Abstract
Sixty genetic loci associated with abdominal obesity, measured by waist circumference (WC) and waist-hip ratio (WHR), have been previously identified, primarily from studies conducted in European-ancestry populations. We conducted a meta-analysis of associations of abdominal obesity with approximately 2.5 million single nucleotide polymorphisms (SNPs) among 53,052 (for WC) and 48,312 (for WHR) individuals of Asian descent and replicated 33 selected SNPs among 3,762 to 17,110 additional individuals. We identified four novel loci near the EFEMP1, ADAMTSL3 , CNPY2 and GNAS genes that were associated with WC after adjustment for body mass index (BMI); two loci near the NID2 and HLA-DRB5 genes associated with WHR after adjustment for BMI and three loci near the CEP120, TSC22D2 and SLC22A2 genes associated with WC without adjustment for BMI. Functional enrichment analyses revealed enrichment of corticotropin-releasing hormone signaling, GNRH signaling, and/or CDK5 signaling pathways for those newly-identified loci. Our study provides additional insight on genetic contribution to abdominal obesity.
Similar content being viewed by others
Introduction
Abdominal obesity, typically measured by waist circumference (WC) and waist-hip ratio (WHR), is more closely associated with metabolic dysfunctions that are related to cardiovascular diseases than is general obesity1, which is generally assessed by body mass index (BMI). Previous studies have identified multiple genetic loci associated with WC and WHR2,3,4,5,6,7,8,9,10,11,12,13. However, the majority of these studies were conducted in populations of European ancestry or included a limited number of East Asians9. East Asians tend to have a higher level of abdominal fat, despite relatively low BMI values; and experience a higher metabolic disease risk than European-ancestry individuals with the same BMI level14. Therefore, it is particularly important to investigate the genetic determinants of abdominal fat, i.e. WC and WHR, in East Asian populations.
We previously reported genetic loci for BMI using data from the Asian Genetic Epidemiology Network (AGEN) Consortium15,16. In this study, we conducted meta-analyses of data from genome-wide association studies (GWAS) of WC and WHR to identify new genetic loci and evaluate associations of previously-identified genetic loci with overall and abdominal obesity in our study populations.
Results
Our initial meta-analysis used two complementary but related measures of abdominal obesity, WC and WHR, as the outcome variables and analyzed the association of WC and WHR with approximately 2.5 million genotyped or imputed SNPs as well as about 50,000 typed exome-chip variants. The total sample sizes in Stage I were 53,052 for WC and 48,312 for WHR. We selected 33 SNPs at 33 independent loci with P < 1.00 × 10−6, based on the GWAS data that were recruited at the first round of Stage I, for a de novo replication (Stage II) of associations with WC or WHR. The replication genotyping was conducted at three study sites (see Supplementary Table 3 online) comprising 3,762 to 17,110 Asian-ancestry individuals based on availability of de novo data for each SNP. Participating studies are described in the Supplementary Information and Supplementary Tables 1 to 3 online.
