Abstract
Since the SOX2 amplification was identified in lung squamous cell carcinoma (lung SCC), SOX2 transcriptional downstream targets have been actively investigated; however, such targets are often cell line specific. Here, in order to identify highly consensus SOX2 downstream genes in lung SCC cells, we used RNA-seq data from 178 lung SCC specimens (containing tumor and tumor-associated cells) and analyzed the correlation between SOX2 and previously-reported SOX2-controlled genes in lung SCC. In addition, we used another RNA-seq dataset from 105 non-small cell lung cancer cell lines (NSCLC; including 4 lung SCC cell lines) and again analyzed the correlation between SOX2 and the reported SOX2-controlled genes in the NSCLC cell lines (no tumor-associated cells). We combined the two analyses and identified genes commonly correlated with SOX2 in both datasets. Among the 99 genes reported as SOX2 downstream and/or correlated genes, we found 4 negatively-correlated (e.g., CDKN1A) and 11 positively-correlated genes with SOX2. We used biological studies to demonstrate that CDKN1A was suppressed by SOX2 in lung SCC cells. G1 cell cycle arrest induced by SOX2 siRNA was rescued by CDKN1A siRNA. These results indicate that the tumorigenic effect of SOX2 in lung SCC cells is mediated in part by suppression of CDKN1A.
Similar content being viewed by others
Introduction
Lung squamous cell carcinoma (lung SCC) is the second most frequent type of non-small cell lung carcinoma (NSCLC) after lung adenocarcinoma (lung AC)1. In contrast to the recent discovery of targeted therapies for lung AC, including EGFR mutant or ALK fusions, there is no effective therapy for lung SCC other than chemotherapy2,3,4. In order to understand the molecular pathogenesis that leads to identifying potential therapeutic molecular targets for lung SCC, extensive genetic analysis, including next-generation sequencing, have been performed, which has revealed amplification of SOX2, PIK3CA, PDGFRA, BRF2 and FGFR1, deletion of CDKN2A/B, PTEN, TP53 and NF1 and mutations of PIK3CA, NRF2, DDR2, PTEN, EPHA2, LKB1 and AKT1 in lung SCC5,6. The genomic amplification of SOX2 is seen in 20% of lung SCC7 while increased expression of SOX2 is seen in 90% of lung SCC8, suggesting that SOX2 mediates a major tumorigenic effect on lung SCC regardless of genetic alterations.
SOX2 plays an oncogenic role not only in lung SCC but also in other cancers, including lung AC, ovarian, breast, esophageal, gastric, colon and pancreatic cancers9,10,11,12,13. SOX2 is a transcription factor, thus SOX2 downstream genes that exert a tumorigenic effect have been actively sought in such different types of cancers (summarized in Table 1). However, due to its relatively recent finding as an oncogene11, consensus SOX2 downstream targets that bear a tumorigenic function have not been established yet. In the present study, we utilized gene expression data from The Cancer Genome Atlas (TCGA) human lung SCC samples (n = 178)14 and determined a correlation in lung SCC between SOX2 and previously-reported SOX2 downstream targets in the multiple cancer cell lines. The limitation of using this TCGA lung SCC dataset is that the expression of each gene in the dataset is comprised of the combined gene expression profiles of tumor cells and tumor-associated endothelial cells, fibroblasts and immune cells, which hampers the identification of tumor cell-specific gene-to-gene correlations. Thus, we also utilized another gene expression dataset from non-small cell lung cancer (NSCLC) cell lines (n = 105), including 4 lung SCC cell lines15 and assessed the correlation between SOX2 and the reported SOX2 downstream targets in the NSCLC cell lines. The limitation of using this NSCLC cell line dataset is that it includes not only lung SCC cell lines but also other lung carcinoma cell lines (e.g., lung AD cell lines). Thus, after we analyzed the two datasets, we selected genes that were commonly correlated with SOX2 in both datasets, which would likely be SOX2-correlated genes specific to lung SCC cells. Among 99 genes identified in previous reports as regulated by or correlated with SOX2 in multiple cancer cell lines, the expression of only 15 genes was positively or negatively correlated with the expression of SOX2 in both the 178 lung SCC specimens and the 105 NSCLC cell lines. Among the 15 genes, CDKN1A (also known as p21[Cip1/Waf1]) that induces G1 cell cycle arrest was determined by RNA interference and adenovirus-mediated ectopic expression experiments to be a negative downstream target of SOX2 in multiple lung SCC cell lines. G1 cell cycle arrest induced by the reduction of SOX2 was reinstated by the reduction of CDKN1A in lung SCC cell lines, indicating that CDKN1A is an intrinsic SOX2 target influencing tumorigenicity in lung SCC cells. Here, we report that CDKN1A is a highly consensus gene target of the oncogenic transcription factor SOX2 in lung SCC cells.
Results
In order to identify highly consensus SOX2 downstream genes in lung SCC cells, we investigated genes previously reported to be regulated by SOX2 in multiple cancer cell lines. As shown in the Table 1, SOX2 regulates cell cycle-related genes positively or negatively. CDKN1A, which induces G1 arrest, is repressed by SOX2 in A549 lung carcinoma cells, pancreatic cancer cells13,16 and gastric cancer cells17. CDKN1B, which also induces G1 arrest, is repressed by SOX2 in pancreatic cancer cells and gastric cancer cells. CCND1, which accelerates cell cycle, is activated by SOX2 in gastric cancer cells and MCF7 breast cancer cells10,17. Overall, SOX2 represses cell cycle inhibitors and activates cell cycle accelerators; however, the pattern of gene regulation is not universal in different cancer cell types. Additionally, genes comprising the WNT, NOTCH, RAS, TGF/BMP and EMT pathways that are involved in cancer progression and metastasis are regulated by SOX2 in multiple cancer cell lines9,16,18,19,20 (summarized in Table 1). In addition to genes identified as regulated by SOX2 in previous reports, we hypothesized that the previously reported top 50 genes positively correlated with SOX2 expression in stage I/II lung SCC11 might be directly regulated by SOX2 in lung SCC cells. In total, we selected 99 genes that might be commonly regulated by SOX2 in lung SCC cells (Table 1).
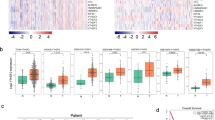
Next, in order to determine which of the possible 99 genes are correlated with SOX2 in lung SCC cells, we used a RNA-seq dataset from TCGA14 to determine whether the expression of any of the 99 genes is correlated with that of SOX2 in 178 lung SCC specimens. The 178 lung SCC specimens were divided into two groups, SOX2-HIGH and SOX2-LOW groups, based on the median level of SOX2 expression in the lung SCC specimens (see Materials and Methods for details). Then, we assessed which genes are correlated with SOX2. As shown in Fig. 1, 57 genes (e.g., BMP7) were positively correlated with SOX2 while 11 genes (e.g., CDKN1A) were negatively correlated with SOX2 in lung SCC, suggesting that these genes might be regulated by SOX2 in lung SCC cells; however, it remains unknown whether the gene-to-gene correlation would be intrinsic or extrinsic since the TCGA lung SCC data contains gene expression from heterogeneous cell populations, including not only tumor cells but also tumor-associated cells. Thus, we used another RNA-seq dataset from 105 non-small cell lung cancer (NSCLC) cell lines (including 4 lung SCC cell lines)15 and performed the same analysis (see Materials and Methods for details). As shown in Fig. 2, 12 genes (e.g., BMP7) were positively correlated with SOX2 while 8 genes (e.g., CDKN1A) were negatively correlated with SOX2 in the NSCLC cells, suggesting that these genes might be regulated by SOX2 in lung SCC cells. Although the NSCLC cell lines do not contain tumor-associated cells, the gene-to-gene correlations might occur in non-lung SCC cells (e.g., lung AD cells). Thus, we combined the two analyses (Figs 1 and 2) and sought to identify most probable gene-to-gene correlation in lung SCC cells. Among the 99 genes, BMP7, CHAC1, DHX9, GPX2, MARK1, MSH6, PRKX, RMND5A, SIAH2, TMPRSS4 and USP39 were positively correlated while BMP2, CDKN1A, SNAI1 and VIM were negatively correlated with SOX2 in both datasets, suggesting that these genes are intrinsically regulated by SOX2 in lung SCC cells.
SOX2 positively or negatively correlated genes in the human lung squamous cell carcinoma (SCC) samples from the TCGA.
The Cancer Genome Atlas (TCGA) human lung squamous cell carcinoma (SCC) samples (n = 178) were divided into two groups that have high RNA expression of SOX2 (HIGH, n = 89) or low expression of SOX2 (LOW, n = 89). Two-tailed Welch’s t-test was used to determine the significance of gene expression between the two groups. Genes with significantly higher expression in the HIGH group (p < 0.05 and average expression > 1) were considered as SOX2 positively correlated genes, while genes with significantly higher expression in the LOW group (p < 0.05 and average expression > 1) were considered as SOX2 negatively correlated genes. The expression was measured in RPKM and quantile normalized. PLA2G4B was not present in the TCGA SCC dataset. Results represent the mean ± S.E.M. *indicates p < 0.05, **indicates p < 0.01 and ***indicates p < 0.001.
SOX2 positively or negatively correlated genes in the human non-small cell lung cancer (NSCLC) cell lines.
Human non-small cell lung cancer (NSCLC) cell lines (n = 105) were separated into two groups that have high RNA expression of SOX2 (HIGH, n = 20) or low expression of SOX2 (LOW, n = 85). The expression of previously reported SOX2 downstream or correlated genes was assessed as to whether the expression of those genes was correlated with that of SOX2. Two-tailed Welch’s t-test was used to determine the significance of gene expression between the two groups. Genes with significantly higher expression in the HIGH group (p < 0.05 and average expression >1) were considered as SOX2 positively correlated genes, while genes with significantly higher expression in the LOW group (p < 0.05 and average expression >1) were considered as SOX2 negatively correlated genes. The expression was measured in RPKM and quantile normalized. Results represent the mean ± S.E.M. *indicates p < 0.05, **indicates p < 0.01 and ***indicates p < 0.001.
Next, in order to confirm whether the RNA-seq data analyses indeed reflect their correlation at the protein level in lung SCC cell lines, we performed western blotting using cell extracts from six lung SCC cell lines and determined the protein expression of SOX2 and two G1 cell cycle inhibitors CDKN1A and CDKN1B. We focused on these cell cycle genes since SOX2 has been shown by several groups to influence cell proliferation in vitro13,16. CDKN1B was repressed by SOX2 in pancreatic cancer cell lines13; however, a negative correlation with SOX2 was not observed in our RNA-seq data analyses (Figs 1 and 2). Consistent with our RNA-seq data analyses (Figs 1 and 2), lung SCC cell lines (EBC2, LK2 and H520) that had a high expression of SOX2 had no expression of CDKN1A at the protein level. Conversely, lung SCC cell lines (EBC1, H226 and SQ5) that had a low expression of SOX2 did express CDKN1A (Fig. 3A), indicating that the expression of SOX2 and CDKN1A is inversely related. In addition to the expression in the cell lines, we also investigated the expression of SOX2 and CDKN1A in human primary lung SCC specimens. As shown in Supplemental Fig. 1, the majority of lung SCC specimens expressing SOX2 did not express CDKN1A. As indicated in the RNA-seq data analysis (Figs 1 and 2), any correlation between SOX2 and CDKN1B in the various lung SCC cell lines was not observed at the protein level (Fig. 3A,C), which suggests that SOX2 regulates CDKN1B in a cell-type specific fashion (pancreatic cancer vs. lung SCC). A similar negative correlation between SOX2 and VIM that was indicated by the RNA-seq data analyses (Figs 1 and 2) was also observed at the protein level in lung SCC cell lines (Supplemental Figure 2A).
SOX2 suppresses CDKN1A in human lung SCC cells.
(A) Immunoblot analysis of SOX2, CDKN1A, CDKN1B and TP53 in the indicated lung SCC cell lines. The expression level of β-actin is shown as a control. (B) qPCR demonstrated that CDKN1A mRNA was significantly increased after SOX2 silencing by SOX2 siRNAs (siSOX2 #1 and siSOX2 #2) in EBC2, LK2 and H520 lung SCC cells. Non-targeting siRNA was used as a control (siCtrl). Detection of GAPDH was used for normalization. Results represent the mean ± SD (n = 3). Statistical significance was defined as p < 0.01 (*). (C) Immunoblot analysis shows increased CDKN1A expression 48hours after SOX2 siRNAs (siSOX2 #1 and siSOX2 #2) transfection in EBC2, LK2 and H520 lung SCC cells. TP53 expression was not changed after SOX2 siRNAs transfection. Non-targeting siRNA was used as control (siCtrl). (D) The qPCR analysis showed that CDKN1A mRNA expression was decreased 48hours after Ad-CMV/SOX2 (Ad-SOX2) infection at a MOI of 100 in EBC1 and at a MOI of 10 in H226 lung SCC cells. Ad-CMV/Luc (Ad-Luc) did not change CDKN1A mRNA expression in both lung SCC cell lines. Non-treated cells were used as a control (Ctrl). Immunoblot analysis demonstrated that CDKN1A protein was suppressed 48hours after Ad-SOX2 infection in EBC1 and H226 lung SCC cells. Ad-Luc did not change CDKN1A protein expression in the cells. TP53 expression was not changed in the cells. Statistical significance was defined as p < 0.01 (*).
Next, in order to determine whether SOX2 indeed functionally regulates the expression of CDKN1A, we employed a RNA interference approach using SOX2 siRNA. Knockdown of SOX2 by RNA interference has been shown to inhibit cell viability and colony formation of lung SCC cells, again indicating that SOX2 sustains cell proliferation11. We were able to reproduce the effect of SOX2 using two independent siRNAs in multiple lung SCC cell lines in vitro and in vivo (Supplemental Figure 3A, 3B and 3C). Then, we investigated using these siRNAs targeting SOX2 to determine whether suppression of SOX2 influences the expression of CDKN1A in lung SCC cells. As shown in Fig. 3B,C, CDKN1A but not CDKN1B was induced by the two independent siRNAs targeting SOX2 in multiple lung SCC cell lines at the mRNA and protein levels, indicating that SOX2 functionally suppresses the expression of CDKN1A. TP53 is known to induce CDKN1A21; however, the expression of TP53 was not influenced by SOX2 siRNAs (Fig. 3C), indicating that SOX2 suppresses CDKN1A in lung SCC cells not through TP53. Additionally, CDKN1A was induced by SOX2 siRNAs in the presence of cycloheximide (Supplemental Fig. 4), suggesting the direct suppression of CDKN1A by SOX2, as has been reported in pancreatic cancer cells13. VIM and ZEB1 expression were also induced by these SOX2 siRNAs in lung SCC cell lines (Supplemental Figure 2B, 2C), indicating that SOX2 functionally suppresses the expression of VIM and ZEB1. The negative correlation between SOX2 and ZEB1 was indicated by the bioinformatical analysis using the 105 NSCLC cell lines dataset but not the 178 lung SCC specimens dataset (Figs 1 and 2); however the biological analysis indicated that SOX2 intrinsically regulates ZEB1 in lung SCC cells, suggesting the requirement of validating bioinformatical analysis by biological experiments. We further confirmed the effect of SOX2 using adenovirus-expressing SOX2. Adenovirus-mediated expression of SOX2 significantly suppressed the expression of CDKN1A at the mRNA and protein levels (Fig. 3D). These loss-of-function and gain-of-function experiments indicate that SOX2 negatively regulates the expression of the G1 cell cycle inhibitor CDKN1A in lung SCC cells.
