Abstract
Ehrlich and demethiolation pathways as two competing branches converted amino acid into alcohols. Controlling both pathways offers considerable potential for industrial applications including alcohols overproduction, flavor-quality control and developing new flavors. While how to regulate ehrlich and demethiolation pathways is still not applicable. Taking the conversion of methionine into methionol and methanethiol for example, we constructed two suppression subtractive cDNA libraries of Clonostachys rosea by using suppression subtractive hybridization (SSH) technology for screening regulators controlling the conversion. E3 ubiquitin-protein ligase gene HUWE1 screened from forward SSH library was validated to be related with the biosynthesis of end products. Overexpressing HUWE1 in C. rosea and S. cerevisiae significantly increased the biosynthesis of methanethiol and its derivatives in demethiolation pathway, while suppressed the biosynthesis of methional and methionol in ehrlich pathway. These results attained the directional regulation of both pathways by overexpressing HUWE1. Thus, HUWE1 has potential to be a key target for controlling and enhancing alcohols production by metabolic engineering.
Similar content being viewed by others
Introduction
Ehrlich and demethiolation pathways, two major branches for catabolizing amino acid, is received much attention for both pathways significantly participated to the production of fuel and valuable flavor alcohols, the quality and uniqueness of many foodstuffs1,2,3,4,5. L-phenylalanine was catabolized into 2-phenylethanol which is widely used in perfumery and cosmetics for its rose-like odor6, valine and leucine were converted into butanol, isobutanol and 3-methyl-1-butanol as fuels via ehrlich pathway3,5. Additionally, catalyzing methionine (Met) into methional and methionol via ehrlich pathway, methanethiol (MTL) and its derivatives dimethyl sulfide (DMS), dimethyl disulfide (DMDS) and dimethyl trisulfide (DMTS) via demethiolation pathway is of prime importance in the overall flavor formation of fermented food and make a significant contribution to their typical flavors4. In order to improve the production of alcohols or control the quality of fermented food, it is necessary to cost-effectively engineer fermentation strains to fine-tune the ehrlich and demethiolation pathways.
The ubiquitin-proteasome system (UPS), regulating cellular homeostasis, was involved in numerous cellular processes including cell cycle progression, gene transcription, protein quality control and signal transduction7,8. Ubiquitination is achieved through the action of three enzyme classes: E1-ubiquitin-activating enzymes, E2-ubiquitin-conjugating enzymes and E3-ubiquitin ligases8. Of which E3-ubiquitin ligases played critical role during post-translational modifications for they polyubiquitinate their substrates with lys48-linked chains of ubiquitin and target the substrates for destruction by the proteasome9. These substrates included transcriptional factors and catabolic enzymes9. Up to now, E3-ubiquitin ligases were found to regulate numerous cellular pathways closely related with carbon and lipid catabolites, stress adaptation10,11,12,13. Therefore, ubiquitination constituents, especially E3-ubiquitin ligases, are potential targets for regulating the complex metabolic switches.
Ehrlich pathway genes including aminotransferase genes ARO8, BAT and α-ketoacid decarboxylase gene PDC and demethiolation pathway gene STR3 are highly conserved in eukaryotes. Their coordinated expression at transcriptional level controlled the degradation of amino acids14,15. The regulators performing key regulation tasks during the coordinated expression of pathway genes have potential to be targets for overproduction of metabolites by metabolic engineering16,17. Up to now, only one regulator ARO80 was found to activate the expression of ehrlich pathway genes encoding aminotransferases and α-ketoacid decarboxylases in response to aromatic amino acids18,19. Demethiolation pathway as competing branch directly impacted the biosynthesis of ehrlich pathway metabolites. Thus how to regulate the two pathways by metabolic engineering is closely related with controlling the metabolites compositions and concentrations which decided the alcohols production and food quality.
In this study, Clonostachys rosea tang 19 was selected for its ability to degrade Met into methionol and MTL via ehrlich and demethiolation pathways, respectively. To achieve rational regulation of the two pathways, we constructed a suppression subtractive cDNA library and screened differentially expressed genes especially regulator genes associated with the biosynthesis of end products. The effects of these regulators on the production of ehrlich and demethiolation pathway metabolites were analyzed in Clonostachys rosea and Saccharomyces cerevisiae. Our findings support the rational regulation of ehrlich and demethiolation pathways for enhancing alcohols production, the quality optimization and fermentation control.
Results
Differentially expressed genes associated with the conversion of Met into methionol and MTL
To enrich the differentially expressed genes associated with alcohols production, suppression subtractive hybridization PCR was adopted in this study. The optimal PCR cycle numbers were 17 for “Driver” cDNA and 14 for “Tester” cDNA. The PCR products of SMARTer cDNA were mainly distributed between 0.7 and 2.0 kb (Fig. S1A–D). Subsequently, subtractive cDNA libraries were constructed to isolate the differentially expressed fragments triggered by Met addition. A total of 2405 clones, ranging from 100 to 1500 bp for the two libraries, were obtained. The forward and reverse libraries containing 1272 clones and 1133 clones were constructed by using suppression subtractive hybridization (SSH) technology, respectively.
