Abstract
The uses of fluorescent reporters derived from green fluorescent protein have proved invaluable for the visualisation of biological processes in bacteria grown under aerobic conditions. However, their requirement for oxygen has limited their application in obligate anaerobes such as Clostridium difficile. Fluorescent proteins derived from Light, Oxygen or Voltage sensing (LOV) domains have been shown to bridge this limitation, but their utility as translational fusions to monitor protein expression and localisation in a strict anaerobic bacterium has not been reported. Here we demonstrate the utility of phiLOV in three species of Clostridium and its application as a marker of real-time protein translation and dynamics through genetic fusion with the cell division protein, FtsZ. Time lapse microscopy of dividing cells suggests that Z ring assembly arises through the extension of the FtsZ arc starting from one point on the circumference. Furthermore, through incorporation of phiLOV into the flagella subunit, FliC, we show the potential of bacterial LOV-based fusion proteins to be successfully exported to the extracellular environment.
Similar content being viewed by others
Introduction
Clostridium difficile is a pathogen of major significance worldwide, being recognised as the leading cause of antibiotic associated diarrhoea in the nosocomial setting. In the U.S. increased rates of C. difficile infection (CDI) and relapse, seen in up to 20.9% of CDI these cases1, incur annual health care costs in excess of $4.8 billion2. Consequently, there is an urgent need to understand the pathogenic traits of this organism and the influence of expression of specific genes during infection. To date, most research has centred on the study of the two major toxins A and B (TcdA & TcdB) whose production and activity are responsible for the majority of symptoms observed during CDI3,4. Whilst experimental vaccines based on these toxins reduce or eliminate symptoms5 they do not prevent germination, colonisation and sporulation of the bacteria, which are all essential steps in transmission from host to host. Greater understanding of these processes is essential if we are to identify new targets for future prophylactic or therapeutic treatments.
One factor limiting insight into the molecular analysis of this pathogen is the inability to visualise and monitor ongoing cellular processes. Fluorophores, derived from green fluorescent protein (GFP) have been used successfully in other species6, but require molecular oxygen for fluorescence maturation, thus preventing their use in anaerobic systems. In addition, many Clostridium species generate a green autofluorescence when excited with blue/ultraviolet (UV) light7 making visualisation with green fluorescent reporters problematic. However, advances in C. difficile bioimaging are beginning to occur, with the oxygen-dependent fluorescent proteins [Cyan Fluorescent Protein (CFP) and mCherry] used to characterise the cellular localisation of MdlA/B/C & ZapA in dividing bacteria7,8. In addition, tags based on O-6-methylguanine-DNA methyltransferase (SNAP-tags) have been used successfully to investigate protein localisation9,10. These elegant studies have significantly extended our understanding of the temporal and spatial interplay between the RNA polymerase sigma subunits during sporulation. However, due to oxygen requirements or cell toxicity of substrates used, they are unsuited for monitoring real time protein dynamics in anaerobic environments. In contrast, small flavin-based photosensory modules such as Light, Oxygen or Voltage sensing (LOV) domains have shown promising potential as real time probes in oxygen depleted situations11,12,13,14,15.
LOV domains were first identified in plants associated with the blue-light receptor kinases16 (phototropins) and bind flavin mononucleotide (FMN) as a light-absorbing chromophore. Upon activation, conformational changes associated with the FMN chromophore induce phototropin kinase activity17. Since their discovery, LOV domains have been found to be present in all forms of life (Archaea, Bacteria & Eukaryota), where they regulate a wide variety of functions including virulence and homeostasis18,19,20. Recently, Christie and co-workers enhanced the fluorescence and the photostability of the LOV domain from Arabidopsis thaliana phototropin 2 (phot2)21,22. The use of improved LOV (iLOV) or photostable iLOV (phiLOV) as fluorescent reporters has three main advantages over GFP as a reporter; phiLOV is smaller (~13 kDa vs ~25 kDa), fluorescence is stable over a greater pH range23 and fluorescence is not dependent on molecular oxygen24.
In addition, the discovery and mutation of LOV fluorescent reporters from diverse sources, each with different attributes25,26, are being increasingly exploited. For example, LOV domains from the freshwater algae Chlamydomonas reinhardtii (CreiLOV) have increased brightness over other LOV fluorophores27 whereas engineered LOV domains from Arabidopsis thaliana (phiLOV) have enhanced photobleaching-resistant properties22. As such LOV-based fluorescent proteins, together with other oxygen-independent reporters, including UnaG28 & mBFP29, are receiving increased attention as GFP alternatives and will also boost the bio-imaging toolkit in such systems.
In this paper, we show that phiLOV can be used as a tool to fluorescently label a number of obligate anaerobes including Clostridium difficile, Clostridium sordellii and Clostridium acetobutylicum. In addition, we reveal the potential of the phiLOV reporter to monitor the real time intracellular localisation and extracellular secretion of proteins within C. difficile through the generation of LOV translational reporter fusions. These data provide evidence of the wider utility of phiLOV technology, particularly in the study of pathogenesis within other anaerobic bacteria.
