstract
Leukemic patients are often immunocompromised due to underlying conditions, comorbidities and the effects of chemotherapy and thus at risk for developing systemic infections. Bloodstream infection (BSI) is a severe complication in neutropenic patients and is associated with increased mortality. BSI is routinely diagnosed with blood culture, which only detects culturable pathogens. We analyzed 27 blood samples from 9 patients with acute leukemia and suspected BSI at different time points of their antimicrobial treatment using shotgun metagenomics sequencing in order to detect unculturable and non-bacterial pathogens. Our findings confirm the presence of bacterial, fungal and viral pathogens alongside antimicrobial resistance genes. Decreased white blood cell (WBC) counts were associated with the presence of microbial DNA and was inversely proportional to the number of sequencing reads. This study could indicate the use of high-throughput sequencing for personalized antimicrobial treatments in BSIs.
Similar content being viewed by others
Introduction
Systemic infections, such as BSIs can develop in hematological malignancies due to inherent immune defects and therapy-related immunosuppression and are associated with increased mortality1. BSIs are routinely diagnosed by blood culture and treated by empirical broad-spectrum antimicrobials. Antimicrobial treatment might be inappropriate due to the lack of coverage of the underlying pathogen(s), or antimicrobial resistance of the causative pathogens2. Blood culture requires relatively large sample volumes, only detects culturable pathogens and represents a narrow spectrum of the microbes present in a sample3. Furthermore, only 10–30% of blood cultures from febrile neutropenia and 50% of blood cultures from septic shock are positive1,4,5. Although bacteria are the most commonly detected pathogens, fungal and viral infections also represent major complications in hematological malignancies4,6,7. Candida and Aspergillus species are the most commonly occurring fungal pathogens, but others, including Fusarium and Trichosporon, can also be detected8.
With the recent advantages of high-throughput sequencing technologies, it has become possible to reconstruct the taxonomic diversity of uncultured microbial communities, even in complex clinical samples. The two main approaches remain the amplicon sequencing (mostly restricted to the 16S rDNA universal gene in bacteria) and shotgun metagenomics, which allows untargeted sequencing of DNA in a given sample. Compared to the 16S rDNA sequencing approach, shotgun metagenomics not only allows the analysis of bacterial diversity in clinical samples, but can also provide information on the presence of parasitic, fungal and viral pathogens. An important clinical aspect is microbial resistance: sequencing of the 16S rDNA gene does not provide information regarding resistance patterns, while shotgun sequencing can identify resistance genes, their point mutations and also chromosomal resistance mechanisms. The latter approach also permits functional annotation of the sequencing data by assigning molecular functions to sequencing reads.
The aim of this study was to characterize the microbial content of blood samples from neutropenic patients with newly diagnosed acute leukemia. Sampling was performed at different time points before and after the initiation of antimicrobial treatments and sequence analysis was combined with functional annotation.
Results
Sequencing reads
In total, 1,005,260,502 reads were generated (excluding spike in samples), with 33.5 million reads being sequenced per sample on average. Seventy-nine percent of the reads were mapped to the human genome, while 20.93% of the reads were unmapped. Microbial reads consisted 0.07% of the total reads (Fig. 1). As shown in previous studies, human DNA exceeds microbial DNA approximately 107–108 –fold in sepsis9,10. The applied background suppression treatments reduced human DNA approximately 10,000-fold as sequencing reads aligned to the human genome outweighed microbial reads by 104 on average.
(A) Distribution of sequencing reads between human and microbial origins. (B) Distribution of the detected microbes. Colored numbers indicate the number of occurrences per sampling category (red numbers: fever onset, blue: persistent fever, green: follow up). Two fever samples did not contain microorganisms and there were no detections of pathogen DNA in 1 persistent fever sample and 6 follow up samples.
Microbial content of blood samples per sample category
Samples from nine patients were included in this study; in two patients, no microbial detection occurred. Samples (n = 27) were analyzed at different time points during antimicrobial treatments: 1) at inclusion (n = 1), 2) at fever onset (before antibiotic treatment started, n = 9), 3) 1 day after antibiotics were given (n = 7) and 4) follow up (1–5 days after the start of antibiotic treatment started, n = 11).
Three out of nine fever onset samples only contained bacteria, while four out of seven persistent fever samples contained bacteria, fungi and viruses (Fig. 1B).