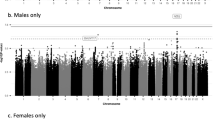
The associations of SNPs with WC or WHR were analyzed with or without adjustment for BMI (see Methods), following the common practice employed in published studies2,3,4,5,6,7,8,9,10,11,12,13. Thus, there were four traits included in this study: WC with adjustment for BMI (WCadjBMI), WHR with adjustment for BMI (WHRadjBMI), WC without adjustment for BMI (WCnoBMI) and WHR without adjustment for BMI (WHRnoBMI). The results of the initial Stage I and Stage II for the selected 33 SNPs are presented in Supplementary Table 4 online. In Table 1 (see also Fig. 1 and Supplementary Table 5), we present the newly-identified loci that were associated with WCadjBMI, WHRadjBMI and WCnoBMI at a genome-wide significance level (P < 5.00 × 10−8) based on Stage I data alone or the combined Stage I and Stage II data in Asian-ancestry populations. For WCadjBMI, we have identified four new loci (index SNPs) near these genes: EFEMP1 (rs3791679, P = 4.86 × 10−14), ADAMTSL3 (rs8030379, P = 1.62 × 10−9), CNPY2 (rs3809128, P = 3.74 × 10−9) and GNAS (rs2057291, P = 4.02 × 10−8); for WHRadjBMI, we have identified two loci near the NID2 (rs1982963, P = 1.07 × 10−14) and HLA-DRB5 (rs5020946, P = 1.30 × 10−9) genes; for WCnoBMI, we have identified three loci near the CEP120 (rs10051787, P = 7.23 × 10−12), TSC22D2 (rs1868673, P = 1.49 × 10−8) and SLC22A2 (rs368123, P = 2.64 × 10−8) genes. In addition, three SNPs near three genes (ADAMTS3, IHH, QSOX2) for WCadjBMI and two SNPs near two genes (PPAP2B, PACSIN3) for WHRadjBMI were found to approach the genome-wide significance level (P < 7.56 × 10−7) (see Supplementary Tables 5,6,7 online). We requested an in silico replication for the 14 SNPs described above in the Genetic Investigation of ANthropometric Traits (GIANT) consortium13. No data were available for rs3809128 (MAF < 0.01 in CEU) near CNPY2 in the GIANT data. As shown in Supplementary Table 6 online, the association directions were consistent for 12 out of the remaining 13 SNPs (P = 0.0034 by the binomial test), although the explained variances were generally smaller than those observed in East Asians (see Supplementary Table 5 online). The SNP rs2057291 near GNAS exhibited an opposite association direction in the GIANT data. Seven of the ten loci for WCadjBMI and WHRadjBMI were oppositely associated with BMI, but all three WCnoBMI loci had a consistent association direction with BMI in the GIANT data. In this study, we found no genetic association with WHRnoBMI at loci other than those previously reported (see Supplementary Table 8 online). The variation explained by each newly-identified SNP ranged from 0.02% to 0.09% (Table 1 and Supplementary Table 5 online). There were no Stage II replication data for some of the loci because they were identified after the second round of Stage I GWAS data were added to the meta-analysis due to the expansion of the AGEN, which occurred after the original 33 replication SNPs were selected.
Regional plots for the newly-identified loci in this study.
SNPs are plotted by their position on the chromosome against their association (−log10 P value) with the trait of interest (as shown in Table 1) using stage I (GWAS meta-analysis) data. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the top SNP are color-coded (see inset) to reflect their LD with the top SNP (using pair-wise r2 values from HapMap CHB + JPT data). Genes and positions of exons, as well as directions of transcription, are shown below the plots (using data from the UCSC Genome Browser, genome.ucsc.edu). The arrows shown in Plots a, b, e indicate SNPs that were previously reported to be associated with height. Plots were generated using LocusZoom.
Additional analyses examined effect sizes for differences across sex and population. Analyses stratified by sex (Table 2 and Supplementary Table 5 online) revealed that association of rs3791679 near the EFEMP1 gene with WCadjBMI was significantly stronger among men than among women (effect size: 4.04 vs 2.43, P for homogeneity test = 0.04) and association of rs1982963 near the NID2 gene with WHRadjBMI was significantly weaker among men than among women (effect size: 2.88 vs 6.26, P for homogeneity test = 0.009). No significant heterogeneity across populations of Chinese, Korean, Japanese, or Filipino was found for the newly-identified loci (data not shown).
Supplementary Table 7 online shows the association of the newly-identified loci with different obesity-related traits. The three novel loci (CEP120, TSC22D2 and SLC22A2) for WCnoBMI were much less significantly associated with WCadjBMI; the newly-identified loci for WCadjBMI or WHRadjBMI were either unassociated with BMI (EFEMP1, CNPY2, GNAS and HLA-DRB5) or negatively associated with BMI (ADAMTSL3 and NID2). Of these 11 loci for WCadjBMI and WHRadjBMI, ten had an opposite association direction with BMI. Using the Wald test of whether the BMI-adjusted effect was equal to its expectation proposed by Aschard et al.17, we found Bonferroni-corrected significant p-values (0.05/11) for rs11103390 at QSOX2 (WCadjBMI) and rs1982963 at NID2 (WHRadjBMI), suggesting that the associations of these two SNPs with WCadjBMI or WHRadjBMI may have been influenced by their direct genetic association with BMI. The newly-identified seven loci for WCadjBMI were all moderately or strongly associated with height and the three novel loci for WCnoBMI were also moderately associated with height. None of 14 newly-identified loci were associated with diabetes at P < 0.004 (0.05/14).
