Abstract
First identified in The Netherlands in 2004, human coronavirus NL63 (HCoV-NL63) was found to cause worldwide infections. Patients infected by HCoV-NL63 are typically young children with upper and lower respiratory tract infection, presenting with symptoms including croup, bronchiolitis and pneumonia. Unfortunately, there are currently no effective antiviral therapy to contain HCoV-NL63 infection. CoV genomes encode an integral viral component, main protease (Mpro), which is essential for viral replication through proteolytic processing of RNA replicase machinery. Due to the sequence and structural conservation among all CoVs, Mpro has been recognized as an attractive molecular target for rational anti-CoV drug design. Here we present the crystal structure of HCoV-NL63 Mpro in complex with a Michael acceptor inhibitor N3. Structural analysis, consistent with biochemical inhibition results, reveals the molecular mechanism of enzyme inhibition at the highly conservative substrate-recognition pocket. We show such molecular target remains unchanged across 30 clinical isolates of HCoV-NL63 strains. Through comparative study with Mpros from other human CoVs (including the deadly SARS-CoV and MERS-CoV) and their related zoonotic CoVs, our structure of HCoV-NL63 Mpro provides critical insight into rational development of wide spectrum antiviral therapeutics to treat infections caused by human CoVs.
Similar content being viewed by others
Introduction
Coronaviruses (CoVs) are a diverse group of enveloped positive-strand RNA viruses in the family Coronaviridae1,2. CoVs have been identified in a wide variety of hosts, including mammals and birds and are shown to cause a number of respiratory and enteric diseases1,3,4. In 2003, the global epidemic of an atypical form of pneumonia named severe acute respiratory syndrome (SARS) led to the discovery of SARS-CoV, a previously unknown CoV, as the etiologic pathogen5,6,7,8. Started in South China, SARS outbreak quickly resulted in more than 800 deaths worldwide9. Patients with SARS-CoV infection developed diffuse alveolar damage with the potential to progress into acute respiratory distress syndrome and eventually death10. Almost 10 years later, another previously unknown CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), was found to cause a new epidemic starting in the Arabian Peninsula in 201211,12,13. MERS infection led to acute pneumonia and renal failure, with mortality rate as high as 50% in hospitalized patients14,15. In addition to the deadly SARS-CoV and MERS-CoV, 4 other human CoVs have been identified so far, namely HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1, which are known to cause comparatively mild common colds9,16,17. According to their genomic sequences, these 6 HCoVs are further classified into alphacoronavirus genus (HCoV-229E and HCoV-NL63) and betacoronavirus genus (HCoV-OC43, HCoV-HKU1, SARS-CoV and MERS-CoV)12,18. The emergence of CoV infection in human beings are believed to begin with zoonotic transmission from animal reservoirs9. For example, high degree of genomic sequence similarity was shown between bovine CoV and HCoV-OC43, suggesting a relatively recent animal-to-human transmission11,19. In the case of human SARS-CoV, recent studies identified several SARS-like bat CoVs with over 95% genomic sequence identity, suggesting bats as the potential zoonotic reservoir20,21,22. While dromedary camels are suspected to be either reservoir or vector for MERS, as genomic sequence of isolated dromedary MERS-CoV was found identical to that of human MERS-CoV23,24,25.
Human coronavirus NL63 (HCoV-NL63) was first isolated in 2004 from a 7-month-old child suffering from bronchiolitis and conjunctivitis in the Netherlands16. HCoV-NL63 has been documented to circulate in human population worldwide26,27,28,29,30,31,32,33 and is considered the causative pathogen for up to 10% of all respiratory illnesses34,35,36,37. Infected patients are typically young children with upper and lower respiratory tract infection, presenting with symptoms including croup, bronchiolitis and pneumonia38,39. Nevertheless, infections in adults have also been reported, though consequences could be more severe in those with compromised immune system or other comorbidities40,41,42. Similar to SARS-CoV, HCoV-NL63 also uses angiotensin-converting enzyme 2 (ACE2) as the receptor for cellular entry43. Full genome sequences of HCoV-NL63 have been determined, revealing a mosaic structure with multiple recombination sites which indicate that possible mutation and recombination could occur when co-infected with other CoVs44,45. Based on molecular clock analysis, HCoV-NL63 shares common ancestry with bat alphacoronavirus sequences, with probable divergence 563–822 years ago46. However, the direct bat ancestor of HCoV-NL63 has not been found yet47.
Currently there are no approved antiviral drugs or vaccines against human CoV infection, though several compounds have been investigated in pre-clinical studies9. From a public health perspective, no effective antiviral strategy is available in face of future CoV emergence, potentially transmitted from the vast and mutable zoonotic reservoir. Previously, we have demonstrated that main protease (Mpro) is a conserved drug target throughout the subfamily Coronavirinae, which is suitable for designing wide-spectrum inhibitors48,49. The 5′ two-thirds of coronaviral genome is consisted of open reading frame 1 (ORF1), which encodes two large polypeptides of the replicase machinery: pp1a and through ribosomal frameshift, pp1ab18. These two polypeptides are cotranslationally cleaved into mature nonstructural proteins (Nsps) through two proteases encoded in the 5′ region of ORF1: papain-like protease (PLP) and 3C-like protease (3CL or Nsp5)50,51. 3CL protease is more commonly known as Mpro because of its dominant role in the posttranslational processing of the replicase polyprotein. The Mpros from different human and animal CoVs are known to share significant homology in both primary amino acid sequence and 3D architecture, providing a strong structural basis for designing wide-spectrum anti-CoV inhibitors48,49,52,53,54,55. They employ a similar substrate-binding pocket, usually with a requirement for glutamine at P1 position and a preference for leucine/methionine at P2 position. Interestingly, in contrast to other HCoVs, only HCoV-NL63 and HCoV-HKU1 exhibit a unique substrate preference of histidine in P1 position at the cleavage site between nsp13 and nsp1437,53. The structural and pharmaceutical significance of the P1 position preference of HCoV-NL63 Mpro remains to be addressed.