In the next experiment, we sought to determine whether the increased expression of CDKN1A by SOX2 siRNA alters the cell cycle. As shown in Fig. 4, SOX2 siRNA induced the G1 cell cycle arrest in lung SCC (EBC2 and LK2) cells, presumably through the induced expression of CDKN1A. Inhibition of CDKN1A by siRNAs did not influence the cell cycle in EBC2 and LK2 cells (Fig. 4) presumably because they do not express endogenous CDKN1A (Fig. 2A). siRNAs targeting CDKN1A were validated using cells expressing endogenous CDKN1A (Supplemental Figure 5). We then investigated whether the G1 arrest induced by the SOX2 siRNA is mediated by the increased expression of CDKN1A using the siRNAs targeting CDKN1A. When both SOX2 siRNA and CDKN1A siRNA were transfected in EBC2 and LK2 cells, the G1 arrest induced by SOX2 siRNA was restored to base level by the presence of the CDKN1A siRNA (Fig. 4), indicating that SOX2 sustains cell cycle by inhibiting the expression of CDKN1A, a G1 cell cycle inhibitor.
CDKN1A siRNAs released G1 cell cycle arrest induced by SOX2 siRNA.
Flow cytometry cell cycle analysis of EBC2 and LK2 lung SCC cells transiently transfected with non-targeting siRNA (siCtrl), SOX2 siRNA (siSOX2), CDKN1A siRNA (siCDKN1A) or the mixture of SOX2 and CDKN1A siRNAs (siSOX2 + siCDKN1A). Cells were harvested at 72hours after siRNA transfection. Indicated in the inset of each panel are the percentages of each phase of cell cycle, where values represent the mean ± SD of triplicate measurements.
Discussion
SOX2 has been recognized as a lineage-specific oncogenic transcription factor in lung SCC cells; however, consensus downstream genes of SOX2 have not been established yet. In the present study, we bioinformatically identified highly consensus downstream genes regulated by SOX2 in lung SCC cells by combining previously-reported SOX2 downstream/correlated genes in multiple cancer cell lines with two RNA-seq datasets from 178 lung SCC specimens and 105 NSCLC cell lines. We further biologically validated CDKN1A, VIM and ZEB1 as intrinsic SOX2 downstream genes in lung SCC cells by loss-of-function and gain-of-function biological experiments.
Unlike SOX2, downstream gene targets of NKX2-1 (also known as TTF-1), another lineage-specific oncogenic transcription factor in lung AC, are well known. Due to a lung defect in Nkx2-1 knockout mice22, NKX2-1 has been implicated in the regulation of lung specific genes, including surfactant protein genes23. Recent in vitro and in vivo studies using heterozygous or conditionally deleted Nkx2-1 mice also indicate that NKX2-1 directly suppresses the expression of mucous genes in asthma and lung cancer24,25,26,27. Since Sox2 knockout mice die in early embryonic development28, the role of SOX2 in lung had not been understood until recently. Conditional deletion of SOX2 in lung epithelium in mice recently revealed that SOX2 is required for proliferation/differentiation of airway epithelial cells29,30. In addition to the mouse data, the amplification of SOX2 in human lung SCC31,32 clearly indicates that SOX2 regulates genes involved in normal lung morphogenesis and lung SCC. Since SOX2 controls tumorigenesis in multiple cancers in addition to lung SCC, multiple groups have reported SOX2 downstream genes that are involved in tumor growth in different cancer types. However, such analyses are often based on a limited number of cancer cell lines, thus the proposed SOX2 targets are not regulated by SOX2 in other cell lines that are not included in the analyses. In order to obtain highly conserved SOX2 downstream targets instead, we decided to investigate gene expression profiles of a large number of lung SCC specimens combined with NSCLC cell lines, which would identify genes positively/negatively correlated with SOX2 universally in lung SCC cells. The RNA-seq data of the 105 NSCLC cell lines, including 4 lung SCC cell lines, were obtained from 675 human cancer cell lines, including non-NSCLC cell lines, reported by Klijn et al.15. Ideally, gene expression profiles of only lung SCC cell lines should be employed; however, due to the limited number of definitively classified lung SCC cell lines (4 cell lines), we include the entire 105 NSCLC cell lines, including not definitively classified cell lines such as “lung carcinoma” and “non-small cell lung carcinoma”, for the analysis. However, in the 4 lung SCC cell lines, the mean expression of CDKN1A (expression = 27.610) in SOX2-LOW group (LOU-NH91 and SK-MES-1) was higher than the mean expression of CDKN1A (expression = 5.787) in the SOX2-HIGH group (NCI-H2170 and NCI-H520), indicating that SOX2 is inversely related with CDKN1A even in this limited number of lung SCC cell lines. The results were further biologically validated using multiple lung SCC cell lines, suggesting that our approach using gene expression profiles of the 105 NSCLC cell lines provide statistical power to identify potential genes correlated with SOX2. In addition to determining SOX2 downstream genes in lung SCC cell, our approach will be useful for identifying genes regulated by SOX2 in other cancer cells or genes regulated by any other transcription factors in multiple cancer cells.
In addition to CDKN1A, BMP2, SNAI1 and VIM were also negatively correlated with SOX2 in our data analysis. VIM and SNAI1 are known to induce Epithelial-Mesenchymal Transition (EMT). The EMT is understood as a process that induces tumor invasion and metastasis33,34. The loss-of-function study showed that SOX2 inhibited VIM and another EMT-related factor ZEB1 in vitro. These results suggest a dual-role of SOX2, in which SOX2 suppresses lung SCC invasion and metastasis (tumor suppressor) while SOX2 also promotes lung SCC proliferation (oncogene). Further investigation in vitro and in vivo is required to elucidate this dual function of SOX2 in lung SCC tumorigenesis. TNFSF10 (also known as TRAIL) that was negatively correlated with SOX2 in the 105 NSCLC cell lines (Fig. 2) but not in the 178 lung SCC specimens is known to induce apoptosis; however, SOX2 siRNA did not induce apoptosis in the lung SCC cell lines that we investigated (Supplemental Figure 6), suggesting that TNFSF10 may be a SOX2 downstream gene in non-lung SCC cells. These results indicate that combining multiple RNA-seq datasets with biological experiments is useful to identify unbiased transcriptional downstream targets and their associated biological functions.
Among the genes whose expression that we identified as positively correlated with SOX2 (BMP7, CHAC1, DHX9, GPX2, MARK1, MSH6, PRKX, RMND5A, SIAH2, TMPRSS4 and USP39) in our bioinformatical analysis using both the 178 lung SCC specimens and 105 NSCLC cell lines, all of them were originally reported as top 50 genes that are positively correlated with SOX2 in stage I/II lung SCC11, indicating that our approach is valid and useful to identify gene correlation and potential downstream targets in lung SCC cells. Further biological analysis is required to determine whether the above 11 genes are functionally relevant to SOX2 in lung SCC cells.
Our and others’ data clearly indicate that inhibition of SOX2 suppresses lung SCC growth in vitro and in vivo. However, our present data indicate that the suppression of lung SCC growth by SOX2 inhibition is mediated by cell cycle arrest (via CDKN1A induction) but not apoptotic cell death, suggesting that inhibition of SOX2 may not eliminate lung SCC. In addition, SOX2 inhibition slows lung SCC growth and may therefore lead to tumor resistance to chemotherapy that targets rapidly dividing cells. Furthermore, SOX2 inhibition induced genes related to EMT, which may cause tumor invasion and metastasis. Thus, targeting of SOX2 as a therapy for lung SCC should be taken cautiously. So far, SOX2 has been reported as a better prognosis marker for lung SCC by three groups and a worse prognosis marker by one group8,35,36,37,38. Identification of therapeutic targets to induce cell death and/or inhibit EMT in addition to targeting SOX2 may be required to eradicate SOX2-expressing lung SCC.
In summary, we bioinformatically combined previous reports on SOX2 gene regulation in multiple cancer cells with two gene expression datasets from 178 lung SCC specimens and 105 NSCLC cell lines and identified SOX2 downstream genes, one of which influences cell growth of multiple lung SCC cells. Further bioinformatical approaches, non-biased by previous reports, will make it possible to identify additional SOX2 downstream targets, which will lead to full understanding of the mechanisms of growth of lung SCC that harbors SOX2 amplification and/or highly expressed SOX2 and identify potential therapeutic molecular targets to eradicate lung SCC.
Materials and Methods
Bioinformatical determination of SOX2 positively or negatively correlated genes in the TCGA human lung squamous cell carcinoma (SCC) samples
The RNA-seq data of human lung squamous cell carcinoma (n = 178) were downloaded from The Cancer Genome Atlas (TCGA) data portal https://tcga-data.nci.nih.gov/docs/publications/lusc_2012/gaf.gene.rpkm.20111213.csv.zip. Expression values were measured in RPKM. For details on the original processing of the data, refer to the Supplemental Information of the original paper14. Expression values were quantile normalized using the “normalize.quantiles” function in the Bioconductor package preprocessCore (http://www.bioconductor.org/packages/release/bioc/html/preprocessCore.html). We divided the 178 samples into two groups based on their SOX2 expression values; the SOX2-HIGH group contained 89 lung SCC samples with their SOX2 expression greater than the median in the 178 samples (expression >55.594); and the remaining 89 lung SCC samples comprised the SOX2-LOW group. Two-tailed Welch’s t-test was used to determine the significance of gene expression between the two groups. Genes with significantly higher expression in SOX2-HIGH group (p < 0.05 and average expression >1) were considered as SOX2 positively correlated genes, while genes with significantly higher expression in SOX2-LOW group (p < 0.05 and average expression >1) were considered as SOX2 negatively correlated genes.
Bioinformatical determination of SOX2 positively or negatively correlated genes in the human non-small cell lung cancer (NSCLC) cell lines
The RNA-seq data of human cancer cell lines (n = 675) were downloaded from ArrayExpress using accession code E-MTAB-2706 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2706). 105 out of 675 samples were selected as non-small cell lung cancer (NSCLC) cell lines for analysis based on the following sample annotations; “OrganismPart” is lung and “Disease” is lung carcinoma, lung adenocarcinoma, lung anaplastic carcinoma, non-small cell lung carcinoma, squamous cell lung carcinoma, large cell lung carcinoma, lung mucoepidermoid carcinoma, lung papillary adenocarcinoma, lung adenosquamous carcinoma, bronchoalveolar adenocarcinoma, or squamous cell carcinoma. Among the 105 NSCLC cell lines, four were lung SCC cell line, including SK-MES-1, NCI-H520, NCI-H2170 and LOU-NH91. Expression values were measured in RPKM. For details on the original processing of the data, refer to the original paper15. The expression values were quantile normalized using the “normalize.quantiles” function in the Bioconductor package preprocessCore. We divided the 105 NSCLC cell lines into two groups based on their SOX2 expression; SOX2-HIGH group contained 20 NSCLC cell lines with their SOX2 expression greater than the mean in the 105 NSCLC cell lines (expression >12.195; e.g., NCI-H2170 and NCI-H520 lung SCC cell lines); and the remaining 85 NSCLC cell lines comprised SOX2-LOW group (e.g., LOU-NH91 and SK-MES-1 lung SCC cell lines). Two-tailed Welch’s t-test was used to determine the significance of gene expression between the two groups. Genes with significantly higher expression in SOX2-HIGH group (p < 0.05 and average expression >1) were considered as SOX2 positively correlated genes, while genes with significantly higher expression in SOX2-LOW group (p < 0.05 and average expression >1) were considered as SOX2 negatively correlated genes.
Cell Lines and Culture Conditions
The human lung SCC cells H520, H226, the human lung carcinoma cells A549 and human foreskin fibroblast HFF1 were obtained from the American Type Culture Collection (Manassas, VA) and grown in RPMI 1640 (H520 and H226) or high glucose Dulbecco’s modified Eagle medium (A549 and HFF1) supplemented with 10% heat-inactivated fetal bovine serum. The lung SCC cells EBC1, EBC2, SQ5, LK2 were kindly provided by Dr. Kiura Katsuyuki (Department of Respiratory medicine, Okayama University Graduate School of Medicine and Dentistry, Okayama, Japan) and grown in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum. All cell lines were cultured in 10% CO2 at 37 °C.
Immunohistochemistry
Sections were sequentially deparaffinized through a series of xylene, graded ethanol and water immersion steps. After being autoclaved in target retrieval solution (Dako, Carpinteria, CA, USA) for 15 minutes, sections were incubated with 3% hydrogen peroxide for 5 minutes to block endogenous peroxidase activity. A primary antibody specific for human SOX2 and CDKN1A was obtained from Cell Signaling Technology (Beverly, MA). Specimens were incubated overnight at 4 °C with a 1:50 dilution of SOX2 or CDKN1A antibody followed by three washes with TBS. The slides were treated with streptavidin-biotin complex (Envision System labeled polymer, horseradish peroxidase [HRP], Dako, Carpinteria, CA) for 60 minutes at room temperature. Immunoreactions were visualized using a 3,3′-diaminobenzidine (DAB) substrate-chromogen solution (Dako Cytomation Liquid DAB Substrate Chromogen System, Dako) and counterstained with hematoxylin. Sections were immersed in an ethanol and xylene bath and mounted for examination. For immunohistochemistry analysis, 40 lung SCC tissue sections were obtained from patients diagnosed with lung SCC who underwent surgical resection at Kawasaki Hospital, Okayama, Japan. The experimental protocol was approved by the Ethics Review Committee of Kawasaki Medical School (Ethics Committee reference number: 1310) and all experiments were performed in accordance with relevant guidelines and regulations of Kawasaki Medical School. The informed consent was obtained from all subjects.
siRNA mediated inhibition of SOX2 and CDKN1A
Cells were plated in a 6-well plate at a density of 3 × 105 per well and cultured overnight at 37 °C. The following day 100 pmol of the different SOX2 siRNA (#1: D-011778-01, #2: D-011778-02), CDKN1A siRNA (#1: D-003471-01, #2: 003471-02) or nontargeting siRNA (siCtrl; Thermo Scientific) were transfected using 7.5 μl of Lipofectamine RNAi MAX reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Incubation time for transfection reagents was 24 hours, at which time the medium was replaced with fresh regular medium. Cells were harvested 48 hours after transfection for immunoblotting or colony formation assays.
Adenoviral vectors
The recombinant adenoviral vector Ad-CMV/SOX2 (Ad-SOX2) was obtained from VECTOR BioLabs (Malvern, PA) and the optimal multiplicity of infection (MOI) was determined by infecting each cell line with Ad-CMV/GFP and assessing the expression of GFP. The experimental protocol was approved by the institutional biosafety committee of Kawasaki Medical School (Reference number: 14-07) and carried out in accordance with the approved guidelines.
Cell viability assay
Cells were plated in 12-well plates at a density of 1 × 105 cells and cultured overnight at 37 °C. The following day 100 pmol of two different SOX2 siRNA described above was transfected using 3.25 μl of Lipofectamine RNAi MAX reagent (Invitrogen, Life Technologies) according to the manufacturer’s instructions. Cells were harvested 48 hours after transfection and viable cells were assessed by a TC20 automated cell counter (Bio-Rad, Hercules, CA)39.
Colony formation assay
Cells were plated in a 6-well plate at a density of 2 to 3 × 105 per well and cultured overnight at 37 °C. The following day 100 pmol of different kinds of SOX2 siRNA described above were transfected using 7.5 μl of Lipofectamine RNAi Max reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Incubation time for transfection reagents was 24 hours, at which time the medium was replaced with fresh regular medium. Cells were harvested 48 hours after transfection, counted, plated in triplicate at a density of 5 × 102 cells in 6-well plates for 14 days. The cells were then stained with Diff-Quik (Sysmex, Kobe, Japan)40. Colonies (a group of aggregated cells numbering at least 50) were then counted. The mean number of the control group was arbitrarily set to 100% and all other numbers were normalized and percentage-specific cytotoxicity compared to colony formation in the control group was calculated.