All PCR products ranged from 100 to 1500 bp were arrayed on 10 nylon membranes in duplicate and screened by cDNA array dot blotting. Apparent differences in hybridization signal intensities were observed when comparing the membrane probed with the forward probe to the membrane probed with the reverse subtracted probe (Fig. S2A–D), which indicated that SSH procedures for screening differentially expressed sequences were succeeded. 100 positive clones from forward and reverse libraries were sequenced and analyzed, respectively.
Bioinformatics based functional inference of differentially expressed genes
The potential functions of expressed sequence tags from the libraries were inferred based on homology by using the basic local alignment search tool (BLASTX). 91 and 96 unigenes were obtained from the forward and reverse libraries (Tables 1 and 2). To validate the efficacy for screening differently expressed genes, 4 genes were selected from the forward or reverse libraries and their expression at transcriptional level was analyzed by using real-time PCR, respectively. After Met addition, the expression of FL346 encoding E3 ubiquitin-protein ligase HUWE1, FL671 encoding aromatic amino acid aminotransferase (ARO8-2), FL666 encoding pyruvate decarboxylase (PDC) and FL819 encoding 3-isopropylmalate dehydrogenase was increased by 1.63, 13.74, 2.60 and 4.86 folds, while the expression of RL186 encoding amino-acid permease inda1, RL323 encoding aminotransferase class I and II, RL272 encoding 5-aminolevulinate synthase and RL138 encoding polyketide synthase were decreased by 83.33, 232.56, 714.29 and 769.23 folds, respectively (Fig. 1). The transcriptional expression pattern of these genes met with the results obtained from SSH, which indicated that SSH library had been successfully constructed.
Gene ontology analysis categorized the gene set into 7 and 10 functional groups (Fig. 2A,B, Tables 1 and 2), respectively. Among these, cell metabolism, transcription and protein fate were three categories closely related with the conversion of Met into methionol and MTL. In cell metabolism, ARO8-2 encoding aromatic amino acid aminotransferase and PDC encoding α-ketoacid decarboxylase were selected from forward library which may contribute to the biosynthesis of methionol via ehrlich pathway. Regulators functioned during the transcription of synthase genes which is key step for controlling methionol and MTL production (Tables 1 and 2). Transcription factor analysis revealed that 5 transcription factors including C6 (Cys6)19, F-box (mediating protein interaction, involved in cell cycle and signal transduction)20, PWWP (mediating nucleus protein interaction)21, Tar1p (related with mitochondrial gene expression and mtDNA stability)22 and Hms1p (repressing citrate biosynthesis)23 (Tables 1 and 2) were associated with the biosynthesis of methionol and MTL. Ubiquitin 3 binding protein But2, ubiquitin conjugating E2 and E3 ubiquitin-protein ligase HUWE1 were only three posttranscriptional regulators selected from protein fate group. Relative to ubiquitin 3 binding protein But2 and ubiquitin conjugating E2, E3 ubiquitin-protein ligase HUWE1 directly controlled ubiquitination process (Tables 1 and 2). E3 ubiquitin-protein ligases regulated the biosynthesis of metabolites by polyubiquitating proteins implicated in transcription, protein folding and degradation24. Thus they are ideal targets for large scale alteration of cellular metabolism by metabolic engineeing. The production of methionol and MTL is resulted from the coordinated transcriptional expression of synthase genes and may be controlled by a hierarchical transcriptional regulatory network15. In view to huge potential regulatory capability of HUWE1, HUWE1 is adopted to rationally regulate ehrlich and demethiolation pathways.
Overexpression of HUWE1 impacted the biosynthesis of pathway metabolites in C. rosea
HUWE1 screened from protein fate group of forward SSH library, may contribute to the production of end products. Thus, we inferred that overexpression of HUWE1 may further enhance the production of methionol and MTL. To overexpress HUWE1 in C. Rosea, a 4140 bp sequence located in the upstream genomic region of HUWE1 was cloned by SEFA PCR and then the full-length ORF encoding a 988 amino acid polypeptide was directly integrated into the genome of C. Rosea. Transcriptional analysis indicated that HUWE1 was overexpressed indeed in overexpression strains U1 and U2 (Fig. S3A,B). E3 ubiquitin-protein ligase HUWE1 was characterized with HECT (homologous to E6-associated protein C-terminus) which has a direct role in catalysis during ubiquitination. To further identify that the two pathways are regulated by HUWE1, the sequence encoding catalysis domain HECT was deleted from HUWE1 and then the residue fragment was overexpressed in C. Rosea.