Previous studies have reported about 60 genetic loci associated with abdominal obesity2,3,4,5,6,7,8,9,10,11,12,13 and about 100 genetic loci associated with overall obesity15,16,18, with the majority of those loci being identified in populations of European ancestry. In those studies, the reported associations with WHR were generally adjusted for BMI, while the reported associations with WC were not adjusted for BMI in most studies. Due to the close correlation (r = 0.83 based on data from the Shanghai genome-wide association studies (SGWAS)) between WC and BMI, there was substantial overlap between loci that were associated with WC and BMI. In the current study, we evaluated the associations of those reported loci with WCadjBMI, WHRadjBMI, WCnoBMI and WHRnoBMI. The associations with at least one trait that achieved a Bonferroni-corrected significance level (P < 0.05/60 ≈1.0 × 10−3) and the associations by sex are shown in Supplementary Tables 8 to 11 online (for WCadjBMI, WHRadjBMI, WCnoBMI and WHRnoBMI, respectively). We found that 23 previously-reported loci for abdominal obesity were significantly associated with WCadjBMI (see Supplementary Table 11 online) and/or WHRadjBMI (see Supplementary Table 10 online) and 18 previously-reported BMI/WC loci were significantly associated with WCnoBMI, among men or women or both at P < 1.0 × 10−3 (see Supplementary Table 9 online). Of note, 17 of those 18 loci associated with WCnoBMI were not significantly associated with WCadjBMI. The only SNP demonstrating significant association with WCadjBMI was rs12229654, at our previously-identified Asian-specific BMI locus MYL2. Consistent with previous findings7,19, we observed that 10 out of 23 replicated genetic loci for abdominal obesity showed significant sex differences (P for homogeneity test < 0.05), 9 of which showed larger effects in women than men (see Supplementary Table 10 online). In contrast, 4 replicated WCnoBMI loci revealed significant sex differences, 3 of which showed larger effects in men (see Supplementary Table 9 online). It is worth noting that the SNP rs12229654 at MYL2 and its related SNP rs671 at ALDH2 were associated with every obesity trait analyzed (WCadjBMI, WHRadjBMI, WCnoBMI and WHRnoBMI), with larger effects observed in men.
We examined the modification effect of alcohol consumption on the association between the two SNPs in the 12q24 region16 (rs12229654 and rs671) and WC/WHR using data from the SGWAS, for which we had direct access to individual data. We found that the effects of the SNP rs12229654 on WC and WHR, with or without adjustment for BMI, were mainly observed among non-drinkers (Supplementary Table 13 online). The sex-difference of rs671 effect on WC and WHR is less evident.
We evaluated putative functional significance for each newly-identified locus using the Encyclopedia of DNA Elements (ENCODE) and expression quantitative trait locus (eQTL) databases20. Three of these novel SNPs, rs3809128 near CNPY2, rs2057291 at GNAS and rs3791679 near EFEMP1, were predicted to be functional as they are located either in promoter or enhancer regions based on epigenomic data from the ENCODE project. The ChromHMM annotation on nine ENCODE cell lines have revealed that the SNP rs3809128 resides in an active promoter of the nearest gene, canopy FGF signaling regulator 2 (CNPY2). The DNase and ChIP-Seq has revealed that the SNP rs3809128 is present in the DNase I hypersensitive site (DHS) and in the binding regions of multiple transcription factors (TFs). In particular, this variant has been shown to be associated with the expression of CNYP2 based on a previous eQTL study19. The SNP rs2057291 is located in intron 2 of the gene GNAS. A search of RegulomeDB21 indicates that this variant is annotated to the TF SRF predicted motif. This variant was also observed to be present in the DHS and multiple TFs peaks. The SNP rs3791679 is located in the first intron of the gene EFEMP1 and resides in an enhancer region. According to RegulomeDB annotation, the SNP rs3791679 lies in the TF POU3F2 predicted motif. In addition, the DHS and the TF STAT3 peak were found to harbor the variant. Other potential functional variants which are in strong linkage disequilibrium (LD)(r2 > 0.6) with the newly-identified SNPs are listed in Supplementary Table 12 online.
We conducted two separate functional enrichment analyses for genes located near the newly- and previously-identified loci for WC or WHR, one for the loci associated with WCadjBMI and/or WHRadjBMI, another for the loci associated with WCnoBMI. The corticotropin-releasing hormone signaling (P = 5.72 × 10−4) pathway and Gonadotropin Releasing Hormone (GNRH) signaling (P = 8.63 × 10−4) pathway, were found to be the most significantly enriched for the loci associated with WCadjBMI and/or WHRadjBMI; and the CDK5 signaling (P = 1.66 × 10−4) and corticotropin-releasing hormone signaling (P = 2.21 × 10−4) pathway were significantly enriched for the loci associated with WCnoBMI.
Discussion
Previously-reported genetic loci associated with WC, mainly from studies conducted in European-ancestry populations, were generally not adjusted for BMI. There was substantial overlapping between loci that were associated with WC and BMI due to high correlation between those two measurements. Three novel SNPs (rs10051787 near CEP120, rs1868673 near TSC22D2 and rs368123 at SLC22A2), newly identified for WCnoBMI in this study, did not reach the genome-wide significance level in our previous meta-analysis for BMI and thus were not identified as the BMI loci. After adjustment for BMI, the association of these three SNPs with WCadjBMI was substantially attenuated. In addition, our study showed that previously-reported genetic loci for WC or BMI were generally not significantly associated with WCadjBMI (see Supplementary Table 11 online), suggesting that these two highly-correlated anthropometrics capture a similar biological phenotype. On the other hand, the genetic loci associated with WCadjBMI and/or WHRadjBMI were not typically associated with BMI or showed an opposite association direction (see Supplementary Table 7 and Supplementary Table 14 online). These findings reveal differences in genetic predisposition to overall as opposed to abdominal obesity, as well as the genetic regulation of body fat distribution versus BMI.
Two newly-identified WCadjBMI-related loci (EFEMP1, ADAMTSL3) and the newly-identified WCnoBMI-related loci (CEP120) were previously reported to affect height as well11,22, suggesting these loci may be related to general body frame size. The SNP rs3791679 near EFEMP1 seems particularly relevant to WC and height. EFEMP1 encodes an extracellular matrix protein containing tandemly-repeated epidermal growth factor-like repeats, which are able to stimulate DNA synthesis and are involved in cell proliferation23. EFEMP1 knockout mice exhibited reduced reproductivity and displayed an early onset of aging-associated phenotypes including reduced lifespan and decreased body mass24. A recent study showed that the EFEMP1 locus affected growth rate in children25.
Another newly-identified WCnoBMI-related SNP, rs1868673, resides near the gene TSC22D2. A recent study indicated that this locus was associated with circulating levels of adiponectin, a hormone produced predominantly by adipocytes26.
We previously reported the association of two related SNPs in the 12q24 region (rs12229654 at gene MYL2 and rs671 at ALDH2, r2 = 0.58)15,16, a specific polymorphic region for East Asian-ancestry populations, with BMI. This association was significantly stronger among men than among women. We observed similar associations of those SNPs with WC and WHR, with or without adjustment for BMI (Supplementary Tables 8 to 11 online). The sex differences of associations were more prominent for WHR. The SNPs rs12229654 and rs671 have been reported to be associated with HDL cholesterol27, levels of gamma glutamyl transpeptidase27, elevated blood pressure28, lower risk of coronary heart disease29 and alcohol consumption30 in Asian-ancestry populations. Our previous study16 suggested an antagonistic effect of alcohol consumption on the ALDH2-BMI association. The ALDH2*1 BMI-increasing effect was mainly observed among non-drinkers. In this study, we found that the effects of the SNP rs12229654 at MYL2 on WC and WHR, with or without adjustment for BMI, were mainly seen among non-drinkers (Supplementary Table 13 online).
Consistent with previous reports from European populations, we found evidence for multiple loci with significant sex differences for abdominal obesity in East Asians, with a generally more prominent effect in women, although larger effects in men than in women were observed for loci EFEMP1, MYL2/ALDH2 and FGFR4. Typically, men have more visceral fat, whereas women have more subcutaneous fat. It is well known that sex hormones play an important role in the regulation of body fat distribution31, but the underlying genetic mechanisms remain unclear. It would be worthwhile to investigate the association between these genetic loci and sex hormone levels.
The corticotropin-releasing hormone signaling (P = 5.72 × 10−4) pathway and Gonadotropin Releasing Hormone (GNRH) signaling (P = 8.63 × 10−4) pathway were found to be significantly enriched for genes associated with WCadjBMI and/or WHRadjBMI loci in this study. The corticotropin-releasing hormone signaling (P = 5.72 × 10−4) pathway was found to be related to BMI in our previous report for BMI loci16. Corticotrophin-releasing hormone causes release of adrenocorticotropic hormone from the pituitary gland. Its main role in the body is as the central driver of the stress hormone system, known as the hypothalamic-pituitary-adrenal axis. Gonadotrophin-releasing hormone plays a key role in coordinating the levels of hormones in the hypothalamic-pituitary-gonadal axis; these hormones act on the testes and ovaries to initiate and maintain their reproductive functions. These results provide additional evidence affirming the involvement of stress and sex hormones in obesity and fat distribution.
Recently, Aschard et al.17 raised a concern that adjusting for BMI may bias genetic effects on WC/WHR and observed enrichment of SNPs that were associated with WC/WHR and BMI in the opposite directions, as shown in the Heid et al. study7 and in our study (see Supplementary Table 5 online). As shown in the results section, the associations of SNPs rs11103390 at QSOX2 with WCadjBMI or rs1982963 at NID2 with WHRadjBMI may be influenced by their direct genetic association with BMI. Further studies are warranted to evaluate this influence.
Another limitation of this study is lack of replication data for some SNPs identified in the expanded Stage I study which will need to be confirmed in future studies.
In conclusion, our study identified at the genome-wide significance level four novel loci near the EFEMP1, ADAMTSL3, CNPY2 and GNAS genes that are associated with WCadjBMI, two loci near the NID2 and HLA-DRB5 genes that were associated with WHRadjBMI and three novel loci near the CEP120, TSC22D2 and SLC22A2 genes that were associated with WCnoBMI. Of about 60 genetic loci previously identified for abdominal obesity in predominantly European populations, a similar association was found in our study for 23 in East Asians, suggesting Asian- and European-ancestry individuals have both a shared and a unique genetic basis for abdominal obesity. Functional analyses suggest that genetic regulation for abdominal fat distribution may occur via the corticotropin-releasing hormone signaling, GNRH signaling, and/or CDK5 signaling pathway.
Materials and Methods
Study design
This study had two stages. Stage I was a meta-analysis of study-specific results on the association between SNPs and WC or WHR from GWAS that participated in the Asian Genetic Epidemiology Network (AGEN) Consortium. Stage II conducted de novo or in silico replication analyses to further examine the association for some promising SNPs selected from the Stage I meta-analysis. Supplementary Tables 1,2,3 online and the Supplementary Information page online summarize the basic data for all participating studies.
Stage I samples and genotyping
In stage I, the participating GWAS were recruited in two rounds because of the expansion of the AGEN. The first round included eight GWAS with a total 27,537 (for WC) or 25,241 (for WHR) individuals of East Asian ancestry and the second round included eight more GWAS with a total 17,072 (for WC) or 14,628 (for WHR) individuals of East Asian ancestry. In addition, Stage I analysis included about 50,000 SNPs with minor allele frequency over 1% from 8,443 subjects with genotyping data by exome-chip. Therefore, the total sample sizes in Stage I were 53,052 for WC and 48,312 for WHR.
The sample sizes of the 16 participating GWAS in Stage I varied from 695 to 9,279, comprising a total of 44,596 individuals. Nine studies used Affymetrix arrays and seven studies used the Illumina platform (detailed information is provided in the Supplementary Information online). To allow for combination of the data derived from different genotyping platforms and to improve coverage of the genome, genotype imputation was performed by each participating study using either MACH or IMPUTE with HapMap CHB+JPT data (release #22, build 36) as the imputation reference panel (see Supplementary Table 2 online).
Stage I statistical analysis
A uniform statistical analysis protocol was followed by each participating study. To improve the normality of the WC and WHR distribution and alleviate the impact of outliers, rank-based inverse normal transformation (INT) was applied to WC and WHR data separately for each sex by each study. INT involves ranking all WC and WHR values, transforming these ranks into quantiles and, finally, converting the resulting quantiles into normal deviates. Associations between SNPs and the inverse normal-transformed WC and WHR were analyzed with a linear regression model, assuming an underlying additive genetic model and adjusting for age (continuous), age-squared and sex (if applicable). Stratified analyses by sex were also performed for each study. To evaluate the genetic influence on body fat distribution, all the analyses were conducted using two separate models, one with adjustment for BMI, another without adjustment for BMI.
Next, we carried out meta-analyses using a weighted average method with inverse-variance weights. The meta-analyses were carried out on all data combined and also stratified by sex using the freely available METAL software. The presence of heterogeneity across studies and between sexes was tested with Cochran’s Q statistics32.
To correct each study for residual population stratification or cryptic relatedness, the meta-analyses were performed with genomic control correction33 by adjusting for the study-specific inflation factor (λ), which ranged from 1.000 to 1.078 in Stage I (see Supplementary Table 2 online). After this adjustment, the estimated inflation factors for the Stage I meta-analysis statistic were 1.053 (WCadjBMI), 1.042 (WHRadjBMI), 1.091 (WCnoBMI) and 1.054 (WHRnoBMI), which were further adjusted for when calculating the Stage I results.
Stage II replication analysis
We selected 33 SNPs at 33 independent loci with P < 1.00 × 10−6 for the associations with WC or WHR for a de novo replication (Stage II), based on the GWAS data that were recruited in the first round of Stage I. The replication genotyping was conducted at three study sites (see Supplementary Table 1 online) comprising 3,762 to 17,110 Asian-ancestry individuals based on availability of de novo data for each SNP. Participating studies are described in the Supplementary Information and Supplementary Tables 1 to 3 online.
Each study individually conducted a similar analysis of the association of WC and WHR with the selected SNPs, using the same protocol used in Stage I. The Stage II data were combined using the same meta-analysis methods as in Stage I. Finally, we used meta-analysis to combine all data from both Stages I and II.
Additional Information
How to cite this article: Wen, W. et al. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci. Rep. 6, 17958; doi: 10.1038/srep17958 (2016).
References
Ritchie, S. A. & Connell, J. M. The link between abdominal obesity, metabolic syndrome and cardiovascular disease. NutrMetab Cardiovasc. 17, 319–326 (2007).
Fox, C. S. et al. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC.Med.Genet. 8 Suppl 1, S18 (2007).
Chambers, J. C. et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat.Genet. 40, 716–718 (2008).
Lindgren, C. M. et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS.Genet. 5, e1000508 (2009).
Heard-Costa, N. L. et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS.Genet. 5, e1000539 (2009).
Speliotes, E. K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat.Genet. (2010). 10.1038/ng.686.
Heid, I. M. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat.Genet. 42, 949–960 (2010).
