Abstract
The Escherichia coli eukaryote-like serine/threonine kinase, encoded by yeaG, is expressed in response to diverse stresses, including nitrogen (N) starvation. A role for yeaG in bacterial stress response is unknown. Here we reveal for the first time that wild-type E. coli displays metabolic heterogeneity following sustained periods of N starvation, with the metabolically active population displaying compromised viability. In contrast, such heterogeneity in metabolic activity is not observed in an E. coli ∆yeaG mutant, which continues to exist as a single and metabolically active population and thus displays an overall compromised ability to survive sustained periods of N starvation. The mechanism by which yeaG acts, involves the transcriptional repression of two toxin/antitoxin modules, mqsR/mqsA and dinJ/yafQ. This, consequently, has a positive effect on the expression of rpoS, the master regulator of the general bacterial stress response. Overall, results indicate that yeaG is required to fully execute the rpoS-dependent gene expression program to allow E. coli to adapt to sustained N starvation and unravels a novel facet to the regulatory basis that underpins adaptive response to N stress.
Similar content being viewed by others
Introduction
Continuous bacterial growth is often limited by the availability of key nutrients. Since numerous cellular macromolecules contain nitrogen (N), including proteins, nucleic acids and cell wall components, Escherichia coli and other members of the Enterobacteriaceae family immediately cease growth upon sensing N starvation and simultaneously initiate two major adaptive responses: the nitrogen regulation (Ntr) stress response and the stringent response1. The Ntr response primarily confers the ability to scavenge for alternative N sources, through the transcriptional activation of genes that encode transport systems and catabolic or biosynthetic operons by the global transcriptional regulator NtrC2. The stringent response is mediated by the alarmone guanosine tetraphosphate (ppGpp), which primarily affects the transcriptional program of the bacterial cell: ppGpp influences the competition between different RNA polymerase-associated promoter-specificity sigma (σ) factors, so that different stress-related σ factors (notably σ38 and σ54) bind to the RNA polymerase at the expense of the housekeeping σ factor, σ70 3. Collectively, this results in the adjustment of cellular metabolism to divert resources away from expression of genes required for normal growth to ones that ensure survival until nutrient conditions improve. We recently demonstrated that NtrC activates transcription of relA, the major gene responsible for the synthesis of ppGpp, in N-starved E. coli4. Thus, NtrC, by coupling the Ntr and stringent responses in N-starved E. coli, facilitates efficient and rapid adaptation to N starvation.
Genome-wide binding analysis in N-starved E. coli revealed that NtrC and RNA polymerase bind the promoter region of the yeaGH operon, thus implying that the products of yeaG and yeaH could have a role in the adaptive response of E. coli to N starvation4. Previous global transcriptome analyses have revealed that the expression of yeaG and yeaH are highly upregulated in E. coli in response to diverse stresses, including low pH, hyperosmotic conditions, entry into stationary phase and sulphur limitation5,6. Moreover, in Salmonella Typhimurium these genes are also required in the response against the antimicrobial peptide Polymyxin B7. Therefore, it is possible that yeaG and yeaH may have an important role in how bacteria adapt to diverse stress conditions. Consistent with this view, the yeaGH operon is highly conserved across bacterial species, especially in Enterobacteria (Figure S1A). Whereas yeaH encodes an uncharacterized protein that displays very little sequence or structural similarity to any known proteins described to date, the product of yeaG, YeaG, is a 75 kDa protein containing a carboxyl terminal domain that shares amino acid sequence homology to eukaryote-like serine/threonine kinase (eSTK) (Figure S1B). eSTKs have emerged as critical components of bacterial signal transduction systems and are central to important and diverse cellular functions, including bacterial stress response8,9. Consistent with their different biological roles, many eSTKs are modular and display a high diversity in domain organization10. Therefore, the kinase domain of many eSTKs is associated with additional domains, which commonly mediate signal perception through ligand binding or protein-protein interactions11. A small number of eSTKs are also associated with enzymatic domains (pectinases, esterases, phosphatases, etc.)11. Intriguingly, YeaG contains an amino-terminal ATPase Associated with diverse cellular Activities (AAA+) domain (Figure S1C). Typically, AAA+ domain-containing proteins are oligomeric mechanochemical enzymes that transform chemical energy derived from ATP binding and hydrolysis into mechanical force to induce conformational changes in their substrates12. Therefore, it seems that YeaG is an atypical, yet highly conserved, eSTK. Although a previous study demonstrated that recombinant YeaG has protein kinase activity in vitro13, surprisingly the biological role of YeaG in bacterial stress responses remains elusive. In this study, we have investigated the role of E. coli YeaG in the adaptive response to N starvation.