The effects of HUWE1 overexpression on the biosynthesis of metabolites at metabolic and transcriptional levels were analyzed dynamically. After parent compound Met was added into the fermentation media, the production of KMBA (α-keto-methylthiobutyric acid), methional, methionol, MTL and DMS was observed for the wild type strain and strain UH1 overexpressing HUWE1 with HECT domain sequence deleted and both strains were with similar metabolite profiles. However, Met addition in fermentation medium made HUWE1 overexpression strain U1 produce trace amount of methional and methionol of ehrlich pathway, but much more KMBA of intermediate, MTL and DMS of demethiolation pathway (Figs 3A–E and S4A–E). These results suggested that HUWE1 with HECT domain mediate the regulation of ehrlich and demethiolation pathways.
Effect of HUWE1 overexpression on the production of (A) KMBA (B) Methional, (C) Methionol, (D) MTL, (E) DMS for C. rosea. ○,● C. rosea was inoculated without or with 5 g/L Met addition; △, ▲ C. rosea strain UH1 overexpressing HUWE1 with HECT sequence deleted was inoculated without or with Met addition; □, ■ C. rosea strain U1 with HUWE1 overexpression plasmid pCAMBIA1302-HUWE1 was inoculated without or with Met addition.
Correspondingly, Met addition upregulated the expression of ehrlich pathway genes ARO8-2, BAT, PDC and downregulated the expression of demethiolation pathway gene STR3 for the wild type strain and strain UH1 overexpressing HUWE1 with HECT domain sequence deleted. Besides the upregulation expression of ehrlich pathway genes ARO8-2 and BAT, Met addition made HUWE1 overexpression strain U1 significantly upregulate the expression of demethiolation pathway gene STR3, but did not impact the expression of ehrlich pathway gene PDC (Fig. 4A,B). Similar metabolite and gene expression profiles were observed from another engineered strain U2 (Figs 4A,B and S4A–E). The transcriptional expression of synthase genes corresponded to metabolites production15,25. HUWE1 overexpression accompanied with Met addition led to unchanged expression of decarboxylase gene PDC and upregulated expression of demethiolase STR3, which may account for that HUWE1 overexpression strains accumulated more KMBA, MTL and DMS, but produced trace amount of methional and methionol. Strains UH1 and UH2 overexpressing HUWE1 with HECT domain sequence deleted lost this capacity, indicating that HECT may play a direct role in catalysis during ubiquitination. Overexpressing HUWE1 enhanced demetiolation pathway metabolites but inhibited ehrlich pathway metabolites, suggesting that HUWE1 was a key factor mediated the regulation of ehrlich and demethiolation pathways.
Transcriptional analysis of synthase genes in HUWE1 overexpression strains U1 and U2, UH1 and UH2 with HECT domain sequence deleted.
Normalized fold expression values for aminotransferase genes ARO8-2 and BAT, decarboxylase gene PDC and demethiolase gene STR3 in engineered strains U1, U2, UH1 and UH2 were relative to those in the wild type strain grown in the medium without Met addition. The error type was standard deviation.
Heterologous expression of HUWE1 impacts the biosynthesis of pathway metabolites in S. cerevisiae
S. cerevisiae as a eukaryotic model and cell factory has been adopted to produce valuable alcohols and food aromatic thiol compounds via ehrlich and demethiolation pathways3,25. Regulators are of importance for metabolic engineering of yeast strains to increase metabolites production and tolerance and control the quality of fermented foods26,27,28,29. Since the catalysis mechanism of HECT domain in HUWE1 was high conserved during ubiquitination, we tested whether heterologous expression of HUWE1 with HECT domain sequence in S. cerevisiae could achieve the same effect under oenological conditions.
The addition of galactose into fermentation medium led to the production of methionol and MTL for the plasmid control strain (containing pYES2) except KMBA which was not detected in Hebert’s studies14,15. As compared to that of the plasmid control strain, HUWE1 overexpressing strain (containing pYES2-HUWE1) produced more MTL and its derivative DMS, but trance amount of methional and methionol of ehrlich pathway during whole fermentation process (Fig. 5A–D). Transcriptional analysis of synthase genes indicated that the production of methionol and MTL was accompanied by upregulation of the expression of aminotransferase genes ARO8, ARO9, BAT1 and pyruvate decarboxylase genes PDC1, PDC5, PDC6, ARO10, downregulation of the expression of demethiolase gene STR3 for plasmid control strain (Fig. 6A). The addition of galactose with different concentrations into SC-U media triggered upregulation of HUWE1 expression (Fig. S5). While overexpressing HUWE1 led to downregulation of the expression of aminotransferase gene ARO8 and pyruvate decarboxylase genes PDC1, PDC5, PDC6, ARO10 and has little effect on the expression of aminotransferase gene ARO9 and demethiolase gene STR3 (Fig. 6B). Relative to the plasmid control, overexpressing HUWE1 significantly suppressed the expression of pyruvate decarboxylase genes of ehrlich pathway and alleviate the inhibition effect on demethiolation pathway gene. These data indicates that overexpression of HUWE1 regulated ehrlich and demethiolation pathway metabolites.
