Abstract
DNMT3L is an important epigenetic regulator in mammals, integrating DNA methylation and histone modification based epigenetic circuits. Here we show DNMT3L to be a part of the machinery that enables inheritance of epigenetic modifications from one generation to the next. Ectopic expression of DNMT3L in Drosophila, which lacks DNMT3L and its normal interacting partners DNMT3A and DNMT3B, lead to nuclear reprogramming that was gradual and progressive, resulting in melanotic tumors that were observed only when these flies were maintained for five generations. This global gene expression misregulation was accompanied by aberrations in the levels of H3K4me3 and H3K36me3, globally as well as at specific gene promoters. The levels of these epigenetic aberrations (epimutations) also increased progressively across successive generations. The accumulation and inheritance of epimutations across multiple generations recapitulates the important role of DNMT3L in intergenerational epigenetic inheritance in mammals.
Similar content being viewed by others
Introduction
For proper functioning of a eukaryotic cell, all the genes must be expressed at their optimum levels. This is achieved by maintaining the epigenetic profile of each genetic loci appropriately made possible by the coordinated cross talk between the various epigenetic components. There are several proteins that can read or write epigenetic modifications in response to cues from other epigenetic modifications associated with a specific locus1. Amongst these, DNMT3L is an epigenetic modifier that interacts with de novo methyltransferases DNMT3A/3B and binds to histone H3 in a region that contains H3K4 to bring about DNA methylation2,3,4. It has also been found to be involved in setting up of methylation imprints in mice during gametogenesis5,6.
Previous work from our laboratory has shown that overexpression of DNMT3L in mammalian cell lines causes nuclear reprogramming7. Importantly, the DNMT3L dependent reprogramming was gradual and the morphological phenotypic changes were observed only 20 passages after transfection7. Along with finding out the reason for this gradual and progressive nuclear reprogramming we were interested in identifying the mechanism by which DNMT3L achieves it. In particular, we wanted to probe whether the interactions of DNMT3L with DNMT3A/3B on one hand and histone H3 on the other were redundantly influencing a common subset of genes or whether each of these interactions influenced a different subset of genes. The interaction of DNMT3L with histone H3 in the absence of de novo DNA methyltransferases could either be examined in mammals (mice or cell lines) deleted for DNMT3A and 3B or in an organism that normally lacks these proteins. Deletion of DNMT3A and 3B causes lethality in mice8,9 and cell lines are prone to epigenetic instability10. Therefore, we decided to examine these questions in Drosophila that lacks DNMT3A/3B as well as DNMT3L. This was important, as it allowed us to probe the interaction of DNMT3L exclusively with histone H3 in transgenic Drosophila that ectopically expressed DNMT3L. Moreover, it also allowed us to use the various genetic tools available for this well characterized model organism.
In this study we show that ectopic expression of DNMT3L in Drosophila leads to nuclear reprogramming even in the absence of de novo DNA methyltransferases. Similar to our previous observation in mammalian cells, the reprogramming was gradual and progressive with the phenotypic consequences being observed only after maintaining ectopic expression of DNMT3L in Drosophila for at least five generations. Reduction in the levels of H3K4me3 and H3K36me3 along with genome-wide misregulation of genes was progressive, indicating accumulation of altered epigenetic modifications (epimutations) across multiple generations. Interestingly, these epimutations were passed onto the next generation only by the mother, emphasizing the importance of DNMT3L in the establishment of parent-of-origin-specific epigenetic inheritance in mammals.
Results
Transgenic Drosophila expressing DNMT3L display mild wing phenotype
To understand the functional consequence of DNMT3L interaction with H3, transgenic Drosophila carrying the DNMT3L transgene (cloned in the pUAST vector under the control of an hsp70 promoter that contained GAL4 binding sites [UAS]) was generated by P-element mediated germline transformation (Methods). After injection into w1118Drosophila embryos, 6 independent transgenic Drosophila lines with DNMT3L (referred to UAS-3L here after) integrated at various loci on different chromosomes were established (Supp Table S1). For constitutive expression of DNMT3L, the different UAS-3L transgenic lines were crossed with flies constitutively expressing GAL4 including Tubulin-GAL4, Actin-GAL4 and daughterless-GAL4. The progeny Tubulin-GAL4>DNMT3L, Actin-GAL4>DNMT3L and daughterless-GAL4>DNMT3L from these crosses are referred to as Tub-3L, Actin-3L and da-3L respectively in this manuscript. Tissue-specific expression of DNMT3L was achieved by crossing these transgenic DNMT3L flies to GMR-GAL4 (eye-specific) and hml-GAL4 (Hemolymph-specific) driver stock flies.
Transgenic flies constitutively expressing DNMT3L (Tub-3L, Actin-3L, da-3L) were scored for possible phenotypic defects. In many progeny, phenotypic defect was observed only in the wings. In 50–100% of the progeny, the defect was the appearance of extra vein(s) (Fig. 1, Supp Table S2). The localization of these extra veins differed in individual progenies (Fig. 1). In approximately 2–3% of the progeny, we also observed broken wing phenotype (Supp. Fig. S1).
Ectopic DNMT3L expression causes wing phenotype in the transgenic Drosophila.
Comparison of wings from control and transgenic DNMT3L flies showing extra veins in the DNMT3L expressing flies. Tub-3L, Actin-3L, da-3L - transgenic DNMT3L flies with Tubulin, Actin or daughterless GAL4 driver respectively. Extra veins in the wings are marked by arrows.
Ectopic expression of DNMT3L causes melanotic tumors in Drosophila but only in the 5th generation
Since we had observed a gradual and cascading nuclear reprogramming over 20 passages in HeLa cell overexpressing DNMT3L7, we decided to examine the effect of ectopic DNMT3L expression in transgenic Drosophila maintained across several generations. Homozygous transgenic Tubulin-DNMT3L lines (Tub-3L) were bred by self-crossing progeny in each generation. Until the 4th generation (G4), extra veins in the wings or broken wings were the only phenotypes observed as a consequence of the ectopic DNMT3L expression. No other morphological abnormalities were observed during any of the embryonic, larval or pupal stages till G4. However, in the 5th generation some of the 3rd instar larvae (~5%) had black masses (Fig. 2, Tub-3L) that resembled melanotic tumors11. None of these larvae with melanotic tumors survived beyond the larval stages. No pupae or adults flies had melanotic tumors and the remaining progeny developed into normal adults (with several showing the mild wing phenotype) and were fertile. In all the subsequent generations (maintained till G20), 5–8% of the 3rd instar larvae consistently showed melanotic tumors and did not survive whereas the rest of the progeny were normal and fertile (Table 1). Real-Time RT-PCR and Western blot analysis showed that the expression of DNMT3L remained constant in all the generations from G1 to G5 (Supp. Figs S2, S3) suggesting that the appearance of the larvae with tumors in 5th generation progeny was not due to an abrupt change in its expression.