Here, we report the crystal structure of HCoV-NL63 Mpro in complex with a synthetic peptidomimetic inhibitor, N3. Structural analysis reveals relative conservation at the P1 pocket. Through comparison with Mpros from other CoVs, we provide structural insight into rational drug design at a conserved target across pathological human coronaviruses and their related zoonotic counterparts.
Results
Structural Overview
There are two protein molecules in an asymmetric unit. The two molecules form a typical homodimer (Fig. 1a), which has been observed in the crystal structures of other CoV Mpros48,52,53. Previous studies have demonstrated the existence of Mpro homodimer in solution which is also the only active form of the enzyme56,57,58,59, supporting the physiological relevance of structural findings. A structural comparison of protomer A in Mpro-N3 complex with that in apo enzyme (PDB ID: 3TLO; C. P. Chuck & K. B. Wong, unpublished work) revealed an overall architecture of three domains (Fig. 1b) in each protomer, a common feature among CoV Mpro structures. Domain I (residues 8–100) and domain II (residues 101–183) together form a chymotrypsin-like fold and the substrate-binding site is located in a cleft formed between domain I and domain II. The catalytic dyad composed of Cys144 and His41 lies in the center of substrate-binding site. Domain III (residues 200–303) of HCoV-NL63 Mpro is composed of a globular antiparallel α-helical cluster, a unique feature of CoV Mpro that is required for homodimer formation. Domain III is connected to domain II through a long loop region of 16 residues. X-ray data-processing and refinement statistics are included in Table 1.
Structural overview of HCoV-NL63 Mpro.
(a) Overview of homodimer in one asymmetric unit (A: slate and B: deep salmon). Protomers are shown in cartoons and N3 inhibitors are shown as green sticks. (b) Structural alignment of protomer A in Mpro-N3 (slate) complex with that in apo enzyme (light orange, PDB ID: 3TLO; C. P. Chuck & K. B. Wong, unpublished work). The backbone is shown in cartoons and N3 inhibitor is presented as green sticks.
Michael acceptor and inhibitor binding at active site
Michael acceptor inhibitors such as N3 (Fig. 2a) undergoes mechanism-based inhibition to achieve covalent irreversible inactivation, as shown in the equation (1):
Structure of inhibitor N3 and its interaction with HCoV-NL63 Mpro.
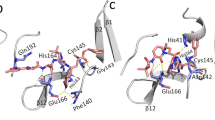
(a) The structure of compound N3. (b) A stereo view of N3 bound to the substrate-binding pocket of HCoV-NL63 Mpro at 2.85 Å. N3 inhibitor is shown in green and covered by an omit map contoured at 1.0 σ. Residues forming the substrate-binding pocket are shown in silver. (c) Detailed view of the interaction between N3 and HCoV-NL63 Mpro. The N3 inhibitor is shown in green. Hydrogen bonds are shown as dashed lines labeled with interaction distances. The covalent bond between N3 and Sγ of Cys144 is labeled in red.

The inhibitor first forms a reversible complex (EI) with the enzyme under the equilibrium-binding constant Ki. It then undergoes nucleophilic attack by the active site Cys of the enzyme, leading to the formation of a stable covalent bond (E-I). This step is governed by the inactivation rate constant, k3. Using a CoV consensus substrate reported previously48,49,53, we first determined the values for Km and kcat of the apo enzyme of HCoV-NL63 Mpro, as 50.8 ± 3.4 μM and 0.098 ± 0.004 s−1, respectively (Table 2). Cross comparison with Mpros from other human CoVs revealed that the kinetic parameters of HCoV-NL63 Mpro is relatively close to those for SARS-CoV (Km = 129 ± 7 μM, kcat = 0.14 ± 0.01 s−1)49. Rather Km of HCoV-NL63 is higher than that for HCoV-229E, while its kcat is approximately ten fold larger than those for HCoV-229E and HCoV-HKU1. We then added inhibitor N3 to the kinetic assay of HCoV-NL63 Mpro and calculated the Ki and k3 as 11.3 ± 1.0 μM and 42.4 ± 5.0 (10−3∙s−1), respectively (Table 2). Although the ki for HCoV-NL63 is higher compared with those for SARS-CoV and HCoV-229E, indicating a lower affinity of inhibitor N3 to the apo enzyme, its k3 is significantly larger, which strongly supports the ability of N3 to achieve mechanism-based irreversible inhibition against HCoV-NL63 Mpro.
Analysis of the complex structure of HCoV-NL63 Mpro bound to inhibitor N3 provides further insight into the inhibition mechanism (Fig. 2b). Since N3 binds to Protomer A and B similarly, we will only look into the binding mode of N3 in Protomer A below. Cβ of vinyl group on inhibitor N3 forms a standard 1.8 Å covalent bond with the Sγ atom of Cys144, as evidenced by clear electron density (Fig. 2b). This indicates that a Michael addition reaction occurred between N3 and the catalytic site Cys144. Compared with the apo enzyme, only one specific conformation is observed due to the formation of covalent bond while there exists a double conformation for Sγ in the apo enzyme. The carbonyl oxygen of ester on N3 is very close to the backbone NH of Gly142 and the backbone of NH of Cys144 (Fig. 2c), which mimics the tetrahedral oxyanion intermediate state formed during serine protease cleavage and provides additional support to anchor the Michael acceptor. The benzyl ester part of N3 further extends into the S1’ pocket, forming Van der Waals interaction with Val26 and Leu27 (Fig. 2b).
Comparison between the molecular surfaces of HCoV-NL63 Mpro complexed with N3 and the apo enzyme reveals that the pocket accommodating N3 undergoes significant conformation changes upon inhibitor binding (Fig. 3a,b) as following: (1) the imidazole group of His41 swifts ~5 Å towards the hydrophobic core of the protein to better accommodate P1’ site; (2) the main chain of residues 138–142 which constitute the outer wall of S1 subsite moves toward the lactam ring, causing the shrinking of S1 pocket; (3) residues 45–51 flip over to act as a lid to cover P2 subsite; (4) residues 164–168 and 187–191 extend into opposite directions to host N3. In the following sections, we will describe the detailed structural features of the subpockets.
Surface representation of native HCoV-NL63 and its complex with inhibitor N3.
(a) Surface representation of substrate-binding pockets from apo enzyme of HCoV-NL63 Mpro (marine, PDB ID: 3TLO). The S1, S2, S4 and S1’ pockets and residues forming the substrate-binding pocket are labeled. (b) Surface representation of HCoV-NL63 Mpro (marine) in complex with N3 inhibitor (green). Water molecules are shown as red spheres. The P1-P5 and P1’ groups of N3 inhibitor are labeled together with residues forming the substrate-binding pocket.
Smaller S1 pocket to accommodate P1 histidine
Coronaviral Mpros are known to have strong preference to glutamine (Q) at P1 site of substrate. Genome sequence analysis of HCoV-NL63 revealed that 10 out of 11 Mpro cleavage sites bear glutamine at P1 position, except that the recognition site between nsp13 (helicase) and nsp14 (exonuclease) where histidine replaces glutamine at P1 site37. This feature is unique only in genomes of HCoV-NL63 (NC_005831) and HCoV-HKU1 (NC_006577). In contrast, glutamine is the exclusive residue in P1 among all cleavage sites for the other four human CoVs, including HCoV-229E (NC_002645), HCoV-OC43 (NC_005147), SARS-CoV (NC_004718) and MERS-CoV (JX869059). Such novel substrate specificity at P1 in HCoV-NL63 and HCoV-HKU1 implies unique structural feature of S1 subpocket. The crystal structure of HCoV-NL63 Mpro shows that the size of its S1 pocket is comparable to that of HCoV-HKU1, but smaller than those of HCoV-229E and SARS-CoV. Clearly, a smaller S1 pocket in HCoV-NL63 could better accommodate the smaller side chain of histidine residue at P1 of nsp13-nsp14 and facilitate cleavage when weakened oxyanion hole is formed.
In our enzyme-inhibitor complex structure, the lactam ring of N3 molecule serves as a structural analog to glutamine or histidine at P1 position (Fig. 2a) and protrudes into the S1 pocket by forming a 2.5 Å hydrogen bond between the lactam oxygen and the imidazole ring NH of His163 (Fig. 2b,c), which is similar to those in the complex structures of SARS-CoV Mpro and HCoV-HKU1 Mpro with inhibitor N349,53. Furthermore, the NH of N3 lactam ring forms two hydrogen bonds with Oε1 of Glu166 (3.1 Å) and the carbonyl oxygen atom of Phe139 (3.2 Å) respectively, which provide additional support to stabilize the lactam ring in S1 pocket. The backbone NH of P1 residue on N3 also forms a 3.0 Å hydrogen bond with the backbone carbonyl oxygen from Gln164, favorably accommodating the S1 pocket.
S2 pocket
The P2 position of natural Mpro cleavage sites across of the genomes of six human CoVs usually prefers a hydrophobic residue (Table 3). It is surprising that the P2 residues are completely identical among all 11 cleavage sites between the two alphacoronaviruses, HCoV-NL63 and HCoV-229E. In a majority of cases, it is leucine residue occupying the P2 position, with valine at nsp6-nsp7 and isoleucine at nsp10-nsp11. On the other hand, more variations are observed in the P2 position among betacoronaviruses, with more alternative residues such as methionine, phenylalanine and proline, which might indicate less stringency of P2 specificity. The P2 site of inhibitor N3 mimics the side chain of leucine, in order to cover the maximum spectrum of natural Mpro substrates for human CoVs49. As observed in the HCoV-NL63 structure with N3, the aliphatic isobutyl side chain of P2 protrudes into the deep S2 pocket via interactions with the alkyl portion of the side chains of Asp187, Pro189 and is well accommodated onto the Van der Waals surface of the pocket (Fig. 2b).