Results
Optimising the conditions for detecting phiLOV expression in C. difficile
The plasmid pRPF185-phiLOV, in which expression of a codon-optimised sequence of phiLOV was placed under the control of a tetracycline-inducible promoter, was introduced into C. difficile R20291 by conjugation (Supplemental Fig. 1a). To assess reporter activity, this strain was grown to mid-logarithmic phase and expression induced by the addition of 500 ng/ml anhydrotetracycline (ATc); a concentration which previously was shown to have limited impact on cell viability (This study &30). Expression of phiLOV by C. difficile was initially confirmed by Western blot analysis using antibodies to the iLOV protein (Supplemental Fig. 2).
Following induction of expression through addition of ATc, these bacteria fluoresced green when excited with light of 470 nm. However, as C. difficile possess an intrinsic green autofluorescence when excited at this wavelength, we considered the relative increase in fluorescence of phiLOV over this background signal. This was achieved by analysis of fluorescence of C. difficile transformed with the plasmid pRPF185-gusA. In addition, the use of the membrane stain FM4-64, which has lipophilic binding activity, allowed visualisation of all bacteria in each field of view. Using the red fluorescence of this stain, it was possible to ensure that the equivalent numbers of LOV or GusA expressing bacteria were compared. Under these conditions, C. difficile containing the pRPF185-phiLOV plasmid, induced for 30 min with optimal concentrations of ATc, showed a relative 2.5-fold increase in fluorescence compared to those cells harbouring pRPF185-gusA alone (Fig. 1). In addition, the strength of the fluorescence signal increased over time reaching a relative 3.5-fold increase 240 min post induction. Figure 1b,c reflect representative images used in the analysis of relative fluorescence from C. difficile cultures expressing either phiLOV or gusA, respectively.
Characterisation of phiLOV expression in Clostridium difficile cells.
(A) Relative fluorescence intensity (left x-axis) mediated by phiLOV (blue line) or gusA (black line) expressing cells. Samples were taken at 0, 30, 60, 120 & 240 min post induction with 500 ng/ml ATc, stained with FM4-64, fixed and viewed by fluorescence microscopy. Results shown are mean ± standard error of mean (SEM) of all bacterial cells from five de-convoluted fields of view and a minimum of two independent experiments. qRT-PCR analysis of phiLOV transcription (right x-axis) used RNA extracted at the same time points following induction from bacteria expressing phiLOV (dashed purple line). Expression represented as the fold change compared to pre-induction levels (0 min). Results shown are mean ± standard deviation (SD) of two independent experiments. Figures (B,C) are representative images of bacteria expressing either phiLOV (B) or gusA (C) following induction of expression for the durations shown in (A). LOV expressing strains generate green fluorescence whilst samples stained with FM4-64 (shown as red fluorescence) allow visualisation of the whole bacterial population. Images shown in (B,C) have undergone post image analysis (deconvolution and fluorescent contrast enhancement) to remove background autofluorescence. Scale bar in (B,C) 5 μm.
Whilst phiLOV expression in C. difficile could be clearly visualised, there was a concern that the detection systems used were not optimal. This is because the optimal excitation maxima (λmax) for phiLOV is 450 nm, however, for microscopic analysis the closest filter set available to us provided excitation at 470 nm. To determine the impact of changing the excitation wavelength on overall fluorescent signal, we induced strains expressing either gusA or phiLOV with 500 ng/ml ATc for 60 min. These samples were then analysed using a variable excitation/emission wavelength FluoroMax-3 spectrofluorometer. Using an excitation wavelength of 450 nm, C. difficile cells expressing phiLOV showed a 2.1-fold fluorescence increase compared to cells expressing gusA (p < 0.0001) whereas excitation at 470 nm showed a 1.7-fold fluorescence increase (p < 0.0001). These data show that whilst a significant difference was observed between C. difficile cells expressing either gusA or phiLOV when exciting at 470 nm, this difference could be further improved using a filter set that excites at 450 nm (Supplemental Fig. 3).
To further investigate the induction kinetics of phiLOV expression, the concentration of ATc was varied. This revealed that whilst an increase in concentration of ATc to 1 μg/ml did not significantly increase fluorescence activity, decreasing the concentration of ATc to 100 ng/ml, reduced the increase in fluorescence (1.9-fold higher fluorescence than control cells at 240 min). Thus under these conditions the optimal induction concentration for ATc was 500 ng/ml.
In parallel, measurement of phiLOV mRNA levels using qRT-PCR indicated a peak in transcript levels (approx. 15-fold higher relative to the control cells), 60 min post induction, after which phiLOV mRNA levels decreased (Fig. 1a dashed line). Interestingly this decrease was not associated with a reduction in fluorescence, providing further evidence to the enhanced stability of phiLOV fluorescence.
Potential of phiLOV fluorescence in other Clostridium species
To confirm that phiLOV could be used as a fluorescent reporter in other Clostridium species, the phiLOV expression plasmid was introduced into C. sordellii and C. acetobutylicum. The strains were grown to A600 nm 0.5 and induced with 500 ng/ml ATc for 60 min, fixed and examined by fluorescence microscopy. Induction of phiLOV expression resulted in a significant increase in fluorescence intensity of the population in each of the Clostridium species tested, compared to plasmid-free controls (p < 0.0001) (Fig. 2). Interestingly, expression of phiLOV in C. difficile saw the smallest fold-change in fluorescence intensity (3.2-fold increase) compared to C. sordellii or C. acetobutylicum (increase of 5.6- & 4.5-fold respectively), which probably reflects the lower levels of autofluorescence displayed by these strains (Fig. 2). This may suggest that this technology may have even greater potential application within these species.