Results
yeaG has a role in the adaptive response to sustained N starvation
Previously we reported that NtrC and RNA polymerase bind to the promoter region of the yeaGH operon in N-starved E. coli4. To establish whether these binding events lead to transcription of the yeaGH operon we measured levels of yeaG mRNA by quantitative real-time PCR. Bacteria were grown in batch cultures in minimal media that was supplemented with a limiting amount (3 mM) of ammonium as the sole source of N. Under these conditions, we previously showed that bacterial growth arrest directly coincides with the acquisition of a bona fide N starved state (indicated as N- in the schematic in Fig. 1A where OD600 is ~0.85 and [NH4Cl] is <0.000625 mM) that occurs 20 min after ammonium has run out in the media (indicated as NRO in the schematic in Fig. 1A)4. As expected, yeaG mRNA levels in N-starved E. coli were 36-fold (±2.3 SD) higher than in bacteria from nitrogen-replete conditions (indicated as N+ in the schematic in Fig. 1A where OD600 is ~0.3 and [NH4Cl] is ~2.5 mM) (Fig. 1A). Having established that yeaG is expressed in N-starved E. coli shortly after sensing N starvation, we next determined the growth characteristics of a ∆yeaG mutant E. coli strain in minimal media containing limiting amount (3 mM) of ammonium as the sole N source. Under these conditions (henceforth referred to as ‘pre-starvation’ growth) no growth difference was detected between the ∆yeaG mutant and wild-type E. coli and both strains ceased growth when ammonium ran out in the media (Fig. 1B). Even though the expression of yeaG occurs shortly after the cells sense N starvation (Fig. 1A), it appears to be continuously expressed for up to 24 h into N starvation (Fig. 1C), we considered whether yeaG could have a role in adaptation to sustained (here defined as 24 h, unless otherwise indicated) N starvation. To investigate this, wild-type and ∆yeaG mutant bacteria were subjected to 24 h of N starvation before being sub-cultured into fresh media and their growth monitored (henceforth referred to as ‘recovery’ growth). As shown in Fig. 1D, the wild-type strain displayed a considerably increased lag phase (by 2.3 h) during ‘recovery’ growth compared to ‘pre-starvation’ growth, which is characteristic of bacterial adaptation to the new growth environment. In marked contrast to the wild-type strain, the ∆yeaG mutant strain displayed a much shorter lag phase (by 32 min or 13% less than the length of wild-type lag phase) during ‘recovery’ growth (Fig. 1D and Table S1). However, once in the exponential phase of ‘recovery’ growth, the doubling times of both strains were comparable (Fig. 1D and Table S1). Similar results were obtained with E. coli strains lacking yeaH or both yeaG and yeaH genes, indicating that yeaG and yeaH are functionally linked (Fig. 1D and Table S1). Control experiments in which we either measured the number of viable cells at different times during ‘recovery’ growth by plate count of colony forming units (CFU) (Fig. 1E) or monitored the rate of ammonium consumption during ‘recovery’ growth (Fig. 1F) independently validated the shorter lag phase of the ∆yeaG mutant compared to the wild-type strain. Importantly, the difference in the length of the lag phase between the wild-type and ∆yeaG strain during ‘recovery’ growth was directly proportional to the length of time spent under N starvation conditions (Fig. 2), which further emphasizes the role for yeaG in sustained N starvation, even though the expression of yeaG occurs shortly after the cells experience N starvation.
yeaG has a role in the adaptive response to sustained N starvation.