To determine the structural diversity in S2 pocket, we then superimposed the backbones of four know Mpro structures from human CoVs. The lid of the S2 pocket in HCoV-NL63 is covered by a tight loop of residues 45–51 (Fig. 4). Interestingly, the same region in SARS-CoV and HCoV-HKU1 adopts a secondary structure of 310 helix to maintain S2 pocket53. Such difference might partially account for the increased variation of natural P2 residues as observed in the genomes of betacoronaviruses.
S1 and S2 binding sites in HCoV-NL63 Mpro.
The main chains of four human CoV Mpro structures (HCoV-NL63: slate, HCoV-229E: cyan, SARS-CoV: magenta and HCoV-HKU1: yellow) are superimposed and displayed in the neightborhood of the substrate-binding site. The S1, S2 and S4 binding sites are labeled. The backbones are represented in worm form and inhibitor is shown in stick format of green color. Residues 45–51 are marked with a red oval (in dash line).
P3, P4 and P5 positions
The P3 side chains of N3 inhibitor are both solvent exposed in the two protomers of HCoV-NL63 Mpro (Figs 2b and 3b). This is consistent with the fact that no specificity for any particular side chains exists at the P3 position of cleavage sites among CoV Mpros55. Further, the NH and carbonyl oxygen of P3 backbone form two hydrogen bonds with the backbone carbonyl oxygen and NH of Glu166 respectively, which help anchor the N3 inhibitor to the HCoV-NL63 Mpro at P3 location.
The P4 position of the inhibitor N3 is alanine and its side chain readily inserts into the relatively shallow P4 pocket (Fig. 3b), forming hydrophobic interactions with Pro189. Also the backbone NH of alanine residue on N3 donates a 2.9-Å hydrogen bond to the carbonyl oxygen of Ser190 (Fig. 2c). The isoxazole at P5 makes Van der Waals interactions with Gly168 and the backbone of residues Leu191 (Fig. 2b). Overall, the pattern of interactions between N3 and HCoV-NL63 Mpro at P3, P4 and P5 positions is similar to that between N3 and SARS-CoV Mpro 49.
Structural conservation of Mpro among clinical HCoV-NL63 isolates
So far, whole genomic sequences of as many as 30 strains of HCoV-NL63 have been deposited into NCBI database16,44,45,60,61. These strains were isolated from patient specimens dated back to the past three decades from several countries, including The Netherlands, United States and China. High percentage of sequence variation among these clinically isolated HCoV-NL63 strains and evidence of in vivo recombination during co-infections with other CoVs have been documented, especially in the N-terminal domain of spike protein and nsp2/nsp3 region44,60. In order to determine the efficacy of inhibitor N3 against sequence variation accumulated during the circulation of HCoV-NL63 in human population, we assessed the level of sequence and structural conservation in the molecular target of this inhibitor among different clinical isolates. Sequence for Mpro (nsp5) was retrieved from these HCoV-NL63 strains with available genomic sequences (Supplementary Table S1) and alignment was performed to determine sequence variations among these clinical isolates. Mpro sequence from strain Amsterdam I (NC_005831) was used as reference16,44. Overall, Mpro is extremely conserved among clinical HCoV-NL63 strains isolated worldwide (Supplementary Fig. S1), evidencing the enzyme’s essential role in viral replication. Sequence variation is only observed in 3 residues: His69 is mutated to Tyr in 13 strains isolated from United States, Cys221 is mutated to Arg among all the strains that have not been cultured in vitro and Met235 is mutated to Ile in only one strain isolated in United States (Fig. 5a). When plotted onto the structure of HCoV-NL63 Mpro, these 3 residues are all located in flexible regions, such as loop and molecular surface (Fig. 5b). None of the 3 residues are directly interacting with inhibitor N3, which provides clear structural evidence to support the effectiveness of N3 against all the clinical strains of HCoV-NL63, in spite of significant genomic sequence variation and potential for recombination in viral transmission.
Structural conservation of Mpro among clinical HCoV-NL63 isolates.
(a) Summary of the 3 residues of variation among a total of 30 HCoV-NL63 strains. Amsterdam I is used as reference strain. Number of strains carrying one particular residue is labeled in bracket. (b) Three-dimensional representation of the 3 nonconserved residues from (a) mapped onto HCoV-NL63 Mpro complex structure with inhibitor N3. Conserved residues are colored light orange, the 3 residues (His69, Cys221 and Met235) are colored cyan and inhibitor N3 is colored green.