Ectopic DNMT3L expression causes melanotic tumors in transgenic flies from the fifth generation.
Images of 3rd instar larvae from DNMT3L transgenic flies in the absence and presence of GAL4 drivers. Melanotic tumors were observed in DNMT3L transgenic flies in the larval stage after maintaining the flies for five generations. The melanotic tumors are marked by arrows. The location of the melanotic tumors varied in different larvae. UAS-3L - transgenic DNMT3L flies without any GAL4 driver. Tub-3L, Actin-3L, da-3L - transgenic DNMT3L flies with Tubulin, Actin or daughterless GAL4 driver respectively.
It was possible that the appearance of melanotic tumors in the DNMT3L transgenic flies only from the 5th generation was due to (i) an unrelated genetic mutation in these flies; (ii) genomic context of the DNMT3L transgene; or (iii) due to the GAL4 driver line used. To rule out the role of an unlinked genetic mutation in the transgenic flies, the Tubulin GAL4 driver was removed from the transgenic DNMT3L flies after 20 generation (G20) by crossing Tub-3L flies to double balancer flies (Pin/Cyo; Tm2/Tm6). No progeny (referred to as UAS-3L* here after) with melanotic tumors was observed in any larvae in subsequent generations (Table 2). This was true for both heterozygous (in G21) and homozygous UAS-3L* (G22 onwards) progeny (Table 2) indicating that the melanotic tumors were due to ectopic expression of DNMT3L.
The role of the GAL4 driver or the genomic context of the DNMT3L transgene in the observed phenotype was ruled out by repeating these crosses with transgenic DNMT3L flies that had DNMT3L transgene integrated at other genomic loci (Suppl Table S1) and crossing them to the constitutive Actin-GAL4 and da-GAL4 driver flies. In all the crosses with both Actin and da-GAL4 drivers, progeny with melanotic tumors were again observed only in G5 and subsequent generations (Fig. 2; Tables 3 and 4).
The melanotic tumors were found to be present in the hemolymph, the circulatory fluid in Drosophila that contains free floating hemocytes. To examine the influence of DNMT3L expression on the number and types of hemocytes in the hemolymph, the hemolymph from control UAS-3L flies was compared with that of 5th generation Tub-3L flies with or without melanotic tumors. The number of hemocytes were increased in Tub-3L flies (both with and without melanotic tumors; Fig. 3A). The number of proliferating cells as indicated by PH3 antibody staining was also found to be significantly more in G5, Tub-3L larvae (Fig. 3B). The types of hemocytes present in the hemolymph was markedly different with the Tub-3L flies showing a large numbers of lamellocytes (Fig. 3C).
Ectopic DNMT3L expression affects the number and types of hemocytes.
(A) Florescent microscopic images of Alexa 488-Phalloidin (stains Actin) stained hemocytes of control UAS-3L and the DNMT3L expressing G5, Tub-3L transgenic flies. The nuclei of the cells was counterstained with DAPI. Note the increase in the number of hemocytes in the DNMT3L expressing Tub-3L and Tub-3L-tumor transgenic flies. (B) PH3 positive (proliferating) cells in control and DNMT3L expressing 3rd instar larvae. (C) Hemocytes stained with Phosphohistone 3 (PH3) from 5th generation control and DNMT3L expressing larvae (with melanotic tumors). Note the increase in the number of lamellocytes in the larvae bearing the tumors. UAS-3L, G5 - 5th generation transgenic UAS-DNMT3L flies (without any GAL4 driver). Tub-3L, G5: 5th generation transgenic flies expressing DNMT3L with Tubulin-GAL4 driver. Tub-3L-tumor, G5P-: 5th generation transgenic Tub-3L flies expressing DNMT3L with melanotic tumors. The error bars represent Standard Deviation (S.D.). *Indicate significant difference (Student’s t test, ***p < 0.001).
Phenotypic defects in transgenic Drosophila expressing DNMT3L in a tissue-specific manner also appear in G5
As the melanotic tumors were found to be present in the hemolymph, we wanted to test whether the effect of DNMT3L was only limited to this tissue in Drosophila. To investigate this UAS-3L flies were crossed with tissue-specific, hemolymph (Hml) and eye (GMR)-specific, GAL4 driver flies. No phenotypic abnormality (not even the wing phenotype) was observed in the progeny of these crosses in G1. For hml-GAL4>DNMT3L lines (hml-3L), 5–7% of the 3rd instar larvae in the 5th and subsequent generations had melanotic tumors as was observed in crosses with the constitutive GAL4 driver lines (Table 5). No melanotic tumors in the hemolymph were observed in the 3rd instar larvae of flies expressing DNMT3L in the eye (GMR-GAL4>DNMT3L and referred to as GMR-3L), but all the progeny in the 5th and subsequent generations showed rough eye phenotype (Fig. 4, Table 6). This indicated that the effect of ectopic DNMT3L expression was not limited to the hemolymph alone. Though DNMT3L was expressed in eyes of Tub-3L, Actin-3L or da-3L flies, its expression was much lower than in GMR-3L line (Supp. Fig. 4). This may be one of the reasons why we did not observe the rough eye phenotype in Tub-3L, Actin-3L or da-3L lines. More importantly, the phenotypic consequence of tissue-specific ectopic DNMT3L expression was also manifested only in G5 and later generations, similar to what was seen with constitutive DNMT3L expression.
Tissue-specific expression of DNMT3L in eyes causes rough eye phenotype in 5th generation flies.
(A) Scanning electron microscope images of eye from control (UAS-3L & GMR-GAL4) and DNMT3L expressing (GMR-3L) transgenic Drosophila. Images were captured at 200X in the upper panel. Zoomed images for the same samples were taken at 1000X and are shown in the lower panel. GMR-GAL4 - Flies containing only the GMR-GAL4 driver. UAS-3L, G5 - 5th generation transgenic UAS-DNMT3L flies (without any GAL4 driver). GMR-3L, G5: 5th generation transgenic flies expressing DNMT3L with GMR-GAL4 driver (Eye-specific).