Expression of phiLOV in different Clostridium species.
Comparison of the relative fluorescent intensities observed between plasmid-free C. difficile R20291 (CD WT), C. sordellii ATCC9714 (CS WT) & C. acetobutylicum ATCC824 (CA WT) and these strains expressing pRPF185-phiLOV. For this experiment, each Clostridium species were grown to A600 nm of 0.5 and induced by the addition of 500 ng/ml ATc for 60 min. Results shown are mean ± standard error of mean (SEM) of all bacterial cells from five de-convoluted fields of view and a minimum of two independent experiments.
Use of phiLOV as a real-time protein reporter
To determine whether phiLOV could be used as a real-time reporter for monitoring protein localization and dynamics within a bacterial cell, we genetically fused this protein to the C-terminus of the cytokinetic protein FtsZ of C. difficile (Supplemental Fig. 1b). FtsZ is involved in the formation of a cytokinetic ring (or Z-ring) at the site of cell division31 and has successfully been tagged with GFP in other bacterial species32. Western blot analysis using anti-iLOV antibody confirmed that C. difficile successfully expressed the FtsZ-phiLOV fusion protein (Supplemental Fig. 2). To determine whether the protein retained functionality, bacteria were grown to mid-logarithmic phase (when cells are most actively dividing) and induced with ATc for 30 minutes. Cell membrane was additionally visualised by co-staining these cells with the lipophilic dye FM4-64. Fluorescent imaging of these organisms revealed a fluorescent signal detected at the cell mid-point in ~77% of bacterial cells undergoing cell division (Fig. 3a). A 3-dimensional recreation of the FtsZ-phiLOV fluorescent signal from z stack images further resolved the mid-cell fluorescent signal into a ring structure (Fig. 3b). Measuring the signal intensity across the diameter of this ring confirmed this structure to be ~1 μm in diameter (Fig. 3c).
Monitoring FtsZ localisation using a FtsZ-phiLOV protein reporter.
(A) Localisation of fluorescence to cell midpoints in C. difficile expressing the FtsZ-phiLOV and grown under standard induction conditions. (Ai) the bacterial population stained with FM4-64; (Aii) green fluorescence as a consequence of LOV expression and (Aiii) merged images. Images underwent post image analysis (deconvolution and fluorescent contrast enhancement) to remove background autofluorescence. Scale bars in 5 μm and, in insert 1 μm. (B) Representation of the FtsZ-phiLOV ring in 3D using compiled data from z-stack images. Z slices were taken at 0.2 μm intervals through the bacterial cell and 3-dimensional analysis completed using max intensity 3D rendering in Volocity. Scale bar, 1 μm. (C) Fluorescent intensity profile through the FtsZ-phiLOV ring shown in (B). This reflects the peak fluorescence intensity plotted against distance.
One of the main advantages of phiLOV over other reporter systems is their potential as real time reporters for protein dynamics. To this end, we induced C. difficile harbouring pRPF185-FtsZ-phiLOV and immediately immobile cells on agarose pads, which were covered and sealed within the anaerobic chamber. Using this technique, C. difficile cells can be transferred without impact on viability to aerobic environments for imaging. In this way we were able to observe the incorporation of FtsZ-phiLOV monomers during assembly of the Z ring in dividing cells and constriction of the Z ring (Supplemental Fig. 4). Time-lapse imaging of a bacterial cell showed accumulation of fluorescent signal at the cell midpoint approximately 70 min post induction. This started at one point on the circumference of the bacterial cell and took ~40 min to increase the length of the FtsZ-phiLOV arc to span the cell (Fig. 4a). Once a complete Z ring had formed, constriction of this ring preceded the narrowing of the cell diameter as membrane biogenesis occurred at the site of division, a process that took ~80 min to complete (Fig. 4b). Constriction of the Z ring, certainly in the later stages of constriction, proceeded at a linear rate (Fig. 4c).
Real time Z ring assembly and constriction in C. difficile using fluorescent microscopy.
(A) FtsZ-phiLOV incorporation during Z-ring assembly in real time using images of the same cell captured every 10 min. (B) Constriction of the Z ring within a bacterial cell during cell division over 120 min. Organisms shown in the images were induced with 500 ng/ml ATc and immobilised on an agarose pad under a sealed cover slip for time-lapse observation. Images have undergone post image analysis (deconvolution and fluorescent contrast enhancement). Scale bar in (A,B) 2 μm. (C) Changes in Z ring diameter over time. These ring diameter measurements were taken from images shown in (B).
Use of phiLOV protein fusions to track exported proteins
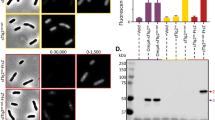
As previously noted, phiLOV is approximately half the size of the GFP-family of fluorescent proteins and as such maybe sufficiently small to be used in protein-membrane translocation studies. To establish whether phiLOV could be used in this context in C. difficile, the sequence was cloned into a central site within the major flagellin protein FliC (Supplemental Fig. 1c). To limit complications of native FliC expression, this construct was transferred into strain C. difficile 630 in which fliC had been deleted (630 ΔfliC) as well as wild-type cells (630). Induced cultures were stained with FM4-64 prior to fixation and examination by fluorescence microscopy. Z stack imaging of these cells showed little to no intracellular fluorescence (Supplemental Fig. 5), but instead an accumulation of fluorescence that appeared to be external to the cytoplasmic membrane of these cells (Fig. 5a). To determine whether the protein was exported into the media, western blot analysis was performed on samples recovered from the cytosolic and membrane fractions of the bacterial cells or from filtered extracts of the culture supernatant (Fig. 5b). Using antibodies to both iLOV and FliC respectively, immunoreactive bands of approximately 44 kDa were identified in samples generated from the cytosolic, membrane and culture supernatant of C. difficile cells expressing fliC-phiLOV. In contrast, an immunoreactive band of 13 kDa was only observed in the cytosolic extract of cells expressing phiLOV alone (Fig. 5c,d). These data help to confirm the observation that phiLOV is able to translocate the membrane when fused to the FliC momomer. In addition, the FliC antibody recognised a protein of approximately 35 kDa, the approximate size of a post-translationally modified FliC monomer. In addition, several breakdown products were observed in the strain expressing fliC-phiLOV. This probably reflects degradation of poorly folded proteins following the relatively unregulated overexpression of the protein from the inducible tetracycline promoter.
Secretion of FliC-phiLOV fusion proteins in C. difficile.
(A) Localisation of fluorescence in C. difficile expressing the fliC-phiLOV fusion protein. Bacteria were grown to A600 nm of 0.5, induced with 500 ng/ml ATc, co-stained with FM4-64 and immobilised on agarose pads as described. (Ai) bacterial population stained with FM4-64; (Aii) green fluorescence as a consequence of LOV expression and (Aiii) the merged images. Extended focus displays a brightest point merge of all planes of representative images of C. difficile expressing fliC-phiLOV fusion protein. (Supplemental Fig. 5 for z slices). Images shown have undergone post image analysis (deconvolution and fluorescent contrast enhancement) to remove background autofluorescence. Scale bar, 5 μm and, in insert, 2 μm. (B) SDS-PAGE of cell lysate, membrane extracts and culture supernatants from C. difficile expressing either gusA, phiLOV or fliC-phiLOV. Western blot analysis of protein expression using either anti-iLOV antibody (C) or anti-FliC antibody (D) as the probes. Predicted protein sizes of phiLOV, FliC and FliC-phiLOV are 13, 30.9 and 43.7 kDa, respectively, however FliC is known to be post-translationally modified35 so size will vary. Molecular weights expressed in kDa.
While expression of fliC-phiLOV in C. difficile appears to result in export from the cells, expression did not restore flagella mediated motility or result in detectable flagella filaments on the cell surface (data not shown). This most likely reflects an inability to correctly fold FliC as a consequence of phiLOV insertion. This could prevent the appropriate polymerization of the FliC monomers into the flagellar structure resulting in their subsequent release and recovery in the filtered media from the culture.
Discussion
In this study, we report the first use of a LOV domain as a fluorescent reporter in the anaerobic bacterium C. difficile. Whilst its relative small size (~13 kDa) makes it attractive as a reporter, its green fluorescence had been thought to limit its potential application in naturally autofluorescent bacteria, such as C. difficile. Further, unlike the GFP/RFP-family of fluorescent proteins, mutational analysis of LOV reporters has largely failed to shift the excitation/emission spectra, with only small spectral blue shifts in fluorescence emission (~10 nm) reported25,26. Also LOV-based FPs engineered thus far exhibit fluorescence quantum yields (QF) that are significantly lower (~0.2–0.5) to that of GFP (0.6)23, which could impact their utility for certain reporter applications. Despite these issues, we report that a differential level of fluorescence due to phiLOV expression is quantifiable with fluorescence appearing stable over time. In addition, the use of phiLOV as a reporter in this context does not appear limited to C. difficile with differential expression also observed in other Clostridium species, including the commercially important C. acetobutylicum in which lower levels of autofluorescence are observed.
Further, we have demonstrated that phiLOV can be used as a reporter of protein expression within C. difficile, using two model proteins FtsZ and FliC. This is the first time that protein expression has been visualised in real time by fluorescence in C. difficile, with previous used GPF-based reporters being restricted by the requirement for oxygen. Using time-lapse fluorescent microscopy, we show the formation of FtsZ-phiLOV z ring and additionally the constriction of the Z ring in real time, taking approximately 130 min to complete the whole process. The genetic fusion of phiLOV to ftsZ appeared to have no detrimental effects on FtsZ function with FtsZ-phiLOV able to polymerise into a ring structure, anchor to the cell membrane, hydrolyse GTP & interact with other proteins in the divisome. Whilst study of this phenomenon in real-time was technically difficult to perform, requiring maintenance of anaerobiosis, the images highlight the integral robust stability of the phiLOV fluorophore. This stability25, is an important feature when considering time lapse measurements.
Resolution of this fluorescence using z-stack analysis, deconvolution and 3D rendering confirmed the formation of a ring-like structure and further underlines the potential of phiLOV as a reporter for protein localisation. Observation of several fields of non-synchronised dividing bacteria also revealed the presence of incomplete rings in some bacteria. This is unsurprising given that these bacteria contain both plasmid-encoded FtsZ-phiLOV and the intact chromosomal ftsZ locus. Consequently, these incomplete rings may reflect structures composed of both labelled and unlabelled FtsZ.