Kraja, A. T. et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes 60, 1329–1339 (2011).
Cho, Y. S. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat.Genet. 41, 527–534 (2009).
Croteau-Chonka, D. C. et al. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity.(Silver.Spring) 19, 1019–1027 (2011).
Berndt, S. I. et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat.Genet. 45, 501–512 (2013).
Liu, C. T. et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS.Genet. 9, e1003681 (2013).
Shungin, D. et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196 (2015).
Deurenberg, P., Deurenberg-Yap, M. & Guricci, S. Asians are different from Caucasians and from each other in their body mass index/body fat per cent relationship. Obes.Rev. 3, 141–146 (2002).
Wen, W. et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat.Genet. 44, 307–311 (2012).
Wen, W. et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum.Mol.Genet. 10.1093/hmg/ddu248 (2014).
Aschard, H., Vilhjálmsson, B. J., Joshi, A. D., Price, A. L. & Kraft, P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 96, 329–339 (2015).
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Randall, J. C. et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS.Genet. 9, e1003500 (2013).
Pickrell, J. K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22, 1790–1797 (2012).
Lango, A. H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Lecka-Czernik, B., Lumpkin, C. K., Jr. & Goldstein, S. An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. MolCell Biol 15, 120–128 (1995).
McLaughlin, P. J. et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum.Mol.Genet. 16, 3059–3070 (2007).
Paternoster, L. et al. Adult height variants affect birth length and growth rate in children. Hum.Mol.Genet. 20, 4069–4075 (2011).
Dastani, Z. et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 8, e1002607 (2012).
Kim, Y. J. et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 43, 990–995 (2011).
Kato, N. et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat.Genet. 43, 531–538 (2011).
Takeuchi, F. et al. Genome-wide association study of coronary artery disease in the Japanese. Eur. J. Hum. Genet. 3, 333–40 (2012).
Baik, I., Cho, N. H., Kim, S. H., Han, B. G. & Shin, C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am. J. Clin. Nutr. 93, 809–816 (2011).
Wells, J. C. Sexual dimorphism of body composition. Best.Pract.Res.Clin.Endocrinol.Metab 21, 415–430 (2007).
Cochran, W. G. The Combination of Estimates from Different Experiments. Biometrics 10, 101–129 (1954).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Acknowledgements
The authors for this work would like to thank the Genetic Investigation of ANthropometric Traits (GIANT) consortium for providing unpublished data for replication of the study findings. The Shanghai Genome Wide Associations Studies (SGWAS) would like to thank the dedicated investigators and staff members from the research teams at Vanderbilt University, the Shanghai Cancer Institute and the Shanghai Institute of Preventive Medicine and most of all, the study participants for their contributions to the studies. The CLHNS investigators thank the Office of Population Studies Foundation research and data collection teams for CLHNS and the Mammalian Genotyping Core at University of North Carolina at Chapel Hill. This work was supported by the National Institutes of Health. Genotyping assays and statistical analyses for the SGWAS were supported primarily by NIH grants [R01 CA064277, R37 CA70867, R01 CA090899, R01 CA118229 and R01 CA122756]; and by the Ingram professorship funds, Allen Foundation funds and Vanderbilt CTSA grant from NCRR/NIH [1 UL1 RR024975]. Grant support for the participating studies includes: the Shanghai Breast Cancer Study [R01 CA064277]; the Shanghai Women’s Health Study [R37 CA70867]; the Shanghai Breast Cancer Survival Study [R01 CA118229]; the Shanghai Endometrial Cancer Study [R01 CA092585]; and the Shanghai Men’s Health Study [UM1 CA173640]. The NHAPC study was supported by the National Natural Science Foundation of China [30930081, 81321062]; the National Key Basic Research Program of Ministry of Science and Technology [2012CB524905; and the Knowledge