Abstract
Gene apterous (ap), chip (chi) and beadex (bx) play important roles in the dorsal-ventral compartmentalization in Drosophila wing discs. Meanwhile, Notch signaling is essential to the same process. It has been reported that Ap and Chi function as a tetramer to regulate Notch signaling. At the same time, dLMO (the protein product of gene bx) regulates the activity of Ap by competing its binding with Chi. However, the detailed functions of Chi and dLMO on Notch signaling and the relevant mechanisms remain largely unknown. Here, we report the detailed functions of Chi and dLMO on Notch signaling. Different Chi protein levels in adjacent cells could activate Notch signaling mainly in the cells with higher level of Chi. dLMO could induce antagonistical phenotypes on Notch signaling compared to that induced by Chi. These processes depend on their direct regulation of fringe (fng) transcription.
Similar content being viewed by others
Introduction
Notch gene was first discovered in Drosophila melanogaster. Misregulation of Notch causes a serrated wing margin phenotype1,2,3. Gene notch encodes a transmembrane surface receptor containing EGF-like repeats4. Meanwhile, most of Notch ligands are also transmembrane proteins5, by which the transduction of Notch signaling is highly depended on the neighbor cellular environment. In the canonical model, the binding of Notch with ligands promotes two proteolytic cleavage events. The first cleavage occurs on the Notch extracellular domain (NECD)6. Truncation of NECD stimulates the second cleavage and releases the Notch intracellular domain (NICD). NICD then enters the nucleus and forms a complex with CSL and Mastermind to promote the transcription of target genes5,7.
Notch signaling activity is also affected by glycosylation. In Drosophila wing discs, fng, a glycosyltransferase, could modify the EGF modules of Notch in the Dorsal (D) compartment8,9,10,11. Its activity enhances the ability of Notch receptor to bind to its ligand Delta, which is expressed by the Ventral (V) cells. It could also decrease the sensitivity of Notch receptor to bind to Serrate, the ligand expressed by the D cells8,10,12,13,14,15. The high levels of Notch signaling are then limited to a narrow band of cells along the D/V boundary12,15,16.
Chip (Chi) is a transcription co-factor. Currently, one of the best studied transcription factors functioning together with Chi is Ap, which is a LIM-homeodomain protein17,18. The relative expression amounts of Ap and Chi are critical for D-V compartmentalization in Drosophila wing discs19,20. The Chi-Ap complex is a tetramer composed by a dimerized Chi and two Ap bridged by the dimer of Chi21,22,23,24. The activation of Chi-Ap complex is negatively regulated by dLMO in vivo, due to that dLMO competes with Ap to bind Chi22. This regulation of Ap activity is essential for the function of the Chi-Ap complex on the D-V compartmentalization during wing imaginal disc development21,22,24,25,26,27. The expression of gene fng mentioned above is initially induced in the D compartment of wing discs in the second instar larvae12. It is reported that fng is the target gene of Ap in the early dorsal. However, late expression of fng does not require Ap activity27.
Although Notch signaling has been studied for a century, some details are still unknown. For example, the details of the phenotypes induced by Chi and dLMO on Notch signaling still remain obscure as well as the mechanisms. Here, we report that the adjacent cells with different Chi protein levels could induce Notch signaling activation along the D/V boundary of wing discs. In the Ap-independent regulation of Notch signaling, dLMO induces opposite phenotypes on Notch signaling relative to Chi. Meanwhile, the functions of chi overexpression and bx RNAi on Notch signaling is limited to the regions far away from A/P boundary and this might be explained by that the interaction between Chi and dLMO is regulated by Dpp signaling. In addition, the function of Chi and dLMO on Notch signaling is dependent on their direct regulation of fng transcription. Our findings uncovered the detailed functions and the mechanism of how Chi and dLMO regulate Notch signaling in the Ap-independent manner. Meanwhile, the finding that Dpp could regulate the interaction between Chi and dLMO implied a potential crosstalk between Dpp and Notch signaling.