The small size of LOV proteins, when compared to GFP, has the potential to be particularly useful when creating translational fusions especially for the study of proteins that are secreted from the cytoplasm. To evaluate the potential of phiLOV in this context we generated a fliC-phiLOV construct, in which phiLOV was inserted at nucleotide 483 of the C. difficile fliC sequence. In other bacterial species, the proposed protein folding and monomer polymerisation of FliC molecules is thought to include the N- & C-terminal sequences of the FliC protein33 with the central portion of the monomer exposed on the flagella surface. Visualisation using Z stack images (Supplementary Fig. 5) taken through the entire bacteria revealed fluorescence on the periphery of those bacteria expressing the fliC/LOV fusion, which suggested that the fusion of phiLOV did not hinder secretion of FliC subunits. This observation was surprising but interestingly also observed at much lower levels in the control strains. Disappointingly, expression and secretion of this this fusion did not result in any detectable flagella filaments or restore motility to the bacterium. These data suggest that the presence of the tag is limiting the opportunity for effective subunit polymerisation rather than secretion. This problem may reflect the chosen location for insertion of the tag. Indeed, whilst the fusion protein was detected in the cytoplasm, membrane fractions and filtered spent growth medium, several related breakdown products were also detected using the specific FliC antibodies. This would suggest that the rate of expression might influence the protein folding and the potential of these proteins to polymerise into definitive flagellar structures. Thus this work shows that although phiLOV can be used to label proteins that are translocated across the membrane, the location of the tag and the regulation of protein expression may influence the incorporation of such fusion proteins into macromolecular structures.
Whilst the examples given here highlight the opportunities offered by the use of phiLOV, the natural autofluoresence of C. difficile is likely to restrict its use for study of low abundance proteins. Whilst this issue cannot be avoided, we have attempted to quantify the level of phiLOV expression needed to detect a significant change in C. difficile fluorescence; in our system, we calculated that a 2.68 fold-increase in phiLOV mRNA levels correlated with a detectable increase in fluorescence. Although based on several assumptions, the calculated expression fold-increase can give an indication of the level of target expression needed before phiLOV expression is observed only further experimentation will allow us to establish the scope of its use. In parallel, the application of fluorophores, which are excited by wavelengths outside the blue/green spectrum, such as SNAP tags (based on O6-alkylguanine-DNA alkyltransferase9), CFP (a GFP-family protein8) or mCherry (a Red Fluorescent Protein (RFP)-family protein7) are important developments. However, the use of these fluorescent systems in anaerobic environments requires either a fixation step before fluorescence detection or exposure to molecular oxygen, limiting their use in the study of protein dynamics in real time. Using the time-lapse experiments described, we show that LOV domains can bridge this limitation, allowing researchers the opportunity to study protein dynamics in anaerobic systems in real-time. As such, we propose that at least for some situations, a combination of both systems is appealing, potentially allowing the amount and location of specific proteins (using reporters systems described above) at specific time points determined by linkage of phiLOV to proteins, for example associated with cell cycle.
In conclusion, this study highlights the use of the phiLOV domain as a fluorescent reporter in the anaerobic genus Clostridium. Furthermore, translational fusions that include these domains can be used to study protein localisation and dynamics in real-time and offer opportunities to increase our understanding of protein dynamics in response to environmental stimuli. In addition, as we advance our capacity to identify and culture a much wider number of strictly anaerobic bacteria, the availability of fluorescent technologies that can be readily applied under hypoxic conditions is attractive. As a result, the potential to apply phiLOV technology is wide ranging and may play a significant role in enhancing our understanding of gene expression and regulation in many important but poorly understood anaerobic organisms.
Materials and Methods
Bacterial strains and growth
Bacterial strains used in this study are listed in Supplemental Table 1. Escherichia coli TOP10 (Invitrogen, U.K.), used as a cloning host and E. coli CA43434 as a conjugal donor were grown aerobically on Luria Bertani medium supplemented with ampicillin (100 μg/ml) or chloramphenicol (15 μg/ml) when required. C. difficile strains R20291 & 630 ΔfliC35 were kind gifts from Prof. B. Wren and Dr Alex Faulds-Pain (London School of Hygiene and Tropical Medicine, London, U.K.). C. acetobutylicum strain ATCC824 was purchased from the American Type Culture Collection (ATCC). C. difficile strains were routinely grown on CCEY agar plates supplemented with cefoxitin-cycloserine, egg emulsion (Lab M Ltd,) and if required 15 μg/ml thiamphenicol, in an anaerobic workstation (Don Whitley Scientific Ltd, U.K.) at 37 °C. Brain-heart infusion (BHI) broth or Tryptone Yeast (TY) broth (supplemented with 15 μg/ml thiamphenicol) were routinely used to grow C. difficile strains during fluorescent experiments. C. sordelli was cultured in TY broth and C. acetobutylicum strain ATCC824 grown using Reinforced Clostridium Media (RCM, Lab M Ltd), both supplemented, when required, with 15 μg/ml thiamphenicol. Plasmids were introduced into C. difficile and C. sordellii by conjugation from E. coli CA434 as previously described34. C. acetobutylicum was transformed with pRPF185-phiLOV2.1 by Dr Liz Jenkinson (Green biologics Ltd, U.K.).