Inhibitor N3 targets a conserved site among human CoVs and their related zoonotic counterparts
The substrate-binding site of Mpro has been shown as a conserved drug target for designing wide spectrum anti-CoV inhibitors48,49,51,53. Given the evidence of repetitive global epidemics (such as SARS and MERS) caused by zoonotic transmission11,19,20,21,22,23,24, it is imperative to examine whether the substrate-binding site and the inhibition mechanism employed by inhibitor N3 are conserved between known human CoVs and their related zoonotic CoVs. We chose three most representative pairs of human CoVs and their zoonotic counterparts: HCoV-229E (NC_002645) and bovine coronavirus (BCoV, NC_003045), SARS-CoV (NC_004718) and SARS-related CoV isolated from bat (BtSARSr-CoV, KC881006), MERS-CoV (JX869059) and dromedary camel MERS-CoV (DcMERS-CoV, KJ713296). We then retrieved their Mpro sequences from NCBI database. The direct bat ancestor of HCoV-NL63 has not been identified47, therefore we only use HCoV-NL63 Mpro sequence and its secondary structure presented in this study as reference. Sequence alignment, shown in Fig. 6a, demonstrates significant homology in primary amino acid sequence among these 7 CoVs.
Inhibitor N3 targets a conserved site among human and related zoonotic CoVs.
(a) Sequence alignment of three pairs of human CoVs and their related zoonotic counterparts: HCoV-229E and bovine coronavirus (BCoV), SARS-CoV and SARS-related coronavirus isolated from bat (BtSARSr-CoV), MERS-CoV and dromedary camel MERS-CoV (DcMERS-CoV). HCoV-NL63 and its secondary structure are used as reference for the alignment. Sequence alignment was performed using ClustalW272 and figure was generated using ESPript 3.073. (b) Surface representation of conserved substrate-binding pockets from 7 CoV Mpros listed in (a). Background is HCoV-NL63 Mpro. Red: identical residues among all seven CoV Mpro; magenta: substitution in one CoV Mpro; orange: substitution in two CoV Mpros. The residues forming the substrate-binding pocket are labeled. S1’: Leu27, Gly142; S1: Phe139, His163, Glu166, His172; S2: His41 Tyr53, Asp187; S4: Leu167, Gln192.
A closer examination at the substrate-binding pocket further reveals a conservative 3D architecture at this drug target (Fig. 6b). Those residues, which are critical for pocket formation, such as Leu27, Gly142 for pocket S1’; Phe139, His163, Glu166, His172 for pocket S1; His41, Tyr53, Asp187 for pocket S2; Leu167, Gln192 for pocket S4, are strictly conserved among various CoVs (Fig. 6a,b). Peptidomimetic inhibitor N3, through its Michael acceptor and well-designed side chains, snugly fits into the conserved substrate-binding pocket. The binding of N3 establishes a concerted interaction network, including a 1.8-Å covalent bond between Cβ of vinyl group on inhibitor N3 and the Sγ atom of Cys144, 7 hydrogen bonds and extensive hydrophobic interactions between N3 and the above residues critical for interplay. These findings demonstrate that inhibitor N3 could exert inhibitory effect towards a conserved site in CoVs both before and after zoonotic transmission. Therefore, in addition to its role in inhibiting known circulating human CoV species, inhibitor N3 might also serve as a lead compound for preclinical and clinical testing against potential future epidemic caused by CoV emerging from zoonotic origin.
Discussion
The world has experienced two global outbreaks of CoV infections since entering the 21st century5,6,7,8,9,11,12,13. Both SARS-CoV and MERS-CoV cause severe respiratory syndrome with high mortality rate10,14,15. In addition, four more human CoVs, namely HCoV-NL63, HCoV-229E, HCoV-HKU1 and HCoV-OC43, have been identified as pathological agents for common cold9,16,17. The lack of effective therapeutic and preventive strategies against human CoVs calls for immediate action of the scientific community9,50. We previously demonstrated that designing wide spectrum inhibitor at a conservative target is a viable method to develop anti-CoV therapeutics, given the high mutation and recombination rates observed in viral replication48,49,51,53. The ability of CoV to cross animal-human boundary provides further support to our strategy. Indeed, several human CoVs, including HCoV-NL63, SARS-CoV and MERS-CoV, have been linked to zoonotic CoVs which naturally infect hosts such as bats or dromedary camels20,21,22,23,24,25,46. In current study, we use the Mpro from HCoV-NL63 as our model molecule and present its crystal structure in complex with an inhibitor. Based on the structural detail at 2.85 Å resolution, the substrate-binding pocket of HCoV-NL63 Mpro is conserved among three pairs of human and zoonotic CoVs (Fig. 6). Several key residues at the subsites for substrate binding are completely identical among these seven coronaviruses, for example Phe139, His163, Glu166, His172 of S1; His41, Tyr53, Asp187 of S2; and Leu167, Gln192 of S4. These findings are consistent with the role of Mpro in viral replication, which is essential to the proteolytic processing and maturation of replicase polyprotein (encoded by ORF1). Analysis of substrate recognition sequence, based on available whole genome sequences of all six human CoVs, provides additional evidence to the conservation of Mpro substrate-binding pocket: the P1 position requires almost exclusively glutamine and the P2 position exhibits strong preference for hydrophobic residues such as Leu and Val (Table 3). Taken together, through designing of wide spectrum inhibitors at a conservative site on Mpro, our study outlines a novel therapeutic approach of containing diseases caused by both existing and possible future emerging human CoVs.