Results
Different Chi protein levels in adjacent cells are essential for Notch signaling activity
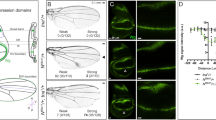
chi RNAi was found to induce serrated wing in a loss-of-function screen (Fig. S1a–S1c). To further study the function of Chi on Notch signaling, we generated a Chi antibody that was proved to work well in both immunofluorescence (IF) and immunoprecipitation (IP) assays (Fig. S2a–S2b). chie5.5 is a widely used chi mutation. To exclude the potential functions of 141 correct amino acids residual in chie5.5, we generated a new chi mutant with 26 amino acids residual named chi26 using Cas9/sgRNA system28 (Fig. S2c–S2e’). Previous study suggested that the functions of Chi on Notch signaling are different at different larvae stages27. To figure out the details, we then did the Chi staining in the early and late third instar stage, respectively. The expression level of chi is higher in the D compartment than that in the V compartment at the early third instar larvae stage. The Notch signaling classical target gene cut expressed in the junctional area between the different Chi level regions (Fig. 1a–a’’’). However, in the late third instar, Chi is equally expressed throughout the wing discs (Fig. S3a). We reasoned that the different Chi levels might contribute to the Notch signaling regulation. So we first employed chi RNAi and chie5.5to induce different Chi levels in wing discs. Since the RNAi efficiency was good (Fig. 1b–b’), Chi protein level should be higher at the outside of chi RNAi and chie5.5clones than that inside. The Notch signaling classical target genes in wing discs, cut and wg, were employed to monitor the Notch signaling activity. Consistent with previous reports, the Notch signaling activity only changed in the clones at the D compartment (Fig. 1c–f’ and Fig. S3b–S3d’). Notch signaling target genes were upregulated along the clone boundary (Fig. 1c–f’ and Figs S3b–S3d’). The statistical analysis showed that around 30% Cut upregulating cells located inside the clones, while around 70% located outside, indicating Notch signaling tends to be activated in the cells with higher Chi protein level (Fig. 1i). We then employed chi26to confirm all the aforementioned observations. All the results gotten from chi RNAi and chie5.5were reproduced in chi26 clones (Fig. 1g–h” and Fig. S3e–S3e’). In the Notch signal upregulating cells along the chi26clones, the majority of them located outside the clones (Fig. 1g–h”). Similar to the results from chi RNAi, the statistical analysis showed that around 30% Cut upregulating cells located inside the clones, while around 70% located outside (Fig. 1i).
We further reasoned that if the different Chi levels are critical for Notch signaling activation, the loss of Chi crossing over the D/V boundary should induce downregulation of Notch signaling along the D/V boundary. To knockdown the endogenous Chi at different regions of wing disc, we then employed ciGal4, hhGal4 or dppGal4 to drive chi RNAi overexpression specifically in the A, P compartment or A/P boundary regions, respectively. In these RNAi overexpression regions, Chi protein should be downregulated and the pattern of different Chi expression levels between D/V compartments should be lost. For example, when chi RNAi was overexpressed in the P compartment, the endogenous Chi was downregulated and the Chi protein should be at a very low level. The pattern of different Chi levels between D/V compartments in P compartment region should then disappear. IF staining showed that Notch target genes along the D/V boundary totally disappeared in the corresponding chi RNAi regions, indicating Notch signaling activation depends on the different Chi protein levels. In addition, since chi RNAi induced a low chi expression region compared to other regions, we also noticed that Notch signaling target genes were activated along the boundary of RNAi regions (Fig. S3f). ciGal4 and dppGal4 induced similar phenotypes as hhGal4, further confirming that the Chi protein level is essential for the Notch signaling activation (Fig. S3g–S3i). To further confirm this conclusion, we induced chi RNAi overexpression in the whole disc with actinGal4 and noticed a total loss of Cut and Wg expressions (Fig. 1j and Fig. S3j–S3k’).
Chi overexpression induces Notch signaling activation mainly inside clones
To further investigate the effect of Chi levels on Notch signaling, we employed AGGal4 to overexpress chi in the clones (Fig. 2a–a’). Compared to the endogenous Chi protein outside the clones, the overexpressed Chi protein level was higher inside the clones. In the experiment, we found that Notch signaling was also upregulated along the clone boundary (Fig. 2b–c”), only that around 70% of the Notch activated cells located inside clones (Fig. 2d). Together with loss-of-Chi results, we concluded that Notch signaling could be activated at the boundary of different Chi protein regions and tends to be activated in the cells with higher Chi protein level (Fig. 2e). In addition, chi overexpression induced by hhGal4 caused loss of Notch target genes along D/V boundary in the P compartment (Fig. S4a–S4b). These phenotypes further verified that different Chi levels are essential for Notch signaling activation.
The Chi overexpression induces Notch signaling activation mainly inside clones.
(a-a’) The overexpression of chi generates the boundary of high and low Chi protein (red) regions. Clones were marked with GFP (green) in all the IF results. (b-c”) Immunostaining of Cut (b-b”; red) and Wg (c-c”; red) in the discs of chi overexpression. Cut and Wg activation did not occur at the clones (arrows) near A/P boundary. (b’)and (c’)are the enlarged pictures of (b,c). (d) Quantitative analysis of Cut activated cells around the clones. Shown are the Means ± s.d. (e) A model for the role of Chi on Notch signaling. Notch signaling could be activated along the boundary of different chi expression regions. Around 70% of activated cells are cells with higher Chi protein level. (f-f”) Immunostaining of NICD (red) in the discs of chi26 discs. Clones are marked with dashed line. (g-h”) Immunostaining of Cut (g; red) and Wg (h; red) in the discs of chi RNAi and UAS-NICD overexpression. Cut and Wg are activated in the clones (green). Chi proteins (blue) are totally lost in the clones.
However, we noticed that phenotypes above could not be induced in the A/P boundary (Fig. 2b–c”). To further investigate this phenotype, we employed dppGal4 to induce chi overexpression. This chi overexpression caused only slight loss of Notch target genes (Fig. S4c–S4d). These phenotypes indicated that the regulation on Notch signaling by Chi levels might be regulated by signals locate in the A/P boundary, such as BMP signaling.
Chi is not involved in the Notch signaling transcription complex directly
Since Chi is a transcription cofactor, we set out to distinguish the genetic relationship between Chi and NICD. In the experiments, we found that NICD level was upregulated outside chi26 clones compared to inside (Fig. 2f–f”). When chi was overexpressed, NICD was upregulated along the clone boundary (Fig. S4e–S4e’). We then overexpressed UAS-NICD and chi RNAi together, the phenotypes caused by UAS-NICD could fully recover that caused by loss-of-Chi (Fig. 2g–h”), indicating Chi is not involved in the transcription complex and genetically locates in the upstream of NICD in Notch signaling.
Chi and dLMO induce opposite functions on Notch signaling
In order to demonstrate the regulation mechanism of Chi on Notch signaling, we set out to seek the potential factor(s) functioning with Chi. Since the phenotypes induced by Chi are limited to the D compartment, we reasoned two candidates, Ap and dLMO, which are reported to share different expression levels between D and V compartment16,18,22. We first confirmed the functions of these two proteins on Notch signaling. The RNAi or overexpression of ap induced slight changes in our experiment condition (Fig. S5a–S5f). However, bx RNAi induced cut and wg upregulation along the clone boundary in the regions away from A/P boundary and a large proportion of the Notch signaling activated cells are located inside clones (Fig. 3a–b’). These phenotypes induced by loss-of-dLMO are similar to that induced by gain-of-Chi. Meanwhile, the bx overexpression induced cut and wg upregulation along the clone boundary excepted that a large proportion of these Notch signaling activated cells located outside clones, similar to those induced by loss-of-Chi (Fig. 3c–d’). Previous study suggested that the expression of bx could downregulate fng and allows the expression of delta in the D compartment27. We then employed delta-lacZ and fng-lacZ. Along the boundary of the chi26and chi RNAi clones, delta-lacZ was upregulated, resembling those induced by the bx overexpression (Fig. 3e–g’). In addition, the chi overexpression induced delta-lacZ downregulation in the whole clones, similar to that induced by overexpression of bx RNAi (Fig. 3h–i’). For fng-lacZ, in chi26and chi RNAi clones, lacZ was downregulated dramatically, similar to that induced by bx overexpression (Fig. 3j–l’). Taken together, Chi and dLMO induce exactly opposite functions on Notch signaling.
Chi and dLMO induce opposite functions on Notch signaling.
(a-b’) Immunostaining of Cut (a-a’; red) and Wg (b-b’; red) in the discs of bx RNAi flies. Cut and Wg activation did not occur at the clones (arrows) near A/P boundary. (a’,b’) are the enlarged pictures of (a,b). In all the IF results, clones are marked with GFP (green) and marked with dashed line. (c-d’) Immunostaining of Cut (c-c’; red) and Wg (d-d’; red) in the discs of bx overexpression flies. HA tag was marked with blue (d). (c’,d’) are the enlarged pictures of (c) and (d). (e-i’) Immunostaining of delta-lacZ (red) in the discs of indicated genotype flies. The delta was activated along the clone boundaries in the bx overexpression (e-e’) and chi loss-of-function flies (f-g’). The delta was downregulated inside the whole clones in the bx RNAi (h-h’) and chi overexpression flies (i-i’). (j-l’) Immunostaining of fng-lacZ (red) in the discs of indicated genotype flies. Compared to control (j-j’), fng was downregulated inside the whole clones in the chi26 (k-k’) and chi RNAi flies (l-l’).
Chi and dLMO form a complex to regulate fng transcription
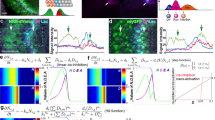
Since fng-lacZ is downregulated inside the clones of loss-of-Chi and gain-of-dLMO, we reasoned that Chi and dLMO might regulate fng transcription. To test this possibility, we first performed Co-IP to confirm whether there was an interaction between them as reported26. Since the functions of Chi and dLMO might be regulated by signals located in the A/P boundary, we performed this experiment with or without Dpp treatment, a morphogen that is expressed by the cells at the A/P boundary29,30. The results showed that Chi and dLMO could interact with each other and this interaction could be weakened by Dpp signal (Fig. 4a,b).
The next question we wanted to figure out was whether Chi and dLMO regulate the transcription of fng directly. We performed chromatin immunoprecipitation (ChIP) assay with Chi antibody and followed by qPCR. We designed 29 pairs of qPCR primers covering about 2,200 bases upstream of the transcription start site (TSS) and 2,000 bases downstream of TSS. Three peaks were observed in the result, indicating there are three binding sites between Chi and fng locus. The highest peak appeared at about 700 bases upstream of TSS. The other two peaks appeared at about 1,500 bases upstream of TSS and 800 bases after TSS (Fig. 4c). These data suggested that Chi directly promotes fng transcription in wing discs.
We also performed ChIP experiment for dLMO. Due to lack of endogenous dLMO antibody, we overexpressed HA-dLMO with ms1096Gal4 and then performed ChIP with HA antibody. The same peaks as those in Chi ChIP were also observed in the result, only that the lowest peak in dLMO ChIP assay was the highest peak in the Chi ChIP assay (Fig. 4d). All these data together with previous biochemical results indicate that Chi and dLMO are in a complex to regulate fng transcription directly. If this conclusion is right, the fng downregulation induced by bx overexpression should be revived by chi overexpression. We then overexpressed chi and bx together and noticed the fng-lacZ downregulation was partially revived (Fig. 4e–f’). In addition, the functions of bx overexpression on Notch target genes were partially revived as well (Fig. 4g–h’), indicating the roles of Chi and dLMO on Notch signaling were dependent on their regulation on fng.