Cloning and plasmid construction
Construction of phiLOV2.1 construct
The phiLOV2.1 gene21 was codon optimized for expression in C. difficile (GenScript USA Inc.) and introduced into vector pUC57 (Supplemental Table 1). Primers 69F & R (Supplemental Table 2) were the used to amplify phiLOV2.1, adding Sac1 & BamH1 restriction enzyme sites to allow ligation of the sequence downstream of the tet promoter, within the pRPF185 vector30. The resultant plasmid, pRPF185-phiLOV2.1, was then transformed into E. coli TOP10 cells (Invitrogen Ltd). To confirm the sequence of the cloned fragment, primers 73F & 72R were used to amplify the region from the plasmid and the resultant product, purified using the Qiagen PCR purification kit and then subjected to sequence analysis.
Construction of C-terminal FtsZ-phiLOV2.1 fusion
phiLOV2.1 was genetically linked to the C-terminus of the cytokinesis protein FtsZ using primers 97F/R & 98F/R (Supplemental Table 2). These primers amplified the ftsZ and phiLOV2.1 sequences respectively, adding Nhe1 restriction enzyme sites to the 3′ and 5′ ends of the ftsZ and phiLOV2.1 sequences respectively. The PCR fragments were then ligated using the complementary NheI sites and the resultant ftsZ-phiLOV2.1 fragment further amplified using primers 99F & R before it was cloned into pRPF185 and sequenced as described.
Construction of fliC-phiLOV2.1 fusion
phiLOV2.1 was linked to the major structural flagella protein encoded by fliC by inserting the phiLOV2.1 sequence between nucleotides 483–484 of fliC. Primers 133F & R and 135F & R were used to independently amplify the first 483 nucleotides and last 390 nucleotides of the C. difficile 630 fliC gene encoded by C. difficile 630. These primers were additionally designed to incorporate complementary sequences to those found in phiLOV2.1 sequences. In parallel, primers 135F &134R (Supplemental Table 2) were designed to amplify phiLOV2.1 whilst additionally removing the stop codon from this protein and add overlapping complementary fliC sequences. The three fragments were then amplified by PCR, purified and assembled using the Gibson Assembly® Mastermix kit (New England Biolabs Inc.). The ligated fliC-phiLOV2.1 construct underwent a further round of PCR amplification using primers 139F & 140R to allow the addition of homologous overlapping sequences allowing subsequent ligation of the sequence into the expression vector pRPF185.
Fluorescence microscopy
To allow the entire bacterial population to be visualised by fluorescence, the membrane dye FM4-64 (Invitrogen) was added to samples taken from each culture at a final concentration of 5 μg/ml (10 min, 4 °C). Stained cells were washed by centrifugation in distilled water and then fixed in 10% formalin (Sigma) for 15 min in the dark. The bacteria were then washed in sterile distilled water before being air-dried onto glass slides. Prepared slides were mounted in Dako Fluorescent Mounting Medium (Dako Ltd) before addition of a glass cover slip. For time-lapse imaging, appropriate C. difficile strains were treated with 500 ng/ml ATc before being immobilised onto thin 1% agarose pads, which were covered with a glass cover slip that was sealed with Dako Fluorescent Mounting Medium. For these experiments, mounting and sealing of the slides was performed within the anaerobic cabinet to ensure viability of the bacteria. Bacterial cells were examined using an Axio-Zeiss Imager M1 light microscope (Carl Zeiss Microscopy GmbH) and images captured using a Hamamatsu ORCA-ER digital camera with motorized excitation and emission filter wheels and Zen 2012 (blue edition) acquisition software (Carl Zeiss Microscopy GmbH). Slides were exposed for a maximum of 500 ms (unless otherwise stated) using an excitation beam of 470 nm, emission was detected at 520 nm to detect phiLOV2.1 fluorescence and an excitation 540 nm, emission 725 nm to detect FM4-64 fluorescence. Z-stack experiments were performed at distances of 0.2 μm. Deconvolution of raw data was performed using a point-spread function with 30 rounds of iteration. Under the excitation/emission spectra used for phiLOV2.1 detection, C. difficile naturally fluoresce green. To determine the impact of this background fluorescence on the data, the mean background control fluorescence of same strain of C. difficile transformed with the control plasmid, pRPF185-gusA, was calculated from a minimum of 5 fields of view in three independently performed experiments. The mean fluorescence intensity of every bacterial cell imaged was determined using Volocity 3D image analysis software version 5.5 (PerkinElmer Inc., U.S.). Results represent the mean population fluorescence intensity ± standard error of mean from five fields of view. 3D image rendering for the FtsZ-phiLOV2.1 Z ring structure was performed using Volocity 3D image analysis software.
Statistical analysis
All statistical analyses were performed using the GraphPad Instat 3.10 (GraphPad Instat Software). A Students T-test was used to determine significant differences in fluorescence emissions between the different strains. P values ≤ 0.05 were considered significant.
Spectroscopic analysis
Fluorescence excitation and emission spectra were recorded using a FluoroMax-3 spectrofluorometer (Horiba Scientific) and a scan speed of 100 nm/min as described previously21.