A close examination of the interaction between HCoV-NL63 Mpro and inhibitor N3 reveals structural details of inhibition mechanism. N3 is a synthetic peptidomimetic compound with Michael acceptor (Fig. 2a), which achieves mechanism-based enzyme inactivation through forming an irreversible covalent bond with Cys144 of catalytic dyad (Fig. 2b,c). The success of two serine protease inhibitors, telaprevir and boceprevir, in the treatment of hepatitis C virus (HCV) has underscored the importance of covalent inhibitors for targeting viral proteases62. Both telaprevir and boceprevir are peptidomimetic inhibitors carry a warhead of α-ketoamide, which forms a covalent yet reversible bond with catalytic triad serine residue of HCV NS3∙4 A protease62,63. Michael acceptor has also been used as a warhead in pharmaceutical targeting against viral protease, for example rupintrivir was developed as an inhibitor for 3C protease of human rhinovirus and enterovirus50,64,65. The backbone of N3 forms 7 hydrogen bonds with residues in the substrate-binding pocket (Fig. 2c). The pockets accommodating N3 undergo gate-regulated switch to facilitate the binding of inhibitor N3 (Fig. 3), an interesting phenomena initially observed in case of Mpro from SARS-CoV49.
In addition, the structure of HCoV-NL63 Mpro exhibits several unique but interesting features. Examination of its natural cleavage sites based on HCoV-NL63 genomic sequence reveals an unusual histidine P1 residue between nsp13 and nsp14, which is unique to HCoV-NL63 and HCoV-HKU1 among all human CoVs37. Such P1 anomaly is partially accommodated by the relatively smaller S1 pocket, as the size of its S1 pocket is comparable to that of HCoV-HKU1, but smaller than those of HCoV-229E and SARS-CoV. The P2 position seems to present genus specificity among alphacoronaviruses, as the P2 residues among all 11 natural cleavage sites are completely identical between HCoV-NL63 and HCoV-229E with a strong dominance of leucine. While large variation on P2 residue is observed in the genomes of human betacoronaviruses.
In summary, the crystal structure of HCoV-NL63 Mpro complexed with inhibitor N3 has provided critical insight into the design of irreversible inhibitor carrying a Michael acceptor warhead. Through detailed sequence and structural comparison, this compound demonstrates feasibility as potential broad spectrum therapeutic agent for both existing and possibly emerging human CoVs. Further pharmaceutical development of such covalent peptidomimetic inhibitors would yield success in clinical management of human coronavirus diseases and public health preparedness for possible future pandemic.
Methods
Protein expression and purification
Protein expression and purification of HCoV-NL63 main protease has been described previously66. The coding sequence was subcloned into pGEX-6P-1 vector and transformed into BL21 (DE3) E. coli cells. Cell culture was first grown in LB medium containing 100 μg ml−1 ampicillin at 37 °C until optical density (OD600) reached 0.6. Protein expression was then induced by adding isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.5 mM and further cultured at 16 °C for 16 hours. Cell pellets were harvested by centrifugation and resuspended in phosphate-buffered saline solution supplemented with 1 mM dithiothreitol (DTT) and 10% glycerol. Cell lysate was prepared using sonication and centrifugation (12,000 g, 50 min, 4 °C). GST-tagged HCoV-NL63 Mpro fusion protein was bound to glutathione sepharose 4B affinity resin and GST tag was removed through on-column cleavage using commercial PreScission protease (GE Healthcare) at 4 °C for 18 hours. Recombinant HCoV-NL63 Mpro protein was subject to an additional step of anion-exchange chromatography using HiTrap Q column (GE Healthcare) and was eluted with a linear gradient of 25 to 250 mM NaCl (20 mM Tris-HCl pH = 8.0, 10% glycerol, 1 mM DTT).
Crystallization and data collection
Purified protein was supplemented with 10% DMSO and concentrated to 1 mg ml−1. Crystals of HCoV-NL63 Mpro in complex with inhibitor N3 were produced by cocrystallization. Inhibitor N3 was added to HCoV Mpro protein at a molar ratio between 3:1 and 5:1 and incubated at 4 °C for 4 hours. The complex was then centrifuged at 12,000 g for 10 min and concentrated to 10 mg ml−1 in a buffer containing 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT. Using hanging-drop vapor diffusion method at 16 °C, best crystals were obtained after 2 days using a reservoir solution containing 0.1 M HEPES pH 5.5, 10%(w/v) polyethylene glycol 8000, 4%(v/v) ethylene glycol (PDB entry 3TLO) and 0.1 M sodium citrate tribasic dehydrate pH 5.6, 1.0 M ammonium phosphate monobasic in a ratio of 80:2067.
Data for HCoV-NL63 Mpro-N3 complex was collected to a 2.85-Å resolution at 100 K on beamline 1W2B of Beijing Synchrotron Radiation Facility (BSRF), using a MAR165 charge-coupled device detector. The cryoprotectant solution contained 20% (v/v) glycerol, 10% (w/v) polyethylene glycol 8000, 4% (v/v) ethylene glycol and 0.1 M HEPES pH 5.5. All data integration and scaling were performed using HKL200068. The Matthews coefficient of the crystal suggested two molecules per asymmetric unit and the solvent content was 59.8%.