Ectopic expression of DNMT3L leads to transcriptional misregulation that progressively increases with each generation
To investigate the reasons for the delay in the phenotypic consequences of ectopic expression of DNMT3L in Drosophila, transcriptional profiling of transgenic DNMT3L larvae from the different generations was performed using Affymetrix Drosophila Gene 1.1 ST array (Methods, Data has been submitted to Gene Expression Omnibus (GEO), NCBI, see Supplementary information for details). RNA isolated from third instar larvae (of approximately same stage) of (i) transgenic Drosophila expressing DNMT3L in various generations (Tub-3L; G1, G2, G4, G5 and G5-phenotype); (ii) control transgenic flies not expressing DNMT3L (UAS-3L; G1 & G5); (iii) G20 transgenic flies after removal of GAL4 driver (UAS-3L*, G20*) (iv) wild type control flies (w1118), were analyzed (see flowchart in Supp. Fig. S5). Expression profiles of larvae in a particular generation was compared to G1 control larvae (UAS-3L; G1) to identify misregulated genes by scatter plots (Fig. 5). We noticed a progressive increase in the number of genes that were misregulated upon DNMT3L expression in successive generation. From 205 in G1, the number of misregulated genes progressively increased to 2873 in G5. This number further increased to 3730 in G5 larvae with melanotic tumors (Supp Table S3). It is interesting to note that in all generations subsequent to G1, the number of genes showing repression were relatively more than the overexpressed genes (Supp Table S3). That the observed misregulation was due to the DNMT3L expression was reiterated by our finding that the numbers of genes that were misregulated dramatically reduced in G20 larvae (UAS-3L*) after the removal of the tubulin-GAL4 driver and only 74 genes were aberrantly expressed, most of which did not show altered expression in flies expressing DNMT3L in any other generations.
Misregulation of gene expression in flies expressing DNMT3L is gradual and progressive.
(A–F) Scatter plots showing the upregulated and downregulated genes in each generation of the DNMT3L expressing transgenic flies. Comparison was made between gene expression levels in 3rd instar Tub-3L larvae from a particular generation (indicated on the Y Axis) with UAS-3L control larvae from G1. Note a progressive increase in the number of genes that were aberrantly expressed in successive generations. Green crosses represent upregulated genes while red crosses represent down regulated genes. Number of genes misregulated was minimal in G20 flies that lacked DNMT3L expression due to the removal of the GAL4 driver. G1, G2, G4, G5, denotes Tub-3L larvae from the particular generation. G5P denotes 5th generation Tub-3L larvae with melanotic tumors. G20*-UAS-3L*- denotes G20 flies after removal of the GAL4 driver by crossing flies with double balancers.
Heat map analysis of the gene expression profiles for various generations provided an important clue about DNMT3L function. Comparison of duplicates (RNA isolated from 2 groups of 3rd instar larvae, Fig. 6A) for each generation showed almost the same gene expression profile. That DNMT3L expression in Drosophila resulted in aberrant expression of the same sets of genes in each generation in each larvae suggested that DNMT3L was acting on a specific set of genes.
Nuclear reprogramming in DNMT3L expressing Drosophila: Microarray data analysis.
(A) Heat map depicting hierarchical clustering of differentially expressed genes in the various samples and examined by microarray analysis. The heat map shows the relative expression levels of each transcript in each samples. Green denotes downregulated, red denotes upregulated and black depicts genes that showed no change with respect to their expression level in control G1 UAS-DNMT3L transgenic larvae. Note the similarity in the heat map profile of the biological replicates for each generation. Sample numbers given above the profile denote: are as follows 1- G1-UAS-3L; 2 to 5 –Tub-3L larvae from generation G1, G2, G4 and G5; 6 – G5-Tub-3L larvae with tumors, 7 – G5-UAS-3L and 8- G20*-UAS-3L* larvae from 20th generation where GAL4 driver was removed. (B,C) Venn diagrams depicting genes upregulated (B) and downregulated (C) in DNMT3L expressing flies across various generations. The number of genes that remain misregulated in successive generations increased progressively (see also Supplementary Table 4). G1 to G5 depict generation number, G5Phen denotes genes misregulated in G5 Tub-3L larvae that had melanotic tumors.
Of the 205 genes that showed altered expressed in G1, 103 genes were upregulated and 102 were down regulated. Only 1 out of the 103 upregulated genes maintained higher than normal expression in the subsequent generation. Similarly, only 3 out of 102 downregulated genes remained so in later generations. (Fig. 6B,C). The gene expression of the remaining 201 misregulated genes was either not altered or was misregulated in the opposite direction. From approximately 2% (4 out of 205) in G1 this percentage increased to 32% for the genes that were misregulated in G2 and continued to be similarly misregulated in subsequent generations. The majority of the genes (approximately 81%) aberrantly expressed in G4 continued to be so in G5 and/or in G5 larvae with melanotic tumors indicating a gradual, but progressively increased influence of ectopic DNMT3L expression.
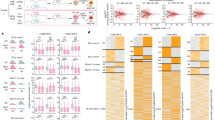
Several genes and gene families were affected by ectopic DNMT3L expression including the Hox gene family, piwi group of proteins, genes involved in Wnt pathway, wing development, cell cycle, cell proliferation, chromosome organization, Polycomb and Trithorax group of proteins, etc. (Fig. 7). Drosophila has 8 Hox genes12 and in our microarray analysis 7 of them (except dfd) were downregulated (Fig. 7A). Similarly, all the 7 Wnt genes13 were expressed at a significantly lower level after G2 (Fig. 7B). The Polycomb (PcG) and Trithorax (TrX) group of genes are epigenetic modulators of gene expression and include readers and writers of the epigenetic circuitry14,15. Several members of the PcG/TrX family were found to be misregulated (Fig. 7C–H). Of these, all the PhoRC subfamily members (involved in tethering these complexes to specific DNA sequences) showed a gradual but significant increase in their expression (Fig. 7C). The formation of melanotic tumors in Drosophila has been associated with hemocyte-mediated immune response16. Several immune response (IR) and immune induced (IM) genes were dysregulated across the various generations (Fig. 7I,J). In addition, the dysregulation of genes involved in wing development, cell cycle and cell proliferation correlated with the phenotypic consequences of DNMT3L ectopic expression in Drosophila (Fig. 7K–M). Interestingly, all the piwi group of proteins, which are involved in transgenerational epigenetic inheritance17, were found to be upregulated (Fig. 7N).
Genes and gene families misregulated across multiple generations in DNMT3L expressing transgenic flies.
Graphical representation of gene expression changes (shown as fold change with respect to G1) for the depicted genes across the various generations. Hox gene family (A), wnt pathway genes (B), Polycomb and Trithorax group of proteins (C–H), Immune response (IR) & Immune induced (IM) genes (I,J), genes involved in wing development (K), cell proliferation (L) and cell cycle (M) and piwi group of proteins (N) were analysed. Y-axis shows fold change with respect to gene expression of the specific gene in control G1 UAS-3L (without Tubulin-GAL4 driver) larvae. G1 and G5-UAS-3L are control larvae are without GAL4 driver from G1 and G5 generation respectively. G1 to G5- Tub-3L larvae from the indicated generation. G5P- G5 Tub3L larvae that had melanotic tumors. G20-UAS-3L* denotes larvae from G20 generation after crossing out of the Tubulin-GAL4 driver. The profile of genes that showed change of gene expression are denoted by blue connecting lines and red connecting line is used to depict genes that do not show a consistent change in gene expression across the various generation. The names of genes examined and their symbols are depicted in the legend for each graph.
During the analysis of gene expression profiles for individual genes and gene families across different generation we found that the alteration in the expression of most genes was not initiated from G1 but after G2. That the escalation of the aberration at molecular level happened after G2 and not in G1 indicated that a secondary event occurred after 2 generations of ectopic DNMT3L expression.
Epigenetic changes upon ectopic DNMT3L expression are inherited across more than one generation
As DNMT3L has been shown to influence DNA methylation in mammalian cells, we first examined the effect of ectopic DNMT3L expression on DNA methylation across successive generations by MeDIP using methyl cytosine antibody. No increase in the level of DNA methylation was detected in any of the generations at the genomic as well as at specific gene promoter level (Supp. Figs S6, S7).
The other known epigenetic partner of DNMT3L is histone H3, particularly when H3 is unmethylated at H3K43. To examine whether ectopic DNMT3L expression had any effect on H3 methylation at H3K4, we probed 3rd instar Drosophila expressing DNMT3L for the level of H3K4me3. As can be seen in the representative western blot (Fig. 8A), the level of H3K4me3 had significantly reduced in tumor bearing Tub-3L (G5) larvae as compared to the control UAS-3L (G5) larvae. This observation was reinforced by immuno-staining of polytene chromosome with H3K4me3 antibody where negligible H3K4me3 staining was observed for the polytene chromosome in DNMT3L expressing Tub-3L flies (Fig. 8B).
Accumulation of aberrant histone methylation in DNMT3L expressing Drosophila across successive generations.
(A) Western blot analysis demonstrating decrease in the levels of the H3K4me3 in 3rd instar larvae or adult flies expressing DNMT3L from the 5th generation as compared to control UAS-3L flies lacking the GAL4 driver. Histone H3 was used as loading control. (B) Immunostaining of Drosophila polytene chromosomes with H3K4me3 antibody. (C) Western blot analysis for the various histone modification as indicated, performed on larvae from the various generations of control and DNMT3L expressing Drosophila larvae. The densitometric scan values for each signal was calculated and is represented graphically in (D) as fold change with respect to the level of the respective histone modification in G1-UAS-3L flies lacking GAL4 driver. G1 and G5-UAS-3L are control larvae without GAL4 driver from G1 and G5 generation respectively. G1 to G5- Tub-3L larvae from the indicated generation. G5P- G5 Tub-3L larvae that had melanotic tumors. G20-UAS-3L* denotes larvae from G20 generation after crossing out of the Tubulin-GAL4 driver. Actin was used as a loading control. The error bars represent Standard Deviation (S.D.). *Indicate significant difference (Student’s t test, *p < 0.05, **p < 0.01).
While the phenotypic consequence of DNMT3L expression was only observed in G5, the aberrant expression of several genes was observed as early as G2. Western blot analysis was performed to investigate whether the H3K4me3 levels were altered in other generations as well. In this analysis, we also tested the levels of other known active (H3K4me2, H3K36me3) and repressive (H3K9me3 and H3K27me3) histone marks18,19. Repressive chromatin marks H3K9me3 and H3K27me3 were unaffected due to DNMT3L expression but the levels of the active histone marks H3K4me3, H3K4me2 and H3k36me3 were dramatically reduced in tumor bearing G5 3rd instar larvae (Fig. 8C). Furthermore, densitometric scanning of the western blots (Fig. 8D) revealed that the reduction in H3K4me3 and H3K36me3 was initiated in G3 and the decrease was progressively more in the successive generations with the maximum decrease being observed in tumor bearing 3rd instar larvae in G5. Thus, decrease in the overall levels of active histone modifications in G3 correlated with the escalation of genome-wide aberrant gene expression after G2.
Western blot analysis provided the genome-wide picture of histone modifications profile. To probe the association of histone marks with the promoters of specific genes, chromatin immunoprecipitation (ChIP) was carried out using antibodies to specific histone modifications. The association of DNMT3L with the promoters of these genes was also examined by ChIP. 9 genes that were either upregulated (kdm4a, piwi, Suv3(3) and esc) or downregulated (abd-A, mamo, wgn, cyr and wg) were chosen for the analysis. ChIP analysis was also done for 3 other genes (bch, hem and sky) whose expression was unaffected. DNMT3L was found to be associated with the promoters of all the genes that were aberrantly expressed in DNMT3L expressing transgenic flies and in all the generations (Fig. 9A). That this association was genuine was supported by the observation that the promoters of these misreulated genes showed loss of DNMT3L association in the G20 UAS-3L* flies where the Tubulin-GAL4 driver had been crossed out (Fig. 9A). We noticed difference in the profile of DNMT3L binding to the promoters of upregulated and downregulated genes across different generations. While all promoters (for both up and down regulated) showed significant increase in DNMT3L association in G1 Tub-3L flies as compared to control UAS-3L flies, the level of DNMT3L associated with the promoters of upregulated genes plateaued in subsequent generations. A slight modification in this trend for upregulated genes was noticed for the piwi promoter where further increase in DNMT3L association was observed in G2 followed by plateauing of its association in subsequent generations (Fig. 9A). On the other hand, DNMT3L association with 3 of the 5 down regulated genes promoters was distinctly increased in the tumor bearing G5 larvae as compared to the other generations (Fig. 9A). Amongst the down regulated genes, the association of DNMT3L with the promoter of abd-A increased progressively with each successive generation.
ChIP analysis for association of DNMT3L and specific Histone modifcations with specific gene promoters.
Graphical representation of the DNMT3L binding (A), association of H3K4me3 (B) and H3K36me3 (C) with the promoter and HeK36me3 within the exonic regions (D) of selected upregulated, downregulated and unaffected genes tested by ChIP using specific antibodies. Results are represented as % input. The names of the specific gene promoters tested are given below the X-axis. U1- control larvae without GAL4 driver from the 1st generation G1, G3 and G5- Tub-3L larvae from the indicated generation. G5P denotes 5th generation Tub-3L larvae having melanotic tumors. G20* denotes larvae from G20 generation after crossing out of the Tubulin-GAL4 driver. The error bars represent Standard Deviation (S.D.). *Indicate significant difference (Student’s t test, *p < 0.05, **p < 0.01).
For the same set of promoters, ChIP analysis was also carried out using H3K4me3, H3K36me3 and H3K27me3 antibodies (Fig. 9A–C). Since H3K36me3 levels in the exonic regions has been associated with regulation of gene expression20, we also examined H3K36me3 levels within the exonic regions of these genes (Fig. 9D). As we had noted in the western blot analysis, the association of H3K27me3 with the various promoters was not altered across the different generations (Supp. Fig. S8) and was in agreement with the microarray data that showed no change in the expression level of E(Z), the H3K27 methyltransferase. At the genome-wide level H3K4me3 and H3K36me3 levels were drastically decreased in tumor bearing G5 larvae (Fig. 8). Two different trends were noticeable for H3K4me3 and H3K36me3 (both promoter and exonic region). While the association of H3K4me3 and H3K36me3 with specific gene promoter was found to have decreased for 3 out 5 the down regulated genes in the tumor bearing G5 larvae (Fig. 9B,C), the promoters of upregulated genes showed an increase in H3K4me3 and H3K36me3 levels in the tumor bearing G5 larvae. This was also true for the association of H3K36me3 with the exonic regions (Fig. 9D). This was a surprising result as association of DNMT3L led to opposite transcriptional outcomes and epigenetic profiles for these two different sets of genes.