Bacterial reads were detected mostly at fever onset and in persistent fever (69% of all reads in both categories), but in less than 1% in follow up samples. The diversity of bacterial taxa did not change during fever, but showed a significant decrease in follow up samples, likely due to antibiotic treatment (Fig. 2A). Viral reads were dominantly detected during persisting fever (Fig. 2) and mostly consisted of bacteriophages. The diversity of fungal and viral taxa did not show significant changes between disease states.
Distribution of bacterial taxa in different time points of sampling
The bacterial content of the samples was dominated by Propionibacterium acnes, Corynebacterium spp and Staphylococcus spp in agreement with a 16S rDNA study on BSIs3. Dolosigranulum pigrum (in follow up samples) and Neisseria spp (during persistent fever) were also frequently detected (Fig. 3A).
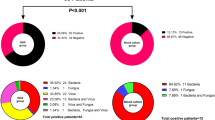
(A) Distributions of bacterial reads are shown in different diseases stages, with Propionibacterium dominating during the initial fever episode, while Corynebacterium, Staphylococcus and Dolosigranulum were also detected in persisting fevers. Bacterial taxa which consisted of <1% of all reads in a sample are not shown. (B) Distribution of fungal taxa detected in the samples used in this study indicate the presence of Fusarium in different disease stages, possibly implying subclinical fungal infections. (C) Distribution of reads belonging viral taxa at different sampling time points. Numbers within the pie charts indicate the number of samples positive for the given pathogen.
The distribution of bacterial taxa per sample type revealed a difference between sampling time points: P. acnes was the dominant taxa only in fever onset samples and was not detected in follow up samples at all. In samples with persisting fever, Corynebacterium spp, D. pigrum and Staphylococcus spp were dominant, while S. spiritivorum was the most prominent taxa in follow up samples.
Distribution of fungal taxa in different time points of sampling
Fusarium oxysporum, a known fungal pathogen in immunocompromised patients, was detected most frequently in every sampling category (in 13 samples out of 27, Fig. 3B). Other detected fungal taxa also represent opportunistic infections (e.g., Aspergillus spp and Malessezia globosa) characteristic for immunocompromised patients.
Distribution of viral taxa in different time points of sampling
Overall, phage related to Propionibacterium were the most dominant viral taxon (Fig. 3C), along with Torque Teno Virus (TTV). TTV was detected in 6 samples from 3 patients, while Propionibacterium phages were detected in 3 samples, from 3 patients. Amongst the known oncoviruses, Merkel cell polyomavirus and Hepatitis C were detected. Human endogenous retroviruses, as part of the human genome were removed from the analysis.
Changes in microbial content in individual samples throughout the disease
As shown, antibiotic treatments were typically efficient against bacterial infections (e.g., P. acnes or Neisseria spp. in patients 1, 4, 5, 7), but less effective against species such as Pseudomonas (patient 2). Viruses and fungi were also often detected and were not affected by the antiviral/-fungal prophylaxis. When the infection was dominantly fungal or viral (patients 3, 5, 6), persistent fever samples contained the most reads, while in bacterial infections, fever onset samples had the highest number of reads.
In patients 3 and 6, fungal and viral taxa were dominant and the number of reads increased with fever (Fig. 4).
Relative abundances of bacterial, fungal and viral taxa are shown in individual samples.
The highest bar for each patient represents the highest number of reads; the heights of other bars of the same patient are proportional to the highest. Wider bars represent taxa on the species level, slim bars show domains. Only taxa with ≥1% relative abundance are shown.
Laboratory data
WBC and ANC were compared between patients in different disease stages (Fig. 5A) and with and without detected microbial DNA. WBC was significantly lower in patients when microbial DNA was identified (p ≤ 0.05, Fig. 5B).
(A) Clinical characteristics of the samples used in this study, measured at the time of sampling. (B) Comparison of white blood cell count and absolute neutrophil count between samples with and without the presence of microbial DNA. (C) Comparison of WBC and ANC between samples with the highest microbial reads (>10,000) versus samples with microbial reads lower than 10,000. *denotes p ≤ 0.05. WBC: white blood cell count, ANC: absolute neutrophil count, CRP: C-reactive protein, MASCC: Multinational Association for Supportive Care in Cancer risk score.
Samples with the highest microbial reads (>10,000 reads, n = 6) had significantly lower WBC (p ≤ 0.05) and lower ANC (p ≥ 0.05) than samples with less microbial reads found (Fig. 5C).
Bacterial resistance
Tetracycline resistance genes were found in 2 samples (patient 4, fever sample and patient 7, persistent fever sample 1). MLS (Macrolide Lincosamide Streptogramin) resistance genes were found in 4 samples (patient 3 (persistent fever), patient 4 (fever and persistent fever samples) and patient 7 (persistent fever sample 1)).
Gene ontology (GO) analysis
In bacterial reads, significant differences were found in molecular processes (binding processes and hydrolase activity) between persistent fever and follow up samples (p ≤ 0.05, Fig. S1a). Metabolic processes also showed a difference between these two groups (p ≤ 0.05, Fig. S1b).
In fungal reads, molecular functions and biological processes did not show changes throughout the disease, assuming the lack of active antifungal drugs (Fig. S2).
The molecular functions and biological processes did not show a significant change in viral reads, although the hydrolase activity was elevated in persistent fever samples (p ≥ 0.05, Fig. S3).
Discussion
Although the blood is considered sterile in physiological state, it may harbor dormant microbes, or microbes can enter the bloodstream due to translocation or pathological conditions11. BSI is treated with empirical broad-spectrum antimicrobials, which are often not efficient against the invading microbes due to the lack of specificity or resistance mechanisms2. Even though detection of microbial DNA in the blood does not necessarily imply the presence of viable pathogens, it is associated with worse clinical outcomes5,12,13; Fig. 5 in present study).
In this study, blood samples from patients with acute leukemia and suspected bloodstream infection were subjected to characterizing their microbiota using shotgun metagenomics. To our knowledge, this is the first investigation of uncultured microbial content and their resistance genes in blood samples from BSIs. Our results corroborate previous findings that the routinely used blood culture only detects a proportion of pathogens3 and our findings confirm the presence of viral and fungal pathogens in immunocompromised patients7,8.
The risk of infection is much greater in leukemic patients due to comorbidities and side effects of chemotherapy. BSI is often caused by bacteria, but can also be caused by fungi and viruses7,8. While bacterial infections are diagnosed with blood culture and treated with antibiotics, fungal and viral infections often go undetected in BSI cases, although they are associated with higher mortality rates in cancer patients8,14. As expected, the employed antibiotic treatment was effective against the majority of bacteria, but dominantly viral (including bacteriophages) and fungal pathogens were detected in 4 patients (Fig. 4). Fungal infections are an important cause of mortality in neutropenic cancer patients8,15, with A. fumigatus and C. albicans being mainly responsible for the infections, along with Fusarium and Trichosporon species. Posaconazole, which was used as fungal prophylactic is effective against most fungal pathogens but resistance can occur against this drug in Fusarium species15,16,17,18. Fusarium oxysporum was the most commonly detected fungal species in the current study (patients 5–7, Fig. 3B) with fungal load changing with fever (Fig. 4), which might indicate low level of persistent, possibly subclinical fungemia.
Viruses commonly occur in leukemic patients with febrile neutropenia6,7. For the patients included in this study, acyclovir was used as a prophylactic drug to prevent Herpesvirus-related infections. Herpesviruses are commonly detected in leukemic patients with neutropenia6,7 and were detected in 2 samples from 1 patient in our study as well, while TTV was the most commonly detected viral species (3 patients, 6 samples). TTV has been detected in leukemic patients but its causative role has not been proven; it was hypothesized however that TTV can be a co-virus in participating in the disease etiology in leukemia19. Bacteriophages are considered as part of the normal microbiota20; in this study, phages have been detected dominantly during persisting fever directly after antibiotic treatment, indicating that the origin of phages were bacteria degraded by antibiotics (Figs 2 and 3C). Additionally, antibiotic treatment is a strong inducer of the release of bacteriophages21,22, which can explain their elevated hydrolase activity (Fig. S3). In case of detection of bacteriophages, their corresponding host bacteria were also detected as phages exert high level of host specificity23.
Bacteria mostly occurred in fever and persistent fever samples, but were almost non-existent in follow up samples, due to the antibiotic treatment (Fig. 2). P. acnes, Staphylococcus and Corynebacterium spp were detected commonly in samples included in this study (Fig. 3A). Dolosigranulum pigrum and Neisseria spp bloodstream infections were also frequently detected, as they can be opportunistic pathogens in patients with immunosuppression24,25,26,27. Many bacteria have been detected and/or have been implied as causative in cancers28. Acinetobacter has been detected in acute leukemia and was postulated to play a role in bacteria-human cell lateral gene transfer29; this genus was detected in 5 samples from 2 patients in this study. Also, mucosal injury or damage in the lumen can also occur as a side effect of chemotherapy and opportunistic bacteria can enter the bloodstream in hematological malignancies3,4,28.
GO analysis of bacteria showed significant changes with antibiotic administration (Fig. S1), implying the effectiveness of antimicrobial drugs and adaptation of bacteria. GO analysis also confers the notion of resistance of the F. oxysporum strains as there were no changes in molecular functions and biological processes throughout the treatment (Fig. S2) despite of antifungal prophylaxis. The elevated ratio of transport processes in fungal reads might indicate resistance to drugs as the rate of transport of antimicrobial agents can be insufficient compared to the efflux pumps, resulting in inefficient doses30. The elevation of hydrolase activity in viral reads (Fig. S3) is most likely due to the release of phages with the degradation of bacteria in persistent fever samples (1 day after AB administration), as no changes were observed between fever onset and follow up samples.
In summary, this work confirms the presence of viral and fungal pathogens alongside bacteria and antibiotic resistance genes in leukemic patients with neutropenic fever. The presence of microbial DNA and the number of sequencing reads were correlated with lower WBC counts in blood samples included in this study. Altogether, our results imply the possible utility of this technology in personalized medicine in the antimicrobial treatment of patients with acute leukemia.
Materials and Methods
Study population and sampling
Eight patients with acute myeloid leukemia and one patient with acute lymphocytic leukemia considered suitable for dose intensive antitumoural treatment at the Hematology Center, Karolinska University Hospital in Stockholm, Sweden, were enrolled for this study upon diagnosis. Included patients were sampled with two 4.5 mL EDTA tubes for venous blood at different time points: 1) at hematological diagnosis, 2) at fever onset during neutropenia before intravenous broad-spectrum antibiotic treatment was initiated, 3) persisting fever during intravenous broad-spectrum antibiotic treatment and 4) follow-up samples 1–5 days after the initiation of antibiotic treatment.
Data on white blood cell count (WBC), absolute neutrophil count (ANC), C-reactive protein (CRP) levels, Multinational Association for Supportive Care in Cancer (MASCC) Risk Scores and hematological diagnoses were extracted retrospectively from the patients’ medical records. Samples were handled anonymously.
Antimicrobial treatments
All patients were treated with empirical broad-spectrum antibiotics at fever onset, according to international IDSA guidelines. Five of the patients received ciprofloxacin prophylaxis. Acyclovir and posaconazole were used as antiviral and antifungal prophylactics for all patients, in accordance with the local hospital guidelines (Table S1).
Ethics statement
All subjects provided written, informed consent. The study and all experimental protocols used in this study was approved by The Regional Ethical Review Board, Stockholm (Regionala Etikprövningsnämnden Stockholm, recordal 2012/1929-31/1) and were carried out in accordance with the approved guidelines.
Definitions
Fever was defined as a single oral temperature of ≥38.5 °C or a temperature of >38.0 °C persisting for >1 hour. Neutropenia was defined as a neutrophil count of ≤0.5 × 109 cells/L, or a higher count with an expected decrease to ≤0.5 × 109 cells/L within 24 hours.
Sample preparation and sequencing
Blood samples for sequencing were drawn into sterile 4.5 ml Vacutainer (Becton Dickinson, Franklin Lakes, NJ USA) tubes, were kept at 4 °C and processed to DNA extraction within 1–24 hrs. MolYsis Complete5 kit (Molzym Life Science, Bremen, Germany) was used to extract bacterial DNA following the manufacturer’s instructions with the following exceptions: 5 minutes were used for the final elution instead of 1 and samples were dissolved in 50 μl water instead of 100 μl. One μg of the eluted DNA was then processed to NebNext microbiome enrichment (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. Ten ng DNA was subjected to multiplex displacement amplification, using the GenomiPhi V2 DNA amplification kit (GE Healthcare, Little Chalfont, United Kingdom), with 90 minutes amplification resulting in 2–4 μg DNA. Two μg DNA was used for library preparation with the Nextera XT kit and libraries were processed to a 2*100 base pair PE sequencing on a HiSeq 2500 instrument. The data generated in this study was uploaded to the NCBI Sequencing Read Archives under experiment SRX1381258.
Validation of the assay
Escherichia coli NCTC 9001 genomic DNA was spiked in to microfiltered human blood (Sigma-Aldrich) in 10-fold dilutions from 1 million CFU to 0. No template controls did not generate sufficient background amplification with the standard protocol for sequencing, therefore the MDA reaction was performed for 3 hours and processed as described above. E. coli DNA spiked into sterile blood was detected down to 10 CFU. Control samples (microfiltered human blood, “Negative blood”) and a sample without added DNA (“No template control”, Fig. 4) were dominated by bacterial species, Achromobacter piechaudii and xylosodans, Pseudomonas species and Massilia timonae, with several of them have already been reported as reagent contaminants31.
Data analysis
Reads shorter than 30 bp and with Phred quality scores below 30 were discarded using the Fastx toolkit. Paired end reads were merged using the Flash software32, while unpaired reads were discarded. RTG Core 3.433 was used to filter reads against the human genome (build Hg_19) and to map the unmapped reads to fungal, bacterial and viral whole genome sequences (downloaded from the NCBI Microbial Genomes Resources (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html)), with minimum e value set to 10−6. Duplicate reads were removed from the analysis using RTG Core 3.4. Taxa detected in the no template control sample, or in the spike in sample with 0 copies of E. coli, or with ≤10 reads/taxon were not included. The ARG-ANNOT database was used to detect antibiotic resistance genes. GO analysis was performed using the Blast2GO v3 software, with e = 10−6. Statistical analysis was performed using the Mann-Whitney U-test, with significance set to 0.05.
Additional Information
How to cite this article: Gyarmati, P. et al. Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci. Rep. 6, 23532; doi: 10.1038/srep23532 (2016).
References
de Naurois, J. et al. Management of febrile neutropenia: ESMO Clinical Practice Guidelines. Ann. Oncol. 21, Suppl 5:v252-6 (2011).
Kalich, B. A. et al. Impact of an Antibiotic-specific Sepsis Bundle on Appropriate and Timely Antibiotic Administration for Severe Sepsis in the Emergency Department. J. Emerg. Med. S0736–4679, 937–943 (2015).
Gyarmati, P. et al. Bacterial Landscape of Bloodstream Infections in Neutropenic Patients via High Throughput Sequencing. PLoS One. 10, 8, 10.1371/pone0135756 (2015).
Hämäläinen, S. et al. Neutropenic fever and severe sepsis in adult acute myeloid leukemia (AML) patients receiving intensive chemotherapy: Causes and consequences. Leuk. Lymphoma. 49, 495–501 (2008).
Liesenfeld, O., Lehman, L., Hunfeld, K. P. & Kost, G. Molecular diagnosis of sepsis: New aspects and recent developments. Eur. J. Microbiol. Immunol. 4, 1–25 (2014).
Öhrmalm, L. et al. Viral findings in adult hematological patients with neutropenia. PLoS One. 7, 5, 10.1371/pone36543 (2012).
Wood, M. J. Viral infections in neutropenia–current problems and chemotherapeutic control. J. Antimicrob. Chemother. 41, Suppl D, 81–93 (1998).
Warnock, D. W. Fungal infections in neutropenia: current problems and chemotherapeutic control. J. Antimicrob. Chemother. 41, Suppl D, 95–105 (1988).
Song, Y. et al. Nuclease-assisted suppression of human DNA background in sepsis. PLoS One. 9, 7, 10.1371/pone103610 (2014).
Zhou, L. & Pollard, A. J. A novel method of selective removal of human DNA improves PCR sensitivity for detection of Salmonella typhi in blood samples. BMC Inf. Dis. 12, 164, 10.1186/1471-2334-12-164 (2012).
Potgieter, M., Bester, J., Kell, D. B. & Pretorius, E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 39, 567–91 (2015).
Lehmann, L. E., Herpichboehm, B., Kost, G. J., Kollef, M. H. & Stüber, F. Cost and mortality prediction using polymerase chain reaction pathogen detection in sepsis: evidence from three observational trials. Crit. Care. 14, R186, 10.1186/cc9294 (2010).
Struelens, M. J. Detection of microbial DNAemia: does it matter for sepsis management? Intensive Care. Med. 36, 193–95 (2010).
Lech-Maranda, E. et al. Infectious complications in patients with acute myeloid leukemia treated according to the protocol with daunorubicin and cytarabine with or without addition of cladribine. A multicenter study by the Polish Adult Leukemia Group (PALG). Int. J. Infect. Dis. 14, e132–140 (2010).
Walsh, T. J. & Gamaletsou, M. N. Treatment of fungal disease in the setting of neutropenia. Hematology Am. Soc. Hematol. Educ. Program. 2013, 423–427 (2013).