(A) yeaG is upregulated in response to N starvation. Levels of yeaG expression in wild-type E. coli were calculated as relative mRNA amounts compared to the 16S control under N-replete (N+) and N-starved (N-) conditions. The insert is a schematic graph showing that E. coli growth arrest corresponds to N run out (NRO) in the media (orange line) and indicates the time points (N+ and N-) at which cells were sampled for total RNA extraction and analysis. (B) ‘Pre-starvation’ growth of ∆yeaG, ∆yeaH and ∆yeaGH mutant strains is similar to that of the wild-type strain. The growth curves were obtained by determining the OD600nm readings at half-hourly time points. (C) Transcription of yeaG occurs during sustained (24 h) N starvation. The level of yeaG expression in wild-type E. coli is expressed as fold change in mRNA levels between N+ and N-24 h starved conditions. In the presence of rifampicin, an inhibitor of RNA synthesis, no yeaG mRNA was detected in the N-24 h sample, which indicates that the yeaG mRNA expression observed in the absence of rifampicin is due to transcription of yeaG over the 24 h period of sustained N starvation. The insert is a schematic representation of the experiment. (D) ∆yeaG, ∆yeaH and ∆yeaGH mutant strains display shorter lag phase during ‘recovery’ growth following 24 h of N starvation compared to a wild-type strain. The ‘recovery’ growth curves were obtained by determining the OD600nm readings at half-hourly time points. The dotted black line indicates the ‘pre-starvation’ growth curve for the wild-type strain. The insert is a schematic representation of the experiment. (E) Measurement of ‘recovery’ growth of wild-type and ∆yeaG mutant strain by colony-forming unit (CFU) count at 0 h, 2 h, 4 h and 6 h. (F) The rate of ammonium consumption by the wild-type and ∆yeaG mutant strain is directly proportional to their respective ‘recovery’ growth characteristics. Error bars on all growth curves and graph represent sd (n = 3). Statistical analyses were performed by one-way ANOVA (**P < 0.01).
The difference in the length of the lag phase between the wild-type and ∆yeaG strain during ‘recovery’ growth is directly proportional to the length of time spent under N starvation.
The ‘recovery’ growth curves of wild-type and ∆yeaG mutant strain were obtained by determining the OD600nm readings at half-hourly time points. The insert is a schematic representation of the experiment. The table summarizes growth parameters of wild-type and ∆yeaG mutant strain in ‘recovery’ growth following 20 min, 12 h, 18 h and 24 h in N starvation. Lag phase defined as the period up to and including OD600 = 0.1. The number in brackets indicates the percentage decrease in the length of the lag phase of the ∆yeaG mutant strain relative to the lag phase of the wild-type strain. The doubling time was determined from the slope of logarithmic growth function.
We were able to revert the growth characteristics of the ∆yeaG mutant strain to that of the wild-type strain by complementing with inducible plasmid-borne yeaG (∆yeaG pBAD-yeaG), thereby demonstrating that the altered growth of the mutant strain is not due to any undesired effects caused by the deletion of the yeaG gene (Fig. 3A,B). To determine if the catalytic activity of the AAA+ and/or STK domains of YeaG are required for function, we constructed mutant variants of YeaG based on amino acid sequence alignments with AAA+ and STK domains of well-characterized proteins (Figure S1B and Figure S1C). These mutants contained catalytically deleterious single amino acid substitutions either in the AAA+ (pBAD18-yeaG K116A and pBAD18-yeaG R232A) or STK (pBAD18-yeaG K426A) domain. We also generated a truncated version of YeaG by introducing a stop codon at amino acid position Y382 (pBAD18-yeaG Y382stop), which results in only the AAA+ domain being expressed. As shown in Fig. 3A–C, even though the mutant variants were produced at 64% (yeaG K116A), 48% (yeaG R232A), 100% (yeaG K426A) and 96% (yeaG Y382stop) of wild-type levels, none of them were able to complement the phenotype of the ∆yeaG mutant strain under our experimental conditions. Overall, the results in Figs 1, 2, 3 clearly demonstrate that yeaG (and yeaH) has a role in the adaptation of E. coli to sustained N starvation and that the catalytic activities of both, the AAA+ and STK domains of YeaG, are required in this process.
The catalytic activities of the AAA+ and eSTK domains of YeaG are required for its role in adaptation to sustained N starvation in E. coli.
(A,B) Growth curves show complementation of the phenotype of the ∆yeaG mutant strain with plasmid-borne yeaG (pBAD18-yeaG), but not with mutant variants of yeaG containing catalytically deleterious mutations in the AAA+ domain (in A. pBAD18-yeaG K116A; pBAD18-yeaG R232A) or eSTK domain (in B. pBAD18-yeaG K426A; pBAD18-yeaG Y382Stop). Error bars on all growth curves represent sd (n = 3). Statistical analyses were performed by one-way ANOVA (**P < 0.01). (C) Protein levels of wild-type and mutant variants of YeaG assessed by anti-His immunoblotting.