Effect of HUWE1 overexpression on the production of (A) Methional, (B) Methionol, (C) MTL, (D) DMS in S. cerevisiae. ○ Control, S. cerevisiae with plasmid pYES2 was innoculated with 5 g/L Met; ● S. cerevisiae with plasmid pYES2 was inoculated with 5 g/L Met and 20 g/L galactose; □, S. cerevisiae with HUWE1 overexpression plasmid was inoculated with 5 g/L Met; ■, S. cerevisiae with HUWE1 overexpression plasmid was inoculated with 5 g/L Met and 20 g/L galactose.
Transcriptional analysis of synthase genes in (A) plasmid control strain, (B) HUWE1 overexpression strain of S. cerevisiae. The controls for A and B were engineered strains grown in SC-U medium without galactose (Gal) addition, respectively. Normalized fold expression values for aminotransferase genes ARO8, ARO9, BAT1, decarboxylase genes PDC1, PDC5, PDC6, ARO10 and demethiolase gene STR3 were relative to those of the controls. The error type was standard deviation.
Discussion
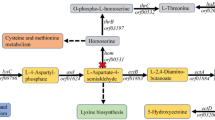
In this study, one regulator E3 ubiquitin-protein ligase HUWE1 was screened from forward library of SSH and is inferred to play positive role during the biosynthesis of methionol and MTL. In Clonostachys rosea and S. cerevisiae, overexpressing HUWE1 significantly increased the biosynthesis of MTL and its derivatives in demethiolation pathway, while suppressed the biosynthesis of methional and methionol in ehrlich pathway (Fig. 7). These results supported the directional regulation of ehrlich and demethiolation pathways.
The overview of HUWE1 mediating the regulation of ehrlich and demethiolation pathways.
Overexpression of HUWE1 increased the expression of ehrlich pathway genes ARO8-2 (ARO9 in S. cerevisiae), BAT (BAT1 in S. cerevisiae) and demethiolation pathway gene STR3, but suppressed the expression of ehrlich pathway gene PDC (PDC1, PDC5, PDC6, ARO10 in S. cerevisiae). The corresponding metabolic products changed accordingly. This indicated that HUWE1 regulated the ehrlich and demethiolation pathways in eukaryotic cell.
Ehrlich pathway has been known for its ability to catabolize amino acid into valuable alcohols, demethiolation pathway as competing branch converted sulfur-containing amino acid into flavor thiols, thus the two pathways controlled the production of fuel and valuable flavor alcohols, the quality and uniqueness of many foodstuffs1,2,3,4,5. Regulators play key roles in metabolites production by controlling the expression of synthase genes. Modifying the behavior of regulators will create new phenotypes important for technological applications, due to that tuning the expression of regulator will reprogram the transcription of pathway genes30,31,32,33,34. This strategy has been successfully applied to optimization of several industrial phenotypes such as tolerance, metabolite over production and so on31,32,34,35,36.
Taken the conversion of Met into methionol and MTL for example, pathway metabolite is complex and under multigenic control, engineering regulators would be a better choice for enhancing or controlling the conversion of Met into methionol and MTL17. One positive regulator ARO80 has been identified thus far for increasing ehrlich pathway metabolites18,19. In this study, a regulator gene HUWE1 was selected, whose overexpression strengthen the expression of ehrlich pathway genes ARO8-2 and BAT encoding aminotransferase, demethiolation pathway gene STR3 encoding demethiolase but had no effect on the expression of ehrlich pathway gene PDC encoding α-ketoacid decarboxylase after Met was added into the fermentation medium. The transcriptional expression pattern of synthase genes corresponded to the production of pathway metabolites. This suggested that HUWE1 may control the biosynthesis of pathway metabolites at transcriptional level. Cellular homeostasis depends on a balance between anabolism and catabolism, which can be regulated by the ubiquitin-proteasome system (UPS). HUWE1, a key constituent of UPS, had a strong link with transcriptional control, cell cycle, lipid metabolism and so on, which was found to be involved in regulation of amino acid metabolism in this study12,37. Thus, HUWE1 is a potential metabolic switch for regulating cell metabolism.
A grand challenge in synthetic biology is to move the design of biomolecular circuits from purely genetic constructs toward systems that integrate different levels of cellular complexity, including regulatory networks and metabolic pathways17,38. The expression of synthase genes is often controlled at the transcriptional level by transcription factors that can sense either the end product or an intermediate metabolite, giving rise to different regulatory architectures15,17. E3 ubiquitin-protein ligase as a key constitution of ubiquitination system controlled the degradation of transcription factors and was located in upstream branch of regulatory architectures. Engineering it can achieve large-scale regulation of metabolites at cell level. Therefore, HUWE1 with HECT as regulatory architecture and motif discovered in this study has a great accomplishment in pathway engineering to improve cellular productivity and yield by exploiting dynamic pathway regulation and metabolic control.
Methods
Microorganism and culture condition
Clonostachys rosea tang 19 (Genbank accession number: KT007105) was grown on potato-agar-dextrose slants and produced methional, methionol, MTL and its derivatives from Met by inoculating it in fermentation medium (Met, 5 g/L; sucrose, 35 g/L; peptone, 2.5 g/L; yeast extract, 2.5 g/L; KH2PO4·H2O, 1 g/L; MgSO4·7H2O 0.5 g/L; vitamin B1, 0.05 g/L). Escherichia coli were grown in Luria-Bertani (LB) media containing 1% (v/w) tryptone, 0.5% yeast extract and 1.5% agar and transformed strains were grown in LB medium supplemented with the appropriate antibiotic (s). Agrobacterium tumefaciens was grown and propagated in LB medium at 28 °C in the presence of appropriate marker selection supplements39. Commercial S. cerevisiae strain INVSc1 (Invitrogen) was used as a host strain for expressing HUWE1 gene cassette. Yeast strains were cultivated at 30 °C in either a rich medium, YPD (Yeast Extract Peptone Dextrose Medium, containing 1% (w/v) yeast extract, 2% peptone and 2% glucose), or a synthetic minimal defined medium, SC (synthetic complete medium, containing 2% glucose or raffinose, 0.67% yeast nitrogen base without amino acids (Biosharp)). Uracil (SC-U) was omitted to make selective plates for growing pYES2 transformants.
Construction of suppression subtractive hybridization library
Total RNA from C. rosea was extracted by using E.Z.N.A.™ Fungal RNA Kit (Omega, USA) and RNA quality was confirmed by visualisation of ethidium bromide-stained bands following agarose gel electrophoresis and quantified by Biotek Synerg Spectrophotometer (OD260/OD280). Total RNAs extracted from C. rosea inoculated in fermentation medium were used as the tester. The deriver RNA was from C. rosea inoculated in fermentation medium without Met addition. Subsequently, the driver or tester RNAs were reversely transcripted to cDNA by SMARTerTM PCR cDNA synthesis kit (Clontech, USA). 5′ PCR Primer II A from the kit was used for PCR amplification to enrich differentially expressed genes (Table S1). The optimal PCR cycles for the driver and tester were determined after dynamical detection of the PCR products by agarose gel electrophoresis (Fig. S1A). SSH was carried out using the PCR-selected cDNA Subtraction Kit (Clontech, USA) and then the cDNAs from tester and driver were digested by RsaI (Fig. S1B). After two-round of SSH (Fig. S1C,D), the resulting PCR products were purified and inserted into pGEM-T vector (Promega, USA), then the product of a ligation reaction was transformed into E. coli DH5α. Recombinant colonies were selected and the length of inserted segments in the library was determined by PCR using nested primers (Table S1.).
Sequencing of subtracted libraries
From the two subtracted cDNA libraries, 2405 recombinant bacterial clones were picked by blue–white selection on plates. These white colonies were grown and then 0.5 μL cultures were used as template for amplifying the exogenous segment by using the nested PCR-primers 1F and 2R (Table S1). PCR products were checked on 2% agarose gel and the remainder was used for screening the differentially expressed genes.
Screened by cDNA array dot blotting, 500 clones with cDNA inserted were selected randomly. The cDNA array dot blotting was conducted using forward- and reverse-subtracted cDNA probes prepared with the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Molecular Biochemicals, Germany). The hybridized signal intensity was analyzed with ChemiDoc Image Lab software (BioRad, USA) and spots that displayed at least threefold higher intensity with the forward subtracted probes than the reverse subtracted probes were scored as positives from forward-subtracted cDNA library. Using the same method, spots that displayed at least threefold higher intensity with the reverse subtracted probes than the forward subtracted probes were scored as positives from forward-subtracted cDNA library (Fig. S2A–D). And 100 clones with strongest hybridization signals were sequenced from forward- and reverse-SSH libraries, respectively.
Relative quantification of mRNAs expression by real-time PCR
Four expressed sequence tags from the forward and the reverse libraries were selected for further analysis of their expression at transcriptional level by using quantitative real-time PCR, respectively. RNA was isolated by using the MasterPure Fungal RNA Purification Kit (Biozym Scientific GmbH). Complementary DNA (cDNA) was synthesized using a PrimeScriptTM 1st Strand cDNA Synthesis Kit (TaKaRa Biotechnology (Dalian) Co., Ltd) with 0.5 μg of total RNA as the template. 18S rDNA served as standard gene for normalization of gene expression. Quantitative real-time PCR was performed on a StepOne Real-Time PCR system in duplicates for at least three independent experiments and the used primers were described in Table S1. SYBR dye (TaKaRa Biotechnology (Dalian) Co., Ltd) was used to visualize gene amplification. Relative quantification of the target gene expression was evaluated using the comparative cycle threshold (CT) method.
Overexpression of HUWE1
A nested PCR-based strategy for genome walking named self-formed adaptor PCR was adopted to obtain the full-length of HUWE140. Three primers including HUWE1-P5SP1, HUWE1-P5SP2 and HUWE1-P5SP3 (Table S1) were designed, which were located upstream of the cDNA sequence of HUWE1. The amplified PCR products were cloned into pGEM-T easy vector and then sequenced. After the whole cDNA was obtained, BLASTX program at the National Center for Biotechnology Information (NCBI) was used for functional inference. ORF Finder program was adopted to predict the ORF (Open Reading Frame) of HUWE1 (Genebank accession number KT762629) and the ORF encoded a 988 amino acid polypeptide which showed 73% identity to its homologue in Fusarium oxysporum f. sp. EXM 30317.1.
The ORF corresponding to the entire coding region of HUWE1 was flanked with XbaI and BstEII sites by using PCR primers HUWE1-5F-XbaI and HUWE1-3R-BstEII (Table S1). After double digestion with XbaI and BstEII, the PCR products were ligated into the linearized vector pCAMBIA1302 containing trpC promoter and terminator to create the expression vector pCAMBIA-HUWE1. pCAMBIA-HUWE1 was transformed into C. rosea with the help of A. tumefaciens strain AGL-139. Transformants survived on the medium containing 50 mg/L hygromycin were identified by PCR with the primers of HPT-F2 and HPT-3 targeting hydromycin resistance gene and then HUWE1 overexpression at transcriptional levels were analyzed by quantitative real-time PCR. The selected HUWE1-overexpressed strains were named U1 and U2.
HUWE1 with HECT sequence deleted was amplified by using PCR primers HUWE1-5F-XbaI and HUWE1-3BstEII (Table S1) and then inserted into fungal expression vector pCAMBIA1302 containing trpC promoter and terminator after double digestion with XbaI and BstEII. The expression vector was transformed into C. rosea with the help of A. tumefaciens strain AGL-1. Overexpression of HUWE1 with HECT sequence deleted was analyzed at transcriptional levels and the targeted strains were named UH1 and UH2.
The effect of HUWE1 overexpression on the biosynthesis of methionol and MTL in C. rosea
HUWE1 overexpression strains U1 and U2, HECT deleted strains UH1 and UH2 and wild type strain Tang 19 were inoculated into the fermentation media. After the cultures were incubated for 0, 1, 3, 5 and 7 days, KMBA, methional, methionol, MTL and its derivatives were analyzed dynamically. Intracellular KMBA was extracted and detected following the previously mentioned method15. Fungal mycelia cultured in fermentation medium with 5 g/L Met were collected by filtration, washed with 20 mL of cold ultrapure water and then filtered again. Mycelial pellets were stored at —80 °C. The pellets were resuspended with 1 mL of 1% (v/v) formic acid, incubated for 10 min at 95 °C and centrifuged for 30 min at 4 °C. The supernatant was lyophilized and stored at −80 °C. Before injection, samples were resuspended in water containing 0.1% formic acid and stored at 4 °C. Chromatographic separation was performed on a Reprosil-Pur Basic C18 column (4.6 mm × 250 mm × 5 μm) from Dr. Maisch GmbH (Germany) using a liquid chromatography 20AD system (Shimadzu Corporation, Japan). The optimized mobile phase was 0.1% formic acid water solution. The column oven temperature was set to 30 °C and the flow rate was 1 mL/min. The detection wavelength was 197 nm. The retention time for KMBA was 12.5 min. Methional, methionol, MTL and its derivatives were extracted and detected as follows. A 5-mL volume of fermentation culture was added into a 15-mL vial, the vial was tightly capped with a silicon septum and pre-equilibrated at 40 °C for 5 min and then solid-phase microextraction (SPME) device was inserted into the headspace of vial to extracte MTL and its derivatives for 20 min at 40 °C. MTL and its derivatives were analyzed by gas chromatography (GC-2010 with a flame ionization detector (FID) from Shimadzu Technologies Inc., Tokyo, Japan) with an oven temperature program. First, the temperature was maintained at 35 °C for 3 min. Subsequently, the temperature reached 100 °C, with an increment of 8 °C/min. Then, the temperature was raised to 220 °C, with an increment of 10 °C for 1 min. Additionally, ehrlich pathway genes including ARO8-2 (Genebank accession number KT157523) and BAT (KT157521) encoding aminotransferase, PDC (KT762630) encoding a-keto acid decarboxylase and demethiolation pathway genes STR3 (KT157524) encoding demethiolase were analyzed at transcriptional level by quantitative real-time PCR.
Effect of HUWE1 overexpession on the biosynthesis of methionol and MTL in S. cerevisiae
The isolation and manipulation of DNA from C. rosea were performed using E.Z.N.A Fungal DNA kit (Omega) according to the manufacturer’s protocol. Primer pairs YHUWE1-5HK and YHUWE1-3XH were designed to amplify the complete ORF of HUWE1 from chromosomal DNA of C. rosea (Table S1). KpnI (in the forward primer) and XbaI (in the reverse primer) restriction sites were introduced to ensure that HUWE1 was correctly inserted into yeast expression plasmid pYES2. Engineered S. cerevisiae strains containing control plasmid pYES2 or HUWE1 overexpression plasmid pYES2-HUWE1 were inoculated into SC-U medium containing 2% (w/v) glucose, respectively. After overnight culture, cells were collected and the pellets were resuspended into SC-U medium (Optical Density of 0.4 at 600 nm) containing 0.5% Met and then 2.0% galactose were added into the SC-U medium for inducing the expression of HUWE1. Methional, methionol, MTL and its derivatives were extracted and dynamically analyzed following the method described above. Additionally, different concentrations of galactose (0, 0.5, 1.0, 2.0% (w/v)) were added into the SC-U media for culturing HUWE1 overexpression strain and plasmid control strain. After 3 days of cultivation, total RNA were extracted and then synthase genes including ARO8, ARO9 and BAT1 encoding aminotransferase, PDC1, PDC5, PDC6 and ARO10 encoding pyruvate decarboxylase and STR3 encoding demethiolase were analyzed by real-time PCR followed the method described above (Table S1).
Additional Information
How to cite this article: Zhang, Q. et al. Regulating ehrlich and demethiolation pathways for alcohols production by the expression of ubiquitin-protein ligase gene HUWE1. Sci. Rep. 6, 20828; doi: 10.1038/srep20828 (2016).
References
Branduardi, P. et al. A novel pathway to produce butanol and isobutanol in Saccharomyces cerevisiae. Biotechnol Biofuels 6, 68 (2013).
Buijs, N. A., Siewers, V. & Nielsen, J. Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol 17, 480–488 (2013).
Generoso, W. C., Schadeweg, V., Oreb, M. & Boles, E. Metabolic engineering of Saccharomyces cerevisiae for production of butanol isomers. Curr Opin Biotechnol 33, 1–7 (2015).
Martinez-Cuesta, M. D. C., Pelaez, C. & Requena, T. Methionine metabolism: major pathways and enzymes involved and strategies for control and diversification of volatile sulfur compounds in cheese. Crit Rev Food Sci Nutr 53, 366–85 (2013).
Si, T., Luo, Y. Z., Xiao, H. & Zhao, H. M. Utilizing an endogenous pathway for 1-butanol production in Saccharomyces cerevisiae. Metab Eng 22, 60–68 (2014).
Kim, B., Cho, B. R. & Hahn, J. S. Metabolic engineering of Saccharomyces cerevisiae for the production of 2-Phenylethanol via ehrlich pathway. Biotech Bioeng 111, 115–124 (2014).
Teixeira, L. K. & Reed, S. I. Ubiquitin ligases and cell cycle control. Annu Rev Biochem 82, 387–414 (2013).
Vittal, V., Stewart, M. D., Brzovic, P. S. & Klevit, R. E. Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J Biol Chem. 290, 21244–51 (2015).
Mund, T., Lewis, M. J., Maslen, S. & Pelham, H. R. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci USA 111, 16736–41 (2014).
Leach, M. D. et al. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol 79, 1574–93 (2011).
Licursi, V. et al. The COP9 signalosome is involved in the regulation of lipid metabolism and of transition metals uptake in Saccharomyces cerevisiae. FEBS J. 281, 175–190 (2014).
Nakatsukasa, K. et al. The ubiquitin ligase SCF (Ucc1) acts as a metabolic switch for the glyoxylate cycle. Mol Cell 59, 22–34 (2015).
Portnoff, A. D., Stephens, E. A., Varner, J. D. & DeLisa, M. P. Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. J Biol Chem 289, 7844–55 (2014).
Hebert, A. et al. Exploration of sulfur metabolism in the yeast Kluyveromyces lactis. Appl Microbiol Biotechnol 91, 1409–1423 (2011).
Hebert, A. et al. New insights into sulfur metabolism in yeasts as revealed by studies of Yarrowia lipolytica. Appl Environ Microbiol 79, 1200–1211 (2013).
Chin, C. S. et al. Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways. PLoS Biol 6, e146 (2008).
Chubukov, V., Zuleta, I. A. & Li, H. Regulatory architecture determines optimal regulation of gene expression in metabolic pathways. Proc Natl Acad Sci USA 109, 5127–32 (2012).
Iraqui, I., Vissers, S., Andre, B. & Urrestarazu, A. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol Cell Biol 19, 3360–71 (1999).
Lee, K. & Hahn, J. S. Interplay of Aro80 and GATA activators in regulation of genes for catabolism of aromatic amino acids in Saccharomyces cerevisiae. Mol Microbiol 88, 1120–34 (2013).
Sandoval, D. et al. Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations. J Biol Chem 290, 1106–18 (2015).
Stec, I., Nagl, S. B., Ommen, G. B. & Dunnen, J. T. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473, 1–5 (2000).
Kermekchiev, M. & Ivanova, L. Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation. Mol Cell Biol 21, 8255–63 (2001).
Chen, L. & Lopes, J. M. Multiple bHLH proteins regulate CIT2 expression in Saccharomyces cerevisiae. Yeast 27, 345–59 (2010).
Avalos, J. L., Fink, G. R. & Stephanopoulos, G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31, 1–7 (2013).
Cholet, O., Henaut, A., Hebert, A. & Bonnarme, P. Transcriptional analysis of L-methionine catabolism in the cheese-ripening yeast Yarrowia lipolytica in relation to volatile sulfur compound biosynthesis. Appl Environ Microbiol 74, 3356–67 (2008).
Alper, H. et al. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314, 1565–8 (2006).
Chen, B., Ling, H. & Chang, M. W. Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol Biofuels 6, 21 (2013).
Kim, I. S. et al. Saccharomyces cerevisiae KNU5377 stress response during high-temperature ethanol fermentation. Mol Cells 35, 210–8 (2013).
An, J. et al. Tolerance to acetic acid is improved by mutations of the TATA-binding protein gene. Environ Microbiol 17, 656–69 (2015).
Bi, Q., Wu, D., Zhu, X. & Turgeon, B. G. Cochliobolus heterostrophus Llm1-a Lae1-like methyltransferase regulates T-toxin production, virulence and development. Fungal Genet Biol 51, 21–33 (2013).
Gao, C. et al. Crp is a global regulator of antibiotic production in streptomyces. Mbio 3 (2012).
Li, L. et al. A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab Eng 29, 12–25 (2015).
Pilsyk, S. et al. The Aspergillus nidulans metZ gene encodes a transcription factor involved in regulation of sulfur metabolism in this fungus and other Eurotiales. Curr Genet 61, 115–25 (2015).
Rachid, S., Gerth, K., Kochems, I. & Muller, R. Deciphering regulatory mechanisms for secondary metabolite production in the myxobacterium Sorangium cellulosum So ce56. Mol Microbiol 63, 1783–96 (2007).
Zhang, F. et al. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab Eng 14, 653–60 (2012).
Zhu, Y. et al. Metabolomic analysis reveals functional overlapping of three signal transduction proteins in regulating ethanol tolerance in cyanobacterium Synechocystis sp. PCC 6803. Mol Biosyst 11, 770–82 (2015).
Qi, L. et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312, 1763–66 (2006).
Xu, P. et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA 111, 11299–11304 (2014).
Groot, M. J., Bundock, P., Hooykaas, P. J. & Beijersbergen, A. G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16, 839–42 (1998).
Wang, S., He, J., Cui, Z. & Li, S. Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl Environ Microbiol 73, 5048–51 (2007).
Acknowledgements
Financial supports from the National Natural Science Foundation of China (NSFC, Project Nos 81503112, 21376066, 21506049 and 31570054) and Hubei Provincial Natural Science Foundation for Innovative Research Team (2015CFA013) are gratefully acknowledged. Prof. Ya-Jie Tang also thanks the National High Level Talents Special Support Plan (“Million People Plan”) by the Organization Department of the CPC Central Committee (2014), Training Program for Top Talents in Hubei Province (2013) and Training Program for Huanghe Talents in Wuhan Municipality (2014).
Author information
Authors and Affiliations
Contributions
Y.J.T. and K.Z.J. conceived the project. Y.J.T. and K.Z.J. designed the experiments. Q.Z., K.Z.J., S.T.X. and Y.H.X. implemented the analysis workflow and conducted the experiments. Y.J.T., K.Z.J., R.S.L. and H.M.L. analysed and interpreted the results. Q.Z. and K.Z.J. prepared all figures and tables. Y.J.T. and K.Z.J. prepared and wrote the manuscript. All authors reviewed, commented on and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, Q., Jia, KZ., Xia, ST. et al. Regulating ehrlich and demethiolation pathways for alcohols production by the expression of ubiquitin-protein ligase gene HUWE1. Sci Rep 6, 20828 (2016). https://doi.org/10.1038/srep20828
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20828
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.