Innovation Program of the Chinese Academy of Sciences [KSCX2-EW-R-10]. The Cardio-metabolic Genome Epidemiology (CAGE) studies were supported by Grants for Scientific Research – Priority Areas “Medical Genome Science (Millennium Genome Project)” and “Applied Genomics” [20018020, 18018030, 12204008]; Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan [24390084, 25293141, 23659352, 23659382, 21390099, 20390185, 19659163, 16790336, 11694285, 17390186, 20390184 and 24390165]; a Health and Labor Sciences Research Grants from the Ministry of Health, Labor and Welfare, Japan [H15-Longevity, H15-genome, H17-longevity, H16-kenko, H18-longevity (kokusai)]; a Science and Technology Incubation Program in Advanced Regions, Japan Science and Technology Agency; National Cardiovascular Research Grants; Research grant from the Japan Atherosclerosis Prevention Found; Research grant from Ehime University. The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) was supported by research grants from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, Maryland [U01HL072507, R01HL087263 and R01HL090682]. The Korea Genome Wide Association Studies, which includes four projects: KARE, HEXA, the Health2 cohort and CAVAS, were supported by grants from the Korea Centers for Disease Control and Prevention (4845-301, 4851-302 and 4851-307) and an intramural grant from the Korea National Institute of Health (2012-N73002-00); The KARE project was supported by grants from the Korea Centers for Disease Control and Prevention (4845-301, 4851-302 and 4851-307) and an intramural grant from the KoreaNational Institute of Health (2012-N73002-00), the Republic of Korea. The Korean Cancer Prevention Study-II (KCPS-II) was supported by a grant from the National R&D Program for Cancer Control; Ministry for Health, Welfare and Family Affairs, Republic of Korea [1220180]. The Singapore Prospective Study program was funded by the Biomedical Research Council of Singapore under the individual research grants scheme; and the National Medical Research Council of Singapore under the clinician scientist award scheme. The Cebu Longitudinal Health and Nutrition Survey (CLHNS) was supported by National Institutes of Health grants [DK078150, TW05596, HL085144, TW008288]; and pilot funds [RR20649, ES10126 and DK56350].
Author information
Authors and Affiliations
Contributions
W.W., N.K., J.-Y.H., X.G., Y.T., H.L., Y.W., R.D., X.Y., F.-J.T., S.L., Y.W., T.W., S.K., X.G. and J.L. analyzed data and prepared study-specific descriptions. L.Q., J.I.R., S.-H.J., D.L., K.L.M., J.H., Z.M., J.-Y.W., E.S.T., X.L., T.M., B.-J.K., F.T., W.Z. and X.-O.S. designed, led or supervised the contributing studies and contributed materials to the current study. They are responsible for the quality of their contributed study data. D.S., L.S.A., K.A., M.A., Q.C., L.-C.C., C.-H.C., Y.-T.C., Y.S.C., B.Y.C., Y.G., M.J.G., D.G., B.-G.H., M.H., J.E.H., Y.H., T.H., M.I., K.J.J., D.K., Y.J.K., Y.K., J.L., N.R.L., J.L., J.-J.L., J.L., S.M., Y.N., M.N., K.O., D.R., J.S., J.W.S., A.T., H.U., C.W., Y.-B.X., K.Y., J.Y., X.Y., M.Y., X.Z. and Y.Z. contributed to field operations or experiments of the contributing studies. W.W., N.K., X.G., E.S.T., W. Z. and X.-O. Shu wrote the manuscript. All authors approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wen, W., Kato, N., Hwang, JY. et al. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci Rep 6, 17958 (2016). https://doi.org/10.1038/srep17958
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17958
This article is cited by
-
Genomics of body fat distribution
Journal of Genetics (2021)
-
Genetic contribution to waist-to-hip ratio in Mexican children and adolescents based on 12 loci validated in European adults
International Journal of Obesity (2019)
-
Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects
Nature Communications (2019)
-
Protein-coding variants implicate novel genes related to lipid homeostasis contributing to body-fat distribution
Nature Genetics (2019)
-
Waist-hip ratio related genetic loci are associated with risk of impaired fasting glucose in Chinese children: a case control study
Nutrition & Metabolism (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.