DNMT3L is known to be an epigenetic reader and binds to H3 only when H3K4 is in the unmethylated state, a state that is associated with inactive chromatin and transcriptional repression3. Our results would indicate that DNMT3L can associate with specific genetic loci even in the presence of H3K4me3. Moreover, as the association of DNMT3L with different gene promoters led to changes in the level of H3K4me3, it would suggest that DNMT3L binding can also influence the level of H3K4me3.
DNMT3L binding within the promoter of the affected genes was observed in G1 and all successive generations but changes in the levels of H3K4me3 and H3K36me3 was noticeable only from G3. This may suggest that DNMT3L likely influences H3K4me3 and H3K36me3 indirectly by modulating the expression of H3K4me3 and H3K36me3 specific histone methyltransferases or demethylases. This contention was backed by our observation that the change in the expression of histone H3K4 and H3K36 methyltransferases and demethylases, especially Su(var)3-3 and Kdm4a, mirrored the decrease in the levels of H3K4me3 and H3K36me3 (Supp. Fig. S9).
The modENCODE database21 for Drosophila has catalogued histone modifications associated with specific genetic loci within the Drosophila genome for the various adult tissue of the adult flies as well as for embryo and larvae at different developmental stages (Comprehensive encyclopedia of genomic functional elements in the model organisms C. elegans and D. melanogaster; http://www.modencode.org/; date of access, 15/9/15). Using the data available for 3rd instar larvae in the modENCODE database, we extracted the normal epigenetic profile of the genes that were found to be misregulated in DNMT3L expressing flies across the various generations. The epigenetic profile of a region 1000 bp on either side of the Transcriptional Start Site (TSS) for each gene was captured and used for the analysis. Of the 205 genes (103 up and 102 down regulated) misregulated in G1, 153 (~75%) were unmethylated at H3K4. For the complete set of Drosophila genes that were represented on the microarray, only 46% of the gene promoters were unmethylated at H3K4. This suggested that in G1, DNMT3L was targeting gene promoters devoid of H3K4me3. However, in successive generations this bias was progressively removed (Table 7). In fact, DNMT3L binding was found to be biased for H3K4me3 promoters in tumor bearing G5 larvae as 63% of the misregulated gene promoters were found to be trimethylated at H3K4 (Table 7). When the same analysis was performed on misregulated genes after categorizing them as up or down regulated, it was noticed that while both sets showed bias for unmethylated H3K4 in G1, the bias favoring H3K4me3 in the following generation was only seen in the down regulated genes. For the upregulated genes the bias for unmethylated H3K4 was removed progressively and by G5 almost equal number of genes were associated or devoid of H3K4me3 (Table 7).
Inheritance of epimutations between two generations in DNMT3L expressing Drosophila is dependent upon piwi
The progressive decrease in H3K4me3 and H3K36me3 indicated that alteration in epigenetic modifications (epimutations) were being inherited from one generation to another. Recent studies have implicated the role of piwi associated piRNAs in transgenerational inheritance of epigenetic characters22,23. Examination of the microarray data indicated that all the members of the piwi group of proteins were upregulated in DNMT3L expressing flies (Fig. 7N). To investigate the role of piwi protein, G5, Actin-3L flies were crossed with piwi mutant flies (piwi06843cn1/CyO; ry506) and the progeny of this cross (piwi-Tub-3L) were scored for melanotic tumors. As shown in Table 8, piwi-Actin-3L flies carrying mutated piwi gene and DNMT3L transgene did not produce any progeny that had melanotic tumors implicating piwi protein in the inheritance of epimutations. Expression of DNMT3L was unchanged in the piwi-Tub-3L flies and similar to what was observed in G5, Tub-3L flies.
Maternal transmission of epimutations
We next investigated whether the epimutations were being inherited through both the germ lines or were being passed on to the progeny by one parent. The phenotypic consequences of ectopic DNMT3L expression in Drosophila appeared only in the 5th generation even though the epimutations (loss of H3K4me3 and H3K36me3) were observed as early as the 3rd generation. Moreover, the extent of these epimutations increased progressively in subsequent generations. Appearance of tumors only in the 5th generation and in each generation thereafter would indicate that from the 5th generation onwards a threshold level of epimutations necessary for the phenotype accumulates and inheritance of which causes tumors in the progeny. If the threshold level of the epimutations was achieved only in G5 and were being passed on to the progeny, then in a cross between G5 Tub-3L and G1 Tub-3L flies, some of the progeny should show tumors. Based on this contention, the following crosses were set up: (i) G5-Tub-3L ♀ X G1-Tub-3L ♂– to check for maternal transmission; (ii) G1-Tub-3L ♀ X G5-Tub-3L ♂ – to check for paternal transmission; (iii) G1-Tub-3L ♀ X G1-Tub-3L ♂– negative control; (iv) G5-Tub-3L ♀ X G5-Tub-3L ♂ – positive control. The results from these crosses are presented in Table 9. As expected G5-Tub-3L ♀ X G5-Tub-3L ♂ produced 5–6.9% larvae that had melanotic tumors. None of the 3rd instar larvae from the crosses G1-Tub-3L ♀ X G1-Tub-3L ♂ had tumors. Progeny from G1-Tub-3L ♀ X G5-Tub-3L ♂ did not show tumors but 2.9–3.7% of the progeny from the G5-Tub-3L ♀ X G1-Tub-3L ♂ developed melanotic tumors indicating maternal inheritance of the epimutations.
Discussion
The inheritance of epigenetic modifications across multiple generations results from events during germ cell development and fertilization that allow epigenetic marks acquired during the life time of an organism to be passed on to the progeny24. Epigenetic inheritance could be intergenerational as has been documented for genomic imprinting25 or transgenerational where epigenetic modification acquired in one generation is inherited unchanged across several generations26,27,28. In both cases, limited knowledge is available on the components and mechanism by which epigenetic modifications cross the meiotic barrier between two generations29. A few recent studies have found piwi group of proteins and piRNA to be involved22,23, however the exact mechanism(s) still needs to be dissected out. Our observation that ectopic expression of DNMT3L in Drosophila resulted in initiation and maternal inheritance of epimutations across several generations not only establishes the role of DNMT3L in intergenerational epigenetic inheritance, but also provides a reason to reevaluate the theories that discuss the need for genomic imprinting in placental mammals.
DNMT3L facilitates nuclear reprogramming even in the absence of de novo DNA methyltransferase
The functional consequence of DNMT3L action is mediated by its interactions with de novo methyltransferases DNMT3A and DNMT3B and histone H32,3,4. Previous work from our laboratory had indicated a role of DNMT3L in nuclear reprogramming in mammalian cell lines7. In the present study, ectopic DNMT3L expression in Drosophila (DNMT3L is not present in the Drosophila genome) caused progressive misregulation of genes and by the 5th generation a very large number of genes (3730) were aberrantly expressed. Some of the transgenic flies expressing DNMT3L also developed melanotic tumors and did not survive. Drosophila has negligible DNA methylation30 and lacks the de novo DNA methyltransferases DNMT3A and DNMT3B as well as the maintenance DNA methyltransferase DNMT1. While the methyltransferase, DNMT2, is present in Drosophila, it has been shown to be a tRNA methyltransferase31. Our results show no change in DNA methylation (globally as well as at specific gene level) in any of the generations indicating that even in the absence of an endogenous functional DNA methyltransferase, the interaction of DNMT3L with histone H3 is sufficient for its nuclear reprogramming capability. It would also suggest that the interaction of DNMT3L with DNMT3A and DNMT3B versus the binding of DNMT3L with histone H3 may be redundant and it would be of interest to determine whether or not both events would influence the same subset of genes.
DNMT3L facilitates intergenerational inheritance of epimutations
Nuclear reprogramming observed in DNMT3L expressing Drosophila (this study) and mammalian cells overexpressing DNMT3L7 was gradual and progressive. However, an important difference exists between the two results. While the nuclear reprogramming in mammalian cells occurred across several mitotic divisions7, the progressive misregulation of genes across several generations in transgenic Drosophila involved germline passage through meiotic divisions. Most epigenetic marks are normally erased and reset in the germline though some studies have shown transgenerational germline passage of epigenetic marks32,33,34,35,36,37. The inheritance of aberrant epigenetic modifications or epimutations across several generations that we observed in this study would fit into the broad definition of transgenerational epigenetic inheritance. However, an important criterion for categorizing inheritance of epigenetic marks as transgenerational is the ability of epigenetic marks to survive germline passage across several generations in the absence of the causative environmental or genetic cue in the subsequent generations32. In this study, inheritance of epimutations across successive generation was observed, but only when the causal DNMT3L expression was present. In G20, when the GAL4 driver was removed from the transgenic flies, the aberrant expression of genes and the epimutations disappeared in the absence of DNMT3L expression. This would indicate that the inheritance of epimutations was intergenerational rather than transgenerational. An important example of intergenerational inheritance is genomic imprinting and DNMT3L has been linked to setting up of methylation imprints in the germ line of mammals (mice)5,6. Genomic imprinting represents a phenomenon wherein epigenetic modifications are inherited to the progeny in a parent-of-origin-specific manner38. In transgenic Drosophila, epimutations were inherited from the mother to her progeny suggesting of a possibility that expression of DNMT3L had allowed a process similar to genomic imprinting to be initiated. Like in mammals, the formation of germ cells in Drosophila, takes place at a time during embryogenesis when the embryo is still under the influence of maternal factors39. Therefore, the possibility also exists that the inheritance of epimutations were due to the influence of maternally produced DNMT3L. While further work is needed to distinguish the two possibilities, a closer examination of our result suggests that the possibility of a process similar to genomic imprinting cannot be ruled out. From the 5th generation onwards, 5–8% of the DNMT3L expressing larvae showed melanotic tumors. However, in our cross where G5 females with G1 males only 3–4% of progeny showed tumors (Table 9). If only the epimutations passed on through the female germline were responsible for the phenotype then in this cross the percentage of larvae showing melanotic tumors should have remained in the 5–8% range. Decrease in this percentage could suggest that some epimutations at specific genomic loci may be being passed on through the paternal germline. Since we were looking only at the gross phenotypic levels examination of the intergenerational inheritance of epimutations at specific loci would help in distinguishing between the two possibilities.
DNMT3L allows accumulation of epimutations across more than one generation
As mentioned earlier, established epigenetic modifications are normally reset in the germ cells before fertilization31. Therefore, any epimutation acquired during one generation would be lost in the germ cells and hence not inherited. Even for imprinted genes, the epigenetic imprints inherited from the previous generation are erased before new epigenetic modifications determined by the sex of the individual are initiated. However, we observed a progressive decrease in the genome-wide levels of H3K4me3 and H3K36me3 across more than one generation. This was also true at specific gene levels. For example, the level of H3K4me3 at the promoter of the hox gene, abd-A, decreased progressively across successive generations. Similar inheritance of reduced H3K4 methylation across several generations in mice was recently reported40. The accumulated decrease in these modifications (or accumulation of epimutations) suggested that in the presence of DNMT3L new epigenetic modifications were being added over and above of what was inherited from the previous generation. Previous studies on DNMT3L have suggested its role in initiating or setting up of methylation imprints in mammalian germ cells5. Based on this, one possible explanation for epimutation accumulation could be incapacitation of protein(s) responsible for erasure or resetting of epigenetic modification, as a secondary event in Drosophila expressing DNMT3L. Hence, in each generation DNMT3L would keep adding new epimutations to the ones inherited from the previous generation without being erased. On the other hand, it is possible that the presence of DNMT3L prevented erasure of epimutations inherited from the previous generation. In this scenario, epigenetic modifiers responsible for setting up of new epigenetic marks in the germ cells would add on to the epigenetic modifications that were not erased due to the presence of DNMT3L. Careful examination of the temporal expression of DNMT3L in mammalian germ cells or derivation of transgenic Drosophila expressing DNMT3L in a very narrow developmental window immediately before or after the time when epigenetic modifications are reset in the germline would help in answering this question. Moreover, further work to understand, (i) how epimutations were inherited in hml-3L and GMR-3L flies where DNMT3L expression was restricted to hemolymph and eyes respectively; and (ii) how epimutations that were accumulated over multiple generations reverted back immediately once the ectopic expression of the DNMT3L transgene was abolished, would help in understanding the mechanism underlying DNMT3L action.
Whether it is in placental mammals, angiosperms or a few species of insects, the phenomenon of genomic imprinting has been invoked as a means to regulate the parasitic relationship that the embryo has with its mother41. If the possibility that DNMT3L allowed parent-of-origin-specific intergenerational inheritance of epigenetic modifications, was found to be true then it would be prudent to reevaluate the reason as to why some species have chosen genomic imprinting as a means of regulating gene expression.
Methods
Fly stocks and crosses
DNMT3L cDNA with the FLAG tag was PCR amplified from pcDNA3.1-DNMT3L7 and sub-cloned into the Drosophila cloning vector pUAST under the UAS containing hsp70 promoter. This vector also contains the reporter mini-white gene for the selection of the transgenic lines. Transgenic flies, using the Drosophila strain W1118, were generated by standard protocol involving P-element mediated germline transformation42. After injection, 6 independent transgenic lines, with integration of DNMT3L transgene on various chromosomes, were established. The induction of the DNMT3L in different lines (UAS-FLAG-DNMT3L) was carried out by crossing these lines with constitutive GAL4 driver lines like Tubulin-GAL4, Actin-GAL4 and daughterless-GAL4 or with tissue specific GAL4 driver like GMR-GAL4 (eye-specific) and hml-GAL4 (haemolymph-specific). All GAL4 driver and the piwi mutant (piwi06843cn1/CyO; ry506) lines were obtained from the Bloomington stock center.
Crosses to get transgenic flies expressing DNMT3L
Initially, the UAS-DNMT3L flies and the GAL4 driver flies were crossed to the double balancer flies to get the desired marker on each chromosomes. The desired flies from both these crosses were further crossed with each other to bring DNMT3L and the GAL4 driver into the same flies. The resulting flies were then self-crossed with each other to obtain homozygous DNMT3L flies with the appropriate GAL4 driver. In case of the crosses with Tubulin-GAL4 and da-GAL4 driver flies (Tubulin-GAL4 and da-GAL4 driver are present on the 3rd chromosome), the DNMT3L line 38.1.1 in which the DNMT3L transgene was on the second chromosome was used. For Actin-GAL4 and hml-GAL4 (Actin-GAL4 and hml-GAL4 driver on 2nd chromosome, the DNMT3L line 42.1.2 with the transgene on the third chromosome was used. For crosses with GMR-GAL4 (GMR driver present on the 1st chromosome), the DNMT3L line 38.1.1 in which the DNMT3L transgene was on the second chromosome was used.
Phenotypic Analysis
Drosophila wing mounting
Wings were removed from both control and transgenic DNMT3L flies and incubated in 10% KOH at 70 °C for 5 minutes followed by washing with PBS. The treated wings were mounted in Canada balsam on a glass slide and photographed using Zeiss Axiocam camera.
Imaging of Drosophila eyes by Scanning Electron Microscope (SEM)
Drosophila heads for imaging were fixed in 2.5% Glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 24 hours followed by incubation in 2% aqueous osmium tetraoxide for 4 hours. The samples were serially dehydrated in alcohol and dried to critical point drying with CPD unit. The processed samples were mounted on the stubs with double sided carbon conductivity tape and coated with gold in an automated sputter coater for 3 minutes. Images were acquired using a Scanning Electron Microscope (SEM- Model JOEL-JSM 5600) at 200X or 1000X at RUSKA Laboratory, College of Veterinary Sciences, Hyderabad, India.
Hemolymph analysis
Third instar larvae were washed with PBS followed by 70% ethanol and blotted dry. The larvae were dissected by gently pulling the epidermis apart with the help of forceps to ooze out the hemolymph on a glass slide. The hemolymph was air dried and further processed. For Phalloidin staining of the hemolymph the cells in the dried hemolymph were fixed in 4% paraformaldehyde for 10 minutes. Excess paraformaldehyde was removed from cells by giving three PBST (PBS with 0.1% triton X-100) washes. A 1:500 dilution of Alexa 488 conjugated anti-Phalloidin antibody (Life Technologies) was added to the sample, the slide was incubated in a humidified chamber for 30 minutes followed by three PBST washes. The slides were mounted with DAPI and observed under florescent microscope. For phospho-histone 3 staining of the hemocytes, 1:100 dilution of mouse anti-PH3 antibody (Millipore) and 1:250 dilution of anti-mouse Alexa Flour 488 (Life Technologies) were used for the immunostaining.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as described previously43. Briefly, 3rd instar larvae was homogenised and then sonicated in Biorupter (Diagenode) biorupter so that DNA was sheared between 200–500 bp. Chromatin immunoprecipitation was performed on the sonicated sample using auto ChIP kit (Diagenode) in the IPStar automated ChIP machine (Diagenode) using specific antibodies.
Antibodies and primers used for the study
The Flag antibody was purchase from Sigma (F1804). H3 (ab1791), H3k4me3 (ab8580), H3K9me3 (ab8898), H3K27me3 (ab6002), H3K36me3 (ab9050) and beta-actin (ab8227) antibodies were purchased from Abcam. Oligonucleotides used in this study are provided as supplementary data.
Polytene chromosome immunostaining
Immunostaining of the polytene chromosome from control and the transgenic larvae was done following established protocol44. Incubation with the primary antibodies against Flag tag (Sigma), histone H3 and H3K4me3 (Abcam) was done overnight at 4 °C.
Microarray analysis
Isolation of RNA from Drosophila
The RNA for the microarray analysis was isolated from third instar larvae using TRI reagent (Sigma). The RNA was further purified using purelink RNA mini kit (Life Technologies). The RNA for the various samples and their duplicates was from 20–30 larvae at approximately same stage of 3rd instar larval development. The total RNA was analysed for gene expression profiling using the Affymetrix Drosophila Gene 1.1 ST array (Affymetrix, Santa Clara, CA) at Imperial Life Sciences, India. This array contained 362,078 probe sets representing more than 15,309 transcripts, including all known genes. Synthesis and labelling of complementary DNA targets, hybridization and scanning of GeneChip were carried out according to the instructions provided by Affymetrix.
Bioinformatic analysis
For initial gene level expression analysis, the Affymetrix CEL-files were first imported into Affymetrix Expression Console Software, version 1.3. The Robust Multichip Analysis (RMA) algorithm was applied to the probe cell intensity data files for all experimental conditions, using default parameters in the RMA-sketch workflow for core gene level analysis. Analysis of differentially expressed genes was performed with Affymetrix Transcriptome Analysis Console (TAC) Software. Significance of the difference for each gene was determined by one-way ANOVA. Genes that were differentially regulated by 2 fold or greater with a p-values ≤ 0.05 were included in the further analysis. The scatter plot for the various upregulated and downregulated genes for the comparison between the transgenic flies without GAL4 driver and the transgenic flies of various generation with Tubulin-GAL4 driver was made using the Transcriptome Analysis Console (TAC) 2.0 software. Pathway analysis was done using DAVID Bioinformatics Resources 6.745 and Gorilla46. The Venn diagrams were created using the web tools provided in the Bioinformatics and Evolutionary Genomics facility website (Van de Peer Y. et al. Calculate and draw custom Venn diagrams; http://bioinformatics.psb.ugent.be/webtools/Venn/; date of acess, 15/9/15).
modENCODE (Comprehensive encyclopedia of genomic functional elements in the model organisms C. elegans and D. melanogaster; http://www.modencode.org/, date of access, 15/9/15) database for Drosophila melanogaster was searched for the availability of experiments for H3K27me3, H3K36me3, H3K4me2 and H3K4me3 histone modifications in 3rd instar larval stage. A total of 89 dataset were found relevant and downloaded for further comparisons. The coordinates for 1000 bases upstream of TSS and 1000 bases downstream of TSS of up and down regulated genes identified using microarray data genes were calculated. BEDtools was used to identify regions that showed presence of a particular modification in at least 50% of the data sets and designated as regions associated with the specific histone modification whereas all other regions were taken be regions lacking that modification.
For validation of the array data, cDNA synthesis was performed on DNase treated RNA and qRT-PCR (ABI 7500) was set up for the indicated misregulated genes.
Additional Information
How to cite this article: Basu, A. et al. DNMT3L enables accumulation and inheritance of epimutations in transgenic Drosophila. Sci. Rep.6, 19572; doi: 10.1038/srep19572 (2016).
Change history
21 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-08888-5
References
Musselman, C. A., Lalonde, M. E., Cote, J. & Kutateladze, T. G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012).
Gowher, H., Liebert, K., Hermann, A., Xu, G. & Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280, 13341–13348 (2005).
Ooi, S. K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
Hu, J. L. et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc. Natl. Acad. Sci. USA 106, 22187–22192 (2009).
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Arima, T. et al. Loss of the maternal imprint in Dnmt3Lmat-/- mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 297, 361–373 (2006).
Gokul, G., Ramakrishna, G. & Khosla, S. Reprogramming of HeLa cells upon DNMT3L overexpression mimics carcinogenesis. Epigenetics. 4, 322–329 (2009).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Dodge, J. E. et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability and spontaneous immortalization. J. Biol. Chem. 280, 17986–17991 (2005).
Humpherys, D. et al. Epigenetic instability in ES cells and cloned mice. Science 293, 95–97 (2001).
Minakhina, S. & Steward, R. Melanotic mutants in Drosophila: pathways and phenotypes. Genetics 174, 253–263 (2006).
Mallo, M. & Alonso, C. R. The regulation of Hox gene expression during animal development. Development 140, 3951–3963 (2013).
Llimargas, M. & Lawrence, P. A. Seven Wnt homologues in Drosophila: a case study of the developing tracheae. Proc. Natl. Acad. Sci. USA 98, 14487–14492 (2001).
Muller, J. & Kassis, J. A. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16, 476–484 (2006).
Schuettengruber, B., Martinez, A. M., Iovino, N. & Cavalli, G. Trithorax group proteins: switching genes on and keeping them active. Mol. Cell. Bio. 12, 799–814 (2011).
Rodriguez, A. et al. Identification of immune system and response genes and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics 143, 929–940 (1996).
Luteijn, M. J. & Ketting, R. F. PIWI-interacting RNAs: from generational to transgenerational epigenetics. Nat. Rev. Genetics 14, 523–534 (2013).
Peterson, C. L. & Laniel, M. A. Histones and histone modifications. Curr. Biol. 14, 546–551 (2004).
Imhof, A. Epigenetic regulators and histone modification. Brief. Funct. Genomics Proteomics 5, 222–227 (2006).
Kimura, H. Histone modifications for human epigenome analysis. J Hum Genet. 58, 439–445 (2013).
Celniker, S. E. et al. Unlocking the secrets of the genome. Nature 459, 927–930 (2009).
Grentzinger, T. et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome research 22, 1877–1888 (2012).
Le Thomas, A., Marinov, G. K. & Aravin, A. A. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell reports 8, 1617–1623 (2014).
Migicovsky, Z. & Kovalchuk, I. Epigenetic memory in mammals. Front Genet. 2, 28 (2011).
Radford, E. J. et al. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 8, e1002605 (2012).
Rakyan, V. K. et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 100, 2538–2543 (2003).
Xing, Y. et al. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 3, 1598–1606 (2007).
Greer, E. L. et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479, 365–371 (2011).
Daxinger, L. & Whitelaw, E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 20, 1623–1628 (2010).
Lyko, F., Ramsahoye, B. H. & Jaenisch, R. DNA methylation in Drosophila melanogaster. Nature 408, 538–540 (2000).
Goll, M. G. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006).
Cavalli, G. & Paro, R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286, 955–958 (1999).
Ruden, D. M., Garfinkel, M. D., Sollars, V. E. & Lu, X. Waddington’s widget: Hsp90 and the inheritance of acquired characters. Semin Cell Dev Biol. 14, 301–310 (2003).
Sollars, V., Lu, X., Xiao, L., Wang, X., Garfinkel, M. D. & Ruden, D. M. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat. Genet. 33, 70–74 (2003).
Heard, E. & Martienssen, R. A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 (2014).
Stern, S. et al. Reduction in maternal Polycomb levels contributes to transgenerational inheritance of a response to toxic stress in flies. The Journal of Physiology 592, 2343–2355 (2014).
Szyf, M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol. Med. 21, 134–144 (2015).
Barlow, D. P. & Bartolomei, M. S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, doi: 10.1101/cshperspect.a018382. (2014).
Togashi, S., Kobayashi, S. & Okada, M. Functions of maternal mRNA as a cytoplasmic factor responsible for pole cell formation in Drosophila embryos. Dev. Biol. 118, 352–360 (1986).
Siklenka, K. et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 350, 651- 665 (2015).
Haig, D. The kinship theory of genomic imprinting. Annual Review of Ecology and Systematics 31, 9–32 (2000).
Voie, A. & Cohen, S. Germ-line Transformation of Drosophila melanogaster. Cell Biology: A Laboratory Handbook, Second Edition 3, 510–517 (1997).
Basu, A., Dasari, V., Mishra, R. K. & Khosla, S. The CpG island encompassing the promoter and first exon of human DNMT3L gene is a PcG/TrX response element (PRE). PloS one 9, e93561 (2014).
James, C., Badenhorst, P. & Turner, B. Preparation and Immunostaining of Polytene Chromosome Squashes (2015) (Prot 1) (Date of access: 15/9/15) Available: http://www.epigenesys.eu/en/protocols/fluorescence-microscopy/186-preparation-and immunostaining-of-polytene-chromosome-squashes.
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 4, 44–57 (2009).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Acknowledgements
We would like to thank Dr. Indira Paddibhatla for help in the Drosophila hemolymph experiments. Help from Srikanth, Rajeev and Dr Rohit Joshi in Drosophila experimentation is acknowledged. A.B. is the recipient of Junior and Senior Research Fellowships of the Council of Scientific and Industrial Research and is registered with the Manipal University for his PhD degree. This work was supported by a grant by Department of Biotechnology, Government of India to SK and RKM.
Author information
Authors and Affiliations
Contributions
A.B. performed the experiments. A.T. and A.B. analysed the microarray and modENCODE data. V.D. helped in some of the Drosophila experiments. The experiments were designed and conceptualized by S.K. and R.K.M. S.K. and A.B. wrote the manuscript and prepared the figures. R.K.M. provided all the Drosophila resources. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Basu, A., Tomar, A., Dasari, V. et al. DNMT3L enables accumulation and inheritance of epimutations in transgenic Drosophila. Sci Rep 6, 19572 (2016). https://doi.org/10.1038/srep19572
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19572
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.