Immunoblot analysis
Cells were lysed in ice-cold M-PER lysis buffer purchased from Thermo Fisher Scientific (Rockford, IL). Cell lysates were clarified by centrifugation (20 min at 15,000 × g at 4 °C) and protein concentration determined using the BCA protein assay (Thermo Fisher Scientific). Equal amounts of protein were separated on an SDS-PAGE gel. The gel was electrophoretically transferred to a Hybond PVDF transfer membrane (GE. Healthcare Ltd., Piscataway, NJ) and incubated with primary and secondary antibodies according to the Supersignal® West Pico chemiluminescence protocol (Pierce, Rockford, IL). Antibody specific for β-actin was obtained from Sigma (St. Louis, MO) and antibody specific for SOX2, CDKN1A, CDKN1B, CDH1 and VIM were obtained from Cell Signaling Technology (Beverly, MA). Antibody specific for ZEB1 and TP53 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary horseradish peroxidase-conjugated antibodies were obtained from Jackson Immunoresearch Laboratories (West Grove, PA).
Real time PCR
Total RNA from the cultured cells was obtained using TRIzol (Invitrogen, Life Technologies, Carlsbad, CA). Two μg of total RNA was used for reverse transcription. Reverse transcription was performed at 37 °C for 15 min using PrimeScript RT reagent Kit (Takara Bio, Inc., Shiga, Japan). Specific probes for SOX2 (Hs01053049_s1), CDKN1A (Hs00355782_m1) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs03929097_g1) for TaqMan Gene Expression Assays were obtained from Applied Biosystems (Life Technologies, CA). The real-time PCR reactions were carried out in a 48-well microtiter plate using the TaqMan Gene Expression Master Mix (Applied Biosystems). The reaction was performed in triplicate for each sample. The fluorescence of the PCR products was detected by TaqMan One-Step RT-PCR. The number of cycles for the amplification plot to reach the threshold limit (Ct value) was used for quantification. GAPDH was used as endogenous control.
Flow cytometric analysis for cell cycle and apoptosis
For cell cycle analysis, cells were plated in 6-well plates at a density of 2 × 105 cells per well and cultured overnight at 37 °C. The following day 100pmol of SOX2 siRNA #1 and/or CDKN1A siRNA #1 was transfected using 7.5 μl of Lipofectamine RNAi Max reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Nontargeting siRNA (siCtrl) was used as a control. 48hours after transfection, cells were harvested and washed once with PBS. Cells were resuspended in PBS containing 0.2% Triton X-100 and 1mg/ml RNase for 5min at room temperature and then stained with propidium iodide at 50μg/ml to determine DNA cell cycle using a FACS Verse (BD Bioscience, San Jose, CA). Doublets, cell debris and fixation artifacts were gated out and DNA cell cycle was determined using FACSuite Version 1.0.2.2238. To detect apoptosis, 100pmol of SOX2 siRNA #1 or #2 was transfected. Cells were analyzed by flow cytometry 48hours after transfection staining with AnnexinV-FITC conjugate (BD Bioscience) and propidium iodide according to manufacturer’s protocol. Non-targeting siRNA (siCtrl) was used as control.
Mouse experiments
LK2 or EBC2 lung squamous carcinoma cells were plated in 10 cm dishes at a density of 4 × 106 per dish and cultured overnight at 37 °C. The following day 580 pmol of the two different SOX2 siRNA described above or nontargeting siRNA (siCtrl; Thermo Scientific) was transfected using 43 μl of Lipofectamine RNAi Max reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Incubation time for transfection reagents was 24 hours, at which time medium was replaced with fresh regular medium. Cells were harvested and washed once with PBS and then resuspended in culture medium. Human lung cancer xenografts were established in 6-wk-old female BALB/c nude mice (Charles River Laboratories Japan, Kanagawa, Japan) by subcutaneous (s.c.) inoculation of siRNA (SOX2 #1, SOX2 #2 or non-targeting) transfected LK2 or EBC2 lung SCC cells (2 × 106 cells/200 μl) into the right dorsal flank. Tumors were measured two times a week and tumor volume was calculated as a x b2 × 0.5, where a and b were large and small diameters, respectively. The experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of Kawasaki Medical School (Ethics Committee reference number: 14-005) and carried out in accordance with the approved guidelines.
Statistical analysis
Statistically significant differences between means and medians of the study groups were evaluated using Student’s t-test except for Figs 1 and 2. Statistical significance was defined as p < 0.01 (*).
Additional Information
How to cite this article: Fukazawa, T. et al. SOX2 suppresses CDKN1A to sustain growth of lung squamous cell carcinoma. Sci. Rep. 6, 20113; doi: 10.1038/srep20113 (2016).
References
Lewis, D. R., Check, D. P., Caporaso, N. E., Travis, W. D. & Devesa, S. S. US lung cancer trends by histologic type. Cancer. 120, 2883–2892 (2014).
Perez-Moreno, P., Brambilla, E., Thomas, R. & Soria, J. C. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 18, 2443–2451 (2012).
Scagliotti, G. V. et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 26, 3543–3551 (2008).
Sandler, A. et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 355, 2542–2550 (2006).
Pikor, L. A., Ramnarine, V. R., Lam, S. & Lam, W. L. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung cancer. 82, 179–189 (2013).
Mantripragada, K. & Khurshid, H. Targeting genomic alterations in squamous cell lung cancer. Front Oncol. 3, 195 (2013).
Hussenet, T. et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PloS One. 5, e8960 (2010).
Sholl, L. M., Long, K. B. & Hornick, J. L. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 18, 55–61 (2010).
Zhang, X. et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 5, 445–457 (2013).
Chen, Y. et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 283, 17969–17978 (2008).
Bass, A. J. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 41, 1238–1242 (2009).