Structure determination and analysis
The structure of HCoV-NL63 Mpro-N3 complex was determined using molecular replacement from that of apo-form HCoV-NL63 Mpro (PDB ID: 3TLO; C. P. Chuck & K. B. Wong, unpublished work). All cross-rotation and translation searches for molecular replacement were performed with Phaser69. Cycles of manual adjustment using Coot70 and subsequent refinement using PHENIX71 led to a final model with a crystallographic R factor (Rcryst) of 19.4% and a free R factor (Rfree) of 24.1% at 2.85-Å resolution.
Enzymatic activity and inhibition assays
Enzymatic assay was performed using a fluorogenic substrate with consensus sequence of CoV Mpro, MCA-AVLQSGFR-Lys(Dnp)-Lys-NH2 (>95% purity, GL Biochem Shanghai Ltd., Shanghai, China), as previously reported48,49,53. Fluorescence intensity was monitored using a Fluoroskan Ascent instrument (Thermo Scientific, USA) with excitation and emission wavelengths of 320 nm and 405 nm, respectively. The assay was performed in a buffer solution consisted of 50 mM Tris-HCl (pH 7.3) and 1 mM EDTA at 30 °C. Kinetic parameters, including Km and kcat of apo HCoV-NL63 Mpro and Ki and k3 of inhibitor N3, were determined using methods described in detail in our previous work49.
Additional Information
How to cite this article: Wang, F. et al. Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design. Sci. Rep. 6, 22677; doi: 10.1038/srep22677 (2016).
References
Weiss, S. R. & Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69, 635–664 (2005).
Masters, P. S. The molecular biology of coronaviruses. Adv Virus Res 66, 193–292 (2006).
Siddell, S., Wege, H. & Ter Meulen, V. The biology of coronaviruses. J Gen Virol 64 (Pt 4), 761–776 (1983).
Weiss, S. R. & Leibowitz, J. L. Coronavirus pathogenesis. Adv Virus Res 81, 85–164 (2011).
Peiris, J. S. et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325 (2003).
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348, 1967–1976 (2003).
Ksiazek, T. G. et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348, 1953–1966 (2003).
Kuiken, T. et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362, 263–270 (2003).
Graham, R. L., Donaldson, E. F. & Baric, R. S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11, 836–848 (2013).
Peiris, J. S., Guan, Y. & Yuen, K. Y. Severe acute respiratory syndrome. Nat Med 10, S88–97 (2004).
Coleman, C. M. & Frieman, M. B. Coronaviruses: important emerging human pathogens. J Virol 88, 5209–5212 (2014).
de Groot, R. J. et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 87, 7790–7792 (2013).
Raj, V. S., Osterhaus, A. D., Fouchier, R. A. & Haagmans, B. L. MERS: emergence of a novel human coronavirus. Curr Opin Virol 5, 58–62 (2014).
Memish, Z. A., Zumla, A. I., Al-Hakeem, R. F., Al-Rabeeah, A. A. & Stephens, G. M. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med 368, 2487–2494 (2013).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367, 1814–1820 (2012).
van der Hoek, L. et al. Identification of a new human coronavirus. Nat Med 10, 368–373 (2004).
Woo, P. C. et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79, 884–895 (2005).
Woo, P. C., Huang, Y., Lau, S. K. & Yuen, K. Y. Coronavirus genomics and bioinformatics analysis. Viruses 2, 1804–1820 (2010).
Vijgen, L. et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79, 1595–1604 (2005).
Drexler, J. F. et al. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol 84, 11336–11349 (2010).
Ge, X. Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013).
He, B. et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J Virol 88, 7070–7082 (2014).
Briese, T. et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio 5, e01146–01114 (2014).
Alagaili, A. N. et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio 5, e00884–00814 (2014).
Azhar, E. I. et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370, 2499–2505 (2014).
Leung, T. F. et al. Epidemiology and clinical presentations of human coronavirus NL63 infections in hong kong children. J Clin Microbiol 47, 3486–3492 (2009).
Zhou, W., Wang, W., Wang, H., Lu, R. & Tan, W. First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect Dis 13, 433 (2013).
Owusu, M. et al. Human coronaviruses associated with upper respiratory tract infections in three rural areas of Ghana. PLoS One 9, e99782 (2014).
Matoba, Y. et al. Detection of the human coronavirus 229E, HKU1, NL63 and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn J Infect Dis 68, 138–141 (2014).
Vabret, A. et al. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J Paediatr Child Health 44, 176–181 (2008).
Suzuki, A. et al. Respiratory viruses from hospitalized children with severe pneumonia in the Philippines. BMC Infect Dis 12, 267 (2012).
Bastien, N. et al. Human coronavirus NL63 infection in Canada. J Infect Dis 191, 503–506 (2005).
Minosse, C. et al. Phylogenetic analysis of human coronavirus NL63 circulating in Italy. J Clin Virol 43, 114–119 (2008).
Forster, J. et al. Prospective population-based study of viral lower respiratory tract infections in children under 3 years of age (the PRI.DE study). Eur J Pediatr 163, 709–716 (2004).
Konig, B. et al. Prospective study of human metapneumovirus infection in children less than 3 years of age. J Clin Microbiol 42, 4632–4635 (2004).
van der Hoek, L. et al. Croup is associated with the novel coronavirus NL63. PLoS Med 2, e240 (2005).
Pyrc, K., Berkhout, B. & van der Hoek, L. The novel human coronaviruses NL63 and HKU1. J Virol 81, 3051–3057 (2007).
Fielding, B. C. Human coronavirus NL63: a clinically important virus? Future Microbiol 6, 153–159 (2011).
Lee, J. & Storch, G. A. Characterization of human coronavirus OC43 and human coronavirus NL63 infections among hospitalized children <5 years of age. Pediatr Infect Dis J 33, 814–820 (2014).
Yu, X. et al. Etiology and clinical characterization of respiratory virus infections in adult patients attending an emergency department in Beijing. PLoS One 7, e32174 (2012).
Hoek, R. A. et al. Incidence of viral respiratory pathogens causing exacerbations in adult cystic fibrosis patients. Scand J Infect Dis 45, 65–69 (2013).
Oosterhof, L., Christensen, C. B. & Sengelov, H. Fatal lower respiratory tract disease with human corona virus NL63 in an adult haematopoietic cell transplant recipient. Bone Marrow Transplant 45, 1115–1116 (2010).
Hofmann, H. et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102, 7988–7993 (2005).
Pyrc, K. et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 364, 964–973 (2006).
Geng, H. et al. Characterization and complete genome sequence of human coronavirus NL63 isolated in China. J Virol 86, 9546–9547 (2012).
Huynh, J. et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86, 12816–12825 (2012).
Drexler, J. F., Corman, V. M. & Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res 101, 45–56 (2014).
Xue, X. et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol 82, 2515–2527 (2008).
Yang, H. et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 3, e324 (2005).
Hilgenfeld, R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J 281, 4085–4096 (2014).
Yang, H., Bartlam, M. & Rao, Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Curr Pharm Des 12, 4573–4590 (2006).
Yang, H. et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100, 13190–13195 (2003).
Zhao, Q. et al. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J Virol 82, 8647–8655 (2008).
Anand, K. et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J 21, 3213–3224 (2002).
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R. & Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300, 1763–1767 (2003).
Graziano, V., McGrath, W. J., Yang, L. & Mangel, W. F. SARS CoV main proteinase: The monomer-dimer equilibrium dissociation constant. Biochemistry 45, 14632–14641 (2006).
Cheng, S. C., Chang, G. G. & Chou, C. Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys J 98, 1327–1336 (2010).
Shi, J., Sivaraman, J. & Song, J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J Virol 82, 4620–4629 (2008).
Tomar, S. et al. Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS. J Biol Chem 290, 19403–19422 (2015).
Dominguez, S. R. et al. Genomic analysis of 16 Colorado human NL63 coronaviruses identifies a new genotype, high sequence diversity in the N-terminal domain of the spike gene and evidence of recombination. J Gen Virol 93, 2387–2398 (2012).
Lednicky, J. A. et al. Isolation and genetic characterization of human coronavirus NL63 in primary human renal proximal tubular epithelial cells obtained from a commercial supplier and confirmation of its replication in two different types of human primary kidney cells. Virol J 10, 213 (2013).
Manns, M. P. & von Hahn, T. Novel therapies for hepatitis C - one pill fits all? Nat Rev Drug Discov 12, 595–610 (2013).
Lin, C., Kwong, A. D. & Perni, R. B. Discovery and development of VX-950, a novel, covalent and reversible inhibitor of hepatitis C virus NS3.4A serine protease. Infect Disord Drug Targets 6, 3–16 (2006).
Matthews, D. A. et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc Natl Acad Sci USA 96, 11000–11007 (1999).
Wang, J. et al. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J Virol 85, 10021–10030 (2011).
Wang, F. et al. Crystallization and preliminary crystallographic study of human coronavirus NL63 main protease in complex with an inhibitor. Acta Crystallogr F Struct Biol Commun 70, 1068–1071 (2014).
Birtley, J. R. & Curry, S. Crystallization of foot-and-mouth disease virus 3C protease: surface mutagenesis and a novel crystal-optimization strategy. Acta Crystallogr D Biol Crystallogr 61, 646–650 (2005).
Otwinowski, Z. & Minor, W. in Macromolecular Crystallography, part A Vol. 276 (eds C. W. Carter, Jr . & R. M. Sweet ) 307–326 (Academic Press, 1997).
McCoy, A. J. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324 (2014).
Acknowledgements
We thank Zengqiang Gao and Tianyi Zhang at beamline 1W2B of Beijing Synchrotron Radiation Facility (BSRF) for their technical support on data collection. This work was supported by National Key Basic Research Program of China (973 program) (No. 2015CB859800), National Natural Science Foundation of China (No. 31300150), Tianjin Marine Science and Technology Program (No. KJXH2014-16), Megapoject on Infectious Disease Control supported by the Ministry of Health of China (2013ZX10004601).
Author information
Authors and Affiliations
Contributions
F.W. and C.C. performed the experiments. F.W., C.C., W.T., K.Y. and H.Y. analyzed the results. K.Y. and H.Y. wrote the manuscript with contributions from the other authors. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, F., Chen, C., Tan, W. et al. Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design. Sci Rep 6, 22677 (2016). https://doi.org/10.1038/srep22677
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22677
This article is cited by
-
Review on development of potential inhibitors of SARS-CoV-2 main protease (MPro)
Future Journal of Pharmaceutical Sciences (2022)
-
Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332
Protein & Cell (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.