Discussion
The Notch signaling plays a very important role in metazoan development. Since Notch was named a century ago, more and more detailed mechanisms are uncovered5,31. However, some important detailed information is still weakly defined. Such as, it is already known that Ap, Chi and dLMO are involved in the regulation of Notch signaling, Ap and Chi function together as a tetramer to regulate Notch signaling and Bx represses Ap’s function by competing binding to Chi22,26. However, the detailed function of Chi and dLMO on Notch signaling remains unknown and the mechanism is also obscure.
Here we demonstrated the detailed functions and the mechanisms of Chi and dLMO in the Notch signaling. Different chi and bx expression levels in the adjacent cells are essential for Notch signaling activation. The roles of Chi and dLMO on Notch signaling depend on their regulation of the fng transcription. Both of them could bind to the same fragments in fng locus: 1.5k bases and 700 bases in the upstream of TSS and 800 bases in the downstream of TSS. In addition, the downregulation of fng expression induced by bx overexpression could be revived by chi overexpression. In our study, we observed that chi overexpression and bx downregulation could not induce the changes of Notch signaling in regions near A/P boundary. We noticed that fng is also not expressed in the A/P boundary region at the late stage of wing disc. In addition, we found that the binding between Chi and dLMO is weakened in the presence of Dpp. All these data indicated that Chi and dLMO function together to directly regulate the transcription of fng, this might be regulated by Dpp signals. It was reported that Notch2 signaling in the outer ciliary epithelium is required for maintaining bone morphogenetic protein (BMP) signaling in mouse eye32. Our results here offered a possibility that the crosstalk between BMP signaling pathway and Notch signaling pathway might partially depend on the interaction of Chi and dLMO.
The overexpression and downregulation of both chi and bx induce similar phenotypes on Notch signaling with just a slight difference in the locations of activated cells, further suggesting the proper amount of Chi and dLMO is essential in the wing discs development. Previous studies showed that both of dLMO and Ap bind with Chi and function in the Notch signaling22,24. Our ChIP assay results here showed that, the highest peak of Chi binding fragments on the fng locus appeared at about 700 bases in the upstream of TSS and this peak is the lowest on the ChIP experiment of dLMO. These results indicate that there might be another transcription factor in the complex with Chi to regulate fng transcription. However, Ap is excluded from the candidates, since in our experiment condition, it did not play an important role and showed only slight effects on Notch signaling. It will be very important to investigate this unknown factor(s) in the future study.
Although both loss-of-Chi/dLMO and gain-of-Chi/dLMO induced fng-lacZ downregulation, their roles on delta-lacZ are opposite and their functions on Notch signaling are not exactly the same. It is normal to reason that there are other mechanisms of how Chi and dLMO are involved in Notch signaling. One of the interesting things we have noticed in our experiments is that delta-lacZ upregulation induced in our experiments is along the clone boundary while downregulation is inside the whole clones. There is a feedback loop in Notch signaling, it might explain why delta-lacZ is upregulated along the clone boundary. However, it is hard to imagine that cells in the middle of the clones could respond to the activated Notch signaling along the clone boundary. These data implied that Chi and dLMO might also regulate delta transcription.
In summary, we uncovered the detailed functions of Chi and dLMO on Notch signaling. Notch signaling tends to be activated along the adjacent cells with different Chi levels in the D compartment of wing discs. The chi overexpression and bx RNAi could not function on Notch signaling in the A/P boundary. This might be explained by that Dpp could regulate the interaction between Chi and dLMO. In addition, the function of Chi and dLMO on Notch signaling is depended on the regulation of fng transcription. The antagonistical regulation of Chi and dLMO on Notch signaling through fng expression is important for wing development.
Materials and Methods
Drosophila stocks and genetics
AGGal4, dppGal4, hhGal4 flies have been previously described (Flybase)33,34,35. chie5.5 (Bloomington, #4541), chi RNAi (VDRC, #43934), bx RNAi (Bloomington, #29454), ap RNAi (Bloomington, #26748, 41673) flies were used in this study. Nanos-Cas9 flies used to generate chi26 are the gifts from Dr. Jianquan Ni at Tsinghua Fly Center, School of Medicine, Tsinghua University, China.
qPCR
Total RNA was extracted from third instar larvae wing discs using Trizol Reagent (Invitrogen) according to manufacturer’s instructions. The acquired RNA was used to synthesize cDNA by ReverTra Ace synthesis kit (Toyobo). Real-time PCR was performed using ABI7500 System with SYBR Green Real-time PCR Master Mix (Toyobo) reagent. rpl32 was used as a normalization control for all of the PCR reactions except ChIP-qPCR.
ChIP-qPCR
ChIP assay was performed as previously described with some modifications (for details, please see Supplemental Information)36,37. Primer pairs and detailed protocols used in this study are listed in the Supplemental Information.
Immunostaining of wing discs
The larvae were heatshock at 37 °C for 30 minutes after birth for 72 hours. The wing discs were dissected for immunostaining with standard protocol as previously described. The antibodies information are listed in the Supplemental Information. Confocal imagings were collected using a Leica TCS system and processed by Adobe Photoshop and Image J.
Cell culture, transfection and Western blotting
S2 cells were cultured in Drosophila Schneider’s Medium with 10% fetal bovine serum, 100 U/ml of penicillin and 100 mg/ml of Streptomycin (Invitrogen). Plasmids were transfected with LipofectAMINE (Invitrogen) according to manufacturer’s instructions. Immunoprecipitation and Western blot were carried out according to standard protocols as previously described (Jin et al., 2012). Antibodies used in this study were as follows: mouse anti-flg (1:5000; Sigma), mouse anti-HA (1:5000; Sigma), rabbit anti-Chi (produced by immunizing rabbits with the whole protein). S2 cells were treated with the Recombinant Drosophila Decapentaplegic (R&D systems) according to the instruction for 6 hours before harvesting. siRNA target of chi is the first 500 bases downstream of TSS. siChi were generated according to the instruction of in vitro transcription T7 kit (Takara).
Additional Information
How to cite this article: Han, H. et al. Chi and dLMO function antagonistically on Notch signaling through directly regulation of fng transcription. Sci. Rep. 6, 18937; doi: 10.1038/srep18937 (2016).
References
Dexter, J. S. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am. Nat. 48, 712–758 (1914).
Morgan, T. H. B. & C. B. Sex-Linked Inheritance in Drosophila. PartII, 63–64 (Press of Gibson Brothers 1916).
Mohr, O. L. Character Changes Caused by Mutation of an Entire Region of a Chromosome in Drosophila. Genetics 4, 275–282 (1919).
Wharton, K. A., Johansen, K. M., Xu, T. & Artavanis-Tsakonas, S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43, 567–581 (1985).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7, 678–689, 10.1038/nrm2009 (2006).
Huovila, A. P., Turner, A. J., Pelto-Huikko, M., Karkkainen, I. & Ortiz, R. M. Shedding light on ADAM metalloproteinases. Trends in biochemical sciences 30, 413–422, 10.1016/j.tibs.2005.05.006 (2005).
Kopan, R. & Ilagan, M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233, 10.1016/j.cell.2009.03.045 (2009).
Munro, S. & Freeman, M. The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr Biol 10, 813–820 (2000).
Bayer, P. & Fanghanel, J. Fringe gives a saccharine to notch. Trends in biochemical sciences 25, 485–486 (2000).
Bruckner, K., Perez, L., Clausen, H. & Cohen, S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415, 10.1038/35019075 (2000).
Moloney, D. J. et al. Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375, 10.1038/35019000 (2000).
Irvine, K. D. & Wieschaus, E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595–606 (1994).
Zhao, D., Clyde, D. & Bownes, M. Expression of fringe is down regulated by Gurken/Epidermal Growth Factor Receptor signalling and is required for the morphogenesis of ovarian follicle cells. Journal of cell science 113 Pt 21, 3781–3794 (2000).
Fleming, R. J., Gu, Y. & Hukriede, N. A. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development 124, 2973–2981 (1997).
Panin, V. M., Papayannopoulos, V., Wilson, R. & Irvine, K. D. Fringe modulates Notch-ligand interactions. Nature 387, 908–912, 10.1038/43191 (1997).
Diaz-Benjumea, F. J. & Cohen, S. M. Interaction between dorsal and ventral cells in the imaginal disc directs wing development in Drosophila. Cell 75, 741–752 (1993).
Rincon-Limas, D. E. et al. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proc Natl Acad Sci USA 96, 2165–2170 (1999).
Cohen, B., McGuffin, M. E., Pfeifle, C., Segal, D. & Cohen, S. M. apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev 6, 715–729 (1992).
Fernandez-Funez, P., Lu, C. H., Rincon-Limas, D. E., Garcia-Bellido, A. & Botas, J. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J 17, 6846–6853, 10.1093/emboj/17.23.6846 (1998).
Herranz, H. & Milan, M. Notch and affinity boundaries in Drosophila. Bioessays 28, 113–116, 10.1002/bies.20366 (2006).
Milan, M. & Cohen, S. M. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Molecular cell 4, 267–273 (1999).
Milan, M., Diaz-Benjumea, F. J. & Cohen, S. M. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev 12, 2912–2920 (1998).
Rincon-Limas, D. E., Lu, C. H., Canal, I. & Botas, J. The level of DLDB/CHIP controls the activity of the LIM homeodomain protein apterous: evidence for a functional tetramer complex in vivo. EMBO J 19, 2602–2614, 10.1093/emboj/19.11.2602 (2000).
van Meyel, D. J. et al. Chip and apterous physically interact to form a functional complex during Drosophila development. Molecular cell 4, 259–265 (1999).
Shoresh, M. et al. Overexpression Beadex mutations and loss-of-function heldup-a mutations in Drosophila affect the 3’ regulatory and coding components, respectively, of the Dlmo gene. Genetics 150, 283–299 (1998).
Weihe, U., Milan, M. & Cohen, S. M. Regulation of Apterous activity in Drosophila wing development. Development 128, 4615–4622 (2001).
Milan, M. & Cohen, S. M. Temporal regulation of apterous activity during development of the Drosophila wing. Development 127, 3069–3078 (2000).
Ren, X. et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA 110, 19012–19017, 10.1073/pnas.1318481110 (2013).
Nellen, D., Burke, R., Struhl, G. & Basler, K. Direct and long-range action of a DPP morphogen gradient. Cell 85, 357–368 (1996).
Lecuit, T. et al. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature 381, 387–393, 10.1038/381387a0 (1996).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Zhou, Y. et al. Notch2 regulates BMP signaling and epithelial morphogenesis in the ciliary body of the mouse eye. Proc Natl Acad Sci USA 110, 8966–8971, 10.1073/pnas.1218145110 (2013).
Percival-Smith, A., Teft, W. A. & Barta, J. L. Tarsus determination in Drosophila melanogaster. Genome/National Research Council Canada=Genome/Conseil national de recherches Canada 48, 712–721, 10.1139/g05-021 (2005).
Zhang, Z. et al. Ter94 ATPase complex targets k11-linked ubiquitinated ci to proteasomes for partial degradation. Dev Cell 25, 636–644, 10.1016/j.devcel.2013.05.006 (2013).
Huang, H. L. et al. Par-1 regulates tissue growth by influencing hippo phosphorylation status and hippo-salvador association. PLoS biology 11, e1001620, 10.1371/journal.pbio.1001620 (2013).
Papp, B. & Muller, J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Gene Dev 20, 2041–2054, 10.1101/Gad.388706 (2006).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods-a Companion to Methods in Enzymology 11, 205–214 (1997).
Acknowledgements
We are grateful to Drs. Hugo Bellen, Dahua Chen, Feng Liu, Haiyun Song and Jianquan Ni for reagents and fly stocks. We are also grateful to DSHB, VDRC, NIG and the Bloomington Stock Center for fly stocks and reagents. This work was supported by the grants from the National Natural Science Foundation of China (31171414, 31371492).
Author information
Authors and Affiliations
Contributions
H.H., wrote the main manuscript text and prepared Figures 1–4, supplementary Figures 1–6; Y.X., revised the article; J.F., W.W., Y.L. and L.Z. analyzed and interpreted the data; Y.Z. analyzed and interpreted the data and revised the article.
Different Chi levels in adjacent cells are essential for Notch signaling activity.
(a–a”’) Immunostaining of Chi (green) in wing discs of early third instar larvae. Cells with high Cut expression (red) locate in the boundary between high and low Chi protein regions. DAPI was used to mark the cell nuclei (blue). (b-b’) Immunostaining of Chi (red) in chi RNAi wing discs. chi RNAi was overexpressed in the clones (green). (c-f’) Immunostaining of Cut (c-d’; red) and Wg (e-f’; red) in yw and chi RNAi wing discs. (d’,f’) are the enlarged pictures of (d,f). Clones are marked with GFP (green). (g-h”) Immunostaining of Wg (g-g”) and Cut (h-h”) in the chi26 wing discs. Wg (g; red) and Cut (h; red) expression are along the D/V boundary. Wg (g-g”) and Cut (h-h”) are activated in the boundary of chi26 clones. (g’) and (h’) are the enlarged pictures of (g,h). Clones are marked with green and circled with dashed line. (i) Quantitative analysis of Cut activated cells around the clones. Shown are the Means ± s.d. (j) qPCR analysis of the Notch target genes. Shown are Means ± s.d. from 3 independent experiments. In every experiment, at least 20 discs were pooled together for analysis.
Chi and dLMO form a complex to directly regulate fng transcription.
(a-b) Co-IP results of endogenous Chi and HA-dLMO with or without Dpp treatment. The images were cropped. The full-length images were presented in Supplementary Figures 6a–b”. (c) ChIP-qPCR analysis using anti-Chi antibody with chi RNAi as control at fng locus. ChIP signal levels are represented as percentage of input chromatin. Chi could bind to fng locus at three regions. (d) ChIP-qPCR analysis using anti-HA antibody from discs of ms1096Gal4 > HA-bx at fng locus. ChIP signal levels are represented as percentage of input chromatin. dLMO could bind to fng locus at three regions. (e-f’) Immunostaining of fng-lacZ (red) in the discs of indicated genotype flies. Compared to the overexpression of HA-bx (e-e’), the fng downregulation could be partially rescued by the chi overexpression (f-f’). In all IF results, clones are marked with GFP (green) and circled with dashed line. (g-h’) Immunostaining of Cut (g-g’; red) and Wg (h-h’; red) in the discs of indicated genotype flies. Compared to the overexpression of HA-bx alone (Figure 3 c–d’), Cut and Wg activation could be partially rescued by the chi overexpression. Clones are marked with GFP (green). (g’,h’) are the enlarged pictures of (g,h).
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Han, H., Fan, J., Xiong, Y. et al. Chi and dLMO function antagonistically on Notch signaling through directly regulation of fng transcription. Sci Rep 6, 18937 (2016). https://doi.org/10.1038/srep18937
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18937
This article is cited by
-
Genomic insights into mite phylogeny, fitness, development, and reproduction
BMC Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.