Cell lysis, fractionation and protein analysis
To determine the expression and location of each recombinant protein, each culture was grown in BHI to an A600 nm of approximately 0.6 O.D. units before induction of protein expression with ATc for 4 h. The cultures were then harvested by centrifugation (500 g, 10 min). The supernatant was removed and immediately filtered using 0.2 nm syringe filter. Proteins within the culture supernatant were concentrated using the TCA/acetone precipitation method as previously described36. The remaining cell pellet was immediately frozen and cytosolic and membrane fractions separated as described30. In brief, samples were prepared to ensure equivalent loading, which was achieved by re-suspending the pellets in PBS containing lysozyme (1.4 mg/ml) and DNAse 1 (120 mg/ml) to provide an OD600 = 20. Membranes and cytoplasmic fractions were then separated by high-speed centrifugation (25,000 g). Whilst the supernatant was removed immediately and mixed with 2× SDS sample buffer, the resultant membrane fractions were then washed twice in PBS before being solubilised in 1% SDS to an equivalent volume of OD600 = 20. These samples were finally mixed with an equal volume of 2× SDS sample buffer. SDS-PAGE and Western blotting analysis were performed using standard methods with protein samples separated using 12% SDS-PAGE gels (Novex, Life technologies). After transfer to Hybond-C nitrocellulose membrane (Amersham Biosciences), blots were either probed using rabbit anti-iLOV antibody (1:1000)21 followed by HRP-linked secondary goat anti-rabbit antibody (1:3000; Sigma) or by chicken anti FliC (a kind gift from Glen Armstrong) (1:5000) detected by goat anti chicken Y-chain HRP conjugate (1:15000). HRP activity was subsequently detected using LumiGLO® (New England Biolabs Inc.).
RNA extraction and purification
qRT-PCR was performed on mRNA samples isolated from mid-log cultures induced with ATc as described previously37. In brief, mRNA was extracted using the FastRNA Pro Blue Kit (MP Biosciences) from bacterial cultures, maintained under anaerobic conditions. Initially, anaerobic cultures were treated with 10 ml RNAlater® (Ambion) for 5 min before the bacteria were collected by centrifugation (5000 × g; 10 min; 4 °C). The bacterial pellets were then re-suspended in 1 ml RNA Pro solution and transferred to matrix tubes in which bacteria were lysed using a FastPrep-24 instrument (MP Biosciences); (2 × 45 s at 6.0 ms−1). To minimise degradation, samples were cooled on ice for 5 min between cycles. The samples were subject to centrifugation (12000 × g; 10 min; 4 °C) and nucleic acid recovered from the supernatant, which was placed into an RNAse free tube to which 300 μl chloroform was added. The resultant mixture was vortexed and then centrifuged (12000 × g; 15 min; 4 °C) and the aqueous phase was separated and transferred to a fresh tube containing 500 μl of cold absolute ethanol. This was inverted and incubated −20 °C for 30 min. After centrifugation (12000 × g; 15 min; 4 °C) the pellet was washed with 500 μl of cold 75% ethanol and air dried for 5 min at room temperature. DNase treatment was carried out using the rigorous Turbo DNase kit (Ambion) according to manufacturer’s instructions. The RNA was further cleaned using the RNeasy Mini kit (Qiagen) and RNA purity and quantity assessed by nanodrop UV spectroscopy.
Analysis of relative levels of mRNA expression
For semi quantitative qRT-PCR, first strand cDNA was synthesized from total RNA using random hexamers as primers and the SuperScriptTM III Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions. qRT-PCR was performed in triplicate in a 20 μl reaction volume containing 200 ng of cDNA, 10 μl Fast SYBR Green Master Mix (5 Prime GmbH) and 500 nM gene specific primers (Supplemental Table 2) in an Illumina EcoTM real-time PCR instrument (Illumina). In each sample, the quantity of LOV specific cDNAs were normalised to the quantity of cDNAs of the rpoA gene. The relative transcript changes were calculated using the 2−ΔΔCt method38.
Additional Information
How to cite this article: Buckley, A. M. et al. Lighting Up Clostridium Difficile: Reporting Gene Expression Using Fluorescent Lov Domains. Sci. Rep. 6, 23463; doi: 10.1038/srep23463 (2016).
References
Lessa, F. C. et al. Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 372, 825–834 (2015).
Dubberke, E. R. & Olsen, M. a. Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 55, 88–92 (2012).
Lyras, D. et al. Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009).
Kuehne, S. a. et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 (2010).
Spencer, J. et al. Vaccination against Clostridium difficile using toxin fragments: Observations and analysis in animal models. Gut Microbes 5, 23–22 (2014).
Levin, P. A. & Losick, R. Transcription factor SpoOA switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10, 478–488 (1996).
Ransom, E. M., Ellermeier, C. D. & Weiss, D. S. Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl. Environ. Microbiol. 81, 1652–1660 (2015).
Ransom, E. M., Williams, K. B., Weiss, D. S. & Ellermeier, C. D. Identification and characterization of a gene cluster required for proper rod shape, cell division and pathogenesis in Clostridium difficile. J. Bacteriol. 196, 2290–2300 (2014).
Pereira, F. C. et al. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet. 9 (2013).
Saujet, L. et al. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 9 (2013).
Drepper, T. et al. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443–445 (2007).
Tielker, D., Eichhof, I., Jaeger, K. E. & Ernst, J. F. Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryot. Cell 8, 913–915 (2009).
Drepper, T. et al. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl. Environ. Microbiol. 76, 5990–5994 (2010).
Choi, C. H., DeGuzman, J. V., Lamont, R. J. & Yilmaz, Ö. Genetic transformation of an obligate anaerobe, P. gingivalis for FMN-green fluorescent protein expression in studying host-microbe interaction. PLoS One 6 (2011).