Alastruey-Izquierdo, A., Cuenca-Estrella, M., Monzón, A., Mellado, E. & Rodríguez-Tudela, J. L. Antifungal susceptibility profile of clinical Fusarium spp. isolates identified by molecular methods. J. Antimicrob. Chemother. 61, 805–809 (2008).
Nucci, M. & Anaissie, E. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704 (2007).
Loeffler, J. & Stevens, D. A. Antifungal drug resistance. Clin. Infect. Dis. 36, S31–41 (2003).
Chu, C. C. et al. Torque teno virus 10 isolated by genome amplification techniques from a patient with concomitant chronic lymphocytic leukemia and polycythemia vera. Mol. Med. 17, 1338–1348 (2011).
De Paepe, M., Leclerc, M., Tinsley, C. R. & Petit, M. A. Bacteriophages: an underestimated role in human and animal health? Front. Cell. Infect. Microbiol. 4, 39, 10.3389/fcimb.2014.00039 (2014).
Allen, H. K. et al. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio. 2, 6, 10.1128/mBio.00260-11 (2011).
McGannon, C. M., Fuller, C. A. & Weiss. A. A. Different classes of antibiotics differentially influence shiga toxin production. Antimicrob. Agents Chemother. 54, 3790–3798 (2010).
Koskella, B. & Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 5, 806–823 (2013).
Domingo, P., Coll, P., Maroto, P., Verger, G. & Prats, G. Neisseria subflava bacteremia in a neutropenic patient. Arch. Intern. Med. 156, 1762–1765 (1996).
Lécuyer, H. et al. Dolosigranulum pigrum causing nosocomial pneumonia and septicemia. J. Clin. Microbiol. 45, 3474–3475 (2007).
Neth, O. W., Bajaj-Elliott, M., Turner, M. W. & Klein, N. J. Susceptibility to infection in patients with neutropenia: the role of the innate immune system. Br. J. Haematol. 129, 713–722 (2005).
Ruoff, K. L. Miscellaneous catalase-negative, gram-positive cocci: emerging opportunists. J. Clin. Microbiol. 40, 1129–1133 (2002).
Cummins, J. & Tangney, M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect. Agent. Cancer. 8, 11, 10.1186/1750-9378-8-11 (2013).
Riley, D. R. et al. Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput. Biol. 9, 6, 10.1371/pcbi1003107 (2013).
Van Acker, H., Van Dijck, P. & Coenye, T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 22, 326–333 (2014).
Salter, S. et al. Reagent contamination can critically impact sequence-based microbiome analyses BMC Biol. 12, 87, 10.1186/s12915-014-0087-z (2014).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27, 2957–2963 (2011).
Cleary, J. G. et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol. 21, 405–419 (2014).
Acknowledgements
The authors would like to thank all participants involved in this study. This work was supported by the Swedish Research Council (Vetenskapsrådet, 2012–3291) to PG, The Swedish Society of Medicine (SLS-408571) to LÖ, Cathrine Everts Research Foundation, Blodcancerfonden to CK. The authors would like to acknowledge support with the sequencing from the Science for Life laboratory.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: P.G. and C.G. Performed the experiments: P.G., C.K., C.A. and Y.S. Analyzed the data: P.G., C.K. and L.Ö. Contributed reagents/materials: P.G., C.K., L.Ö. and C.G. Contributed to the writing of the manuscript: P.G., C.K., C.A., Y.S., L.Ö. and C.G.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gyarmati, P., Kjellander, C., Aust, C. et al. Metagenomic analysis of bloodstream infections in patients with acute leukemia and therapy-induced neutropenia. Sci Rep 6, 23532 (2016). https://doi.org/10.1038/srep23532
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep23532
This article is cited by
-
Evaluation of antibiotic resistance, toxin-antitoxin systems, virulence factors, biofilm-forming strength and genetic linkage of Escherichia coli strains isolated from bloodstream infections of leukemia patients
BMC Microbiology (2023)
-
Rapid DNA visual detection of polymicrobial bloodstream infection using filter paper
Scientific Reports (2022)
-
The Potential Role of Clinical Metagenomics in Infectious Diseases: Therapeutic Perspectives
Drugs (2021)
-
The Microbiota in Hematologic Malignancies
Current Treatment Options in Oncology (2020)
-
Critical steps in clinical shotgun metagenomics for the concomitant detection and typing of microbial pathogens
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.