Tian, Y. et al. SOX2 oncogenes amplified and operate to activate AKT signaling in gastric cancer and predict immunotherapy responsiveness. J Cancer Res Clin Oncol. 140, 1117–1124 (2014).
Herreros-Villanueva, M. et al. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2, e61 (2013).
Comprehensive genomic characterization of squamous cell lung cancers. Nature. 489, 519–525 (2012).
Klijn, C. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 33, 306–312 (2015).
Chen, S. et al. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PloS One. 7, e36326 (2012).
Otsubo, T., Akiyama, Y., Yanagihara, K. & Yuasa, Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer. 98, 824–831 (2008).
Chen, S. et al. SOX2 regulates apoptosis through MAP4K4-Survivin signaling pathway in human lung cancer cells. Carcinogenesis. 35, 613–623 (2014).
Fang, W. T. et al. Downregulation of a putative tumor suppressor BMP4 by SOX2 promotes growth of lung squamous cell carcinoma. Int J Cancer. 135, 809–819 (2014).
Han, X. et al. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PloS One. 7, e41335.
el-Deiry, W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 75, 817–825 (1993).
Kimura, S., Ward, J. M. & Minoo, P. Thyroid-specific enhancer-binding protein/thyroid transcription factor 1 is not required for the initial specification of the thyroid and lung primordia. Biochimie. 81, 321–327 (1999).
Maeda, Y., Dave, V. & Whitsett, J. A. Transcriptional control of lung morphogenesis. Physiol Rev. 87, 219–244 (2007).
Jonckheere, N. et al. The mouse Muc5b mucin gene is transcriptionally regulated by thyroid transcription factor-1 (TTF-1) and GATA-6 transcription factors. FEBS J. 278, 282–294 (2011).
Maeda, Y. et al. Airway epithelial transcription factor NK2 homeobox 1 inhibits mucous cell metaplasia and Th2 inflammation. Am J Respir Crit Care Med. 184, 421–429 (2011).
Maeda, Y. et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest. 122, 4388–4400 (2012).
Snyder, E. L. et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 50, 185–199 (2013).
Avilion, A. A. et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 (2003).
Que, J., Luo, X., Schwartz, R. J. & Hogan, B. L. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 136, 1899–1907 (2009).
Tompkins, D. H. et al. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated and goblet cells. PloS One. 4, e8248 (2009).
Yuan, P. et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PloS One. 5, e9112 (2010).
Gontan, C. et al. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol. 317, 296–309 (2008).
Thiery, J. P., Acloque, H., Huang, R. Y. & Nieto, M. A. Epithelial-mesenchymal transitions in development and disease. Cell. 139, 871–890 (2009).
Lim, J. & Thiery, J. P. Epithelial-mesenchymal transitions: insights from development. Development (Cambridge, England) 139, 3471–3486 (2012).
Velcheti, V. et al. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PloS One. 8, e61427 (2013).
Toschi, L. et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PloS One. 9, e95303 (2014).
Wilbertz, T. et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 24, 944–953 (2011).
Chen, Y. et al. The prognostic value of SOX2 expression in non-small cell lung cancer: a meta-analysis. PloS One. 8, e71140 (2013).
Kosmac, K. et al. Glucocorticoid treatment of MCMV infected newborn mice attenuates CNS inflammation and limits deficits in cerebellar development. PLoS Pathog. 9, e1003200 (2013).
Menges, C. W. et al. Group I p21-activated kinases (PAKs) promote tumor cell proliferation and survival through the AKT1 and Raf-MAPK pathways. Mol Cancer Res. 10, 1178–1188 (2012).
Chou, Y. T. et al. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells. 31, 2607–2619 (2013).
Watanabe, H. et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 124, 1636–1645 (2014).
Santini, R. et al. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene. 33, 4697–4708 (2014).
Acknowledgements
This study was supported by the Ministry of Education, Science and Culture, Japan to T. Fukazawa and Y. Naomoto and by the American Lung Association and Cincinnati Children’s Hospital Medical Center to Y. Maeda. The authors thank M. Durbin (University of California, Irvine) for discussions and M. Iwai, A. Yuge and Y. Kishimoto for technical assistance.
Author information
Authors and Affiliations
Contributions
E.Y., N.M., T.I. and M.G. performed the experiments. T.F. and Y.M. designed the study. T.F., Y.M. and M.G. wrote the manuscript. T.Y., M.T., M.H. and T.O. provided technical help. M.G., N.I. and T.F. performed statistical analysis. N.T. and Y.N. provided scientific discussion. All authors contributed to the discussion and review of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Fukazawa, T., Guo, M., Ishida, N. et al. SOX2 suppresses CDKN1A to sustain growth of lung squamous cell carcinoma. Sci Rep 6, 20113 (2016). https://doi.org/10.1038/srep20113
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20113
This article is cited by
-
USP13 drives lung squamous cell carcinoma by switching lung club cell lineage plasticity
Molecular Cancer (2023)
-
Strategies for understanding the role of cellular heterogeneity in the pathogenesis of lung cancer: a cell model for chronic exposure to cigarette smoke extract
BMC Pulmonary Medicine (2022)
-
SOX2 knockdown with siRNA reverses cisplatin resistance in NSCLC by regulating APE1 signaling
Medical Oncology (2022)
-
Clinical application of a lung cancer organoid (tumoroid) culture system
npj Precision Oncology (2021)
-
G1-phase progression in pluripotent stem cells
Cellular and Molecular Life Sciences (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.