Lobo, L., Smith, C. J. & Rocha, E. R. Flavin mononucloetide (FMN)-based fluorescent protein (FbFP) as reporter for gene expression in the anaerobe Bacteroides fragilis. FEMS Microbiol. Lett. 317, 997–1003 (2011).
Huala, E. et al. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278, 2120–2123 (1997).
Harper, S. M., Neil, L. C. & Gardner, K. H. Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003).
Gaidenko, T. a., Kim, T. J., Weigel, A. L., Brody, M. S. & Price, C. W. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188, 6387–6395 (2006).
Herrou, J. & Crosson, S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 9, 713–723 (2011).
Swartz, T. E. et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317, 1090–1093 (2007).
Chapman, S. et al. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA 105, 20038–20043 (2008).
Christie, J. M. et al. Structural tuning of the fluorescent protein iLOV for improved photostability. J. Biol. Chem. 287, 22295–22304 (2012).
Mukherjee, A., Walker, J., Weyant, K. B. & Schroeder, C. M. Characterization of flavin-based fluorescent proteins: An emerging class of fluorescent reporters. PLoS One 8 (2013).
Christie, J. M., Gawthorne, J., Young, G., Fraser, N. J. & Roe, A. J. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol. Plant 5, 533–544 (2012).
Wingen, M. et al. The photophysics of LOV-based fluorescent proteins - new tools for cell biology. Photochem. Photobiol. Sci. 13, 875–83 (2014).
Buckley, A. M., Petersen, J., Roe, A. J., Douce, G. R. & Christie, J. M. LOV-based reporters for fluorescence imaging. Curr. Opin. Chem. Biol. 27, 39–45 (2015).
Mukherjee, A. et al. Engineering and characterization of new LOV-based fluorescent proteins from Chlamydomonas reinhardtii and Vaucheria frigida. ACS Synth. Biol. 4, 371–377 (2015).
Kumagai, A. et al. A bilirubin-inducible fluorescent protein from eel muscle. Cell 153, 1602–1611 (2013).
Hwang, C. S., Choi, E. S., Han, S. S. & Kim, G. J. Screening of a highly soluble and oxygen-independent blue fluorescent protein from metagenome. Biochem. Biophys. Res. Commun. 419, 676–681 (2012).
Fagan, R. P. & Fairweather, N. F. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286, 27483–27493 (2011).
Erickson, H. P., Anderson, D. E. & Osawa, M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010).
Ma, X., Ehrhardt, D. W. & Margolin, W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93, 12998–13003 (1996).
Samatey, F. a et al. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410, 331–337 (2001).
Emerson, J. E. et al. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74, 541–556 (2009).
Faulds-Pain, A. et al. The post-translational modification of the Clostridium difficile flagellin affects motility, cell surface properties and virulence. Mol. Microbiol. 94, 272–289 (2014).
Görg, a. et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053 (2000).
Dembek, M., Stabler, R. a., Witney, A. a., Wren, B. W. & Fairweather, N. F. Transcriptional analysis of temporal gene expression in germinating Clostridium difficile 630 endospores. PLoS One 8 (2013).
Saujet, L., Monot, M., Dupuy, B., Soutourina, O. & Martin-Verstraete, I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism and virulence factor expression in Clostridium difficile. J. Bacteriol. 193, 3186–3196 (2011).
Acknowledgements
This research was supported by two awards made by The Wellcome Trust to GRD. The first Grant number 086418, awarded collaboratively to G.R.D. and Prof Brendan Wren, LSHTM, UK) and the second, Grant number 090969, awarded to G.R.D. and Prof Neil.Fairweather (Imperial, UK). The authors also acknowledge the help of Jan Petersen in analysis of excitation and emission spectra of LOV expressing strains, Prof Glen Armstrong for the provision of anti-FliC antibody, Dr James Connelly with Western blot analysis and Dr Alex Faulds-Pain for the gift of the C. difficile ΔfliC strain.
Author information
Authors and Affiliations
Contributions
G.R.D., A.B., A.R., J.C. and N.F. designed and coordinated the study. R.F., N.F. and J.C. provided the plasmid backbones and antibody used in this study. A.B. and C. J. conducted the molecular approach to this study. A.B., D.C. and J.I. prepared the bacterial samples for microscopy. A.B., J.S. and A.R. were responsible for fluorescence microscopy and interpretation of data. A.B. and G.R.D. wrote the manuscript with contributions from all co-authors.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Buckley, A., Jukes, C., Candlish, D. et al. Lighting Up Clostridium Difficile: Reporting Gene Expression Using Fluorescent Lov Domains. Sci Rep 6, 23463 (2016). https://doi.org/10.1038/srep23463
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep23463
This article is cited by
-
A monoclonal antibody collection for C. difficile typing ?
Gut Pathogens (2024)
-
Practical observations on the use of fluorescent reporter systems in Clostridioides difficile
Antonie van Leeuwenhoek (2022)
-
Production of the biocommodities butanol and acetone from methanol with fluorescent FAST-tagged proteins using metabolically engineered strains of Eubacterium limosum
Biotechnology for Biofuels (2021)
-
Lessons from Clostridial Genetics: Toward Engineering Acetogenic Bacteria
Biotechnology and Bioprocess Engineering (2021)
-
Visualizing the dynamics of exported bacterial proteins with the chemogenetic fluorescent reporter FAST
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.