∆yeaG mutant strain displays increased metabolic activity than the wild-type strain following sustained N starvation
Since many bacteria adjust their metabolism as an adaptive response to prolonged conditions of nutritional adversity, we considered whether the short lag phase displayed by the ∆yeaG mutant strain during ‘recovery’ growth was indicative of an altered metabolic state ergo adaptive response between ∆yeaG mutant and wild-type strains to sustained N starvation. Accumulation of GFP has been widely used as a reporter for bacterial metabolic status14,15. Therefore, to investigate how E. coli adjust their metabolic state in response to N sustained starvation and to determine if this differs between the wild-type and ∆yeaG mutant strains, we assessed the rate of production of green fluorescent protein (GFP) at the single-cell level as a measure for metabolic activity during recovery growth in the presence of the inducer. Bacterial cells were recovered at hourly time points and fluorescence levels were determined by flow cytometry. Results in Fig. 4A,B, show that GFP production in the wild-type strain first became apparent at 3 h, although, strikingly, only in a subset (25%) of the population. By 4 h into ‘recovery’ growth the majority (77%) of the wild-type population contained GFP-producing bacteria and the increase in fluorescence levels continued progressively. However, a small proportion of the population remained non-fluorescent even after 5 h into ‘recovery’ growth. In marked contrast to the wild-type strain, GFP production could be detected in the ∆yeaG mutant strain population after only 2 h into ‘recovery’ growth and fluorescence levels increased at a constant rate over time. Strikingly, in the ∆yeaG population, the N starvation-induced metabolic heterogeneity clearly evident in the wild-type population was not detected. Overall, results are consistent with the ‘recovery’ growth characteristics of both strains (Fig. 1D) and indicate that the short lag phase observed for the ∆yeaG mutant strain during ‘recovery’ growth is likely due to an increased metabolic activity despite the mutant cells having experienced sustained N starvation. Importantly, the results also reveal differences between ∆yeaG mutant and wild-type strains at the population level: Whereas the wild-type population displayed heterogeneity in its metabolic activity and contained a small sub-population that appeared to be metabolically inactive, this was clearly not the case in the ∆yeaG mutant population.
∆yeaG mutant strain displays increased metabolic activity than the wild-type strain following sustained N starvation.
(A) Representative histograms of GFP fluorescence in wild-type (blue) and ∆yeaG mutant strain (red) populations at selected time points following induction of GFP expression during ‘recovery’ growth (after 24 h N starvation). The insert is a schematic representation of the experiment. (B) Quantification of GFP production over 8 h of ‘recovery’ growth (insert) was obtained from the geometric means of GFP fluorescence. Error bars represent sd (n = 3).
The yeaG-dependent adaptive response to sustained N starvation involves the transcriptional repression of mqsR/mqsA and dinJ/yafQ toxin-antitoxin genes
Toxin-Antitoxin (TA) genes, which are ubiquitous in bacteria, are key effectors that link bacterial growth to the nutritional status of the bacterial cell16. Most widely studied TA modules consist of two genes in an operon, which encode for a stable toxin that disrupts essential cellular processes, leading to reversible growth arrest and a labile antitoxin that forms a tight complex with the toxin and neutralises its effect17. The ratio between toxin and antitoxin has important implications in how bacterial cells adapt to stress. During nutritional starvation the antitoxin is degraded, thus generating sufficient free toxin to influence the metabolic status and induce population heterogeneity17. Since results in Fig. 4 reveal that E. coli adjusts its metabolism in response to sustained N starvation, resulting in a metabolically heterogeneous population, we decided to investigate the relative expression levels of ten known E. coli TA gene pairs in ∆yeaG and wild-type cells following 24 h N starvation. Results showed that the mRNA levels of the mqsR/mqsA TA module to be increased by 2.0 ± 0.9/1.8 ± 0.7 fold in wild-type bacteria following 24 h of N starvation compared to N+ conditions (Fig. 5A). Notably, we detected an even larger increase in mRNA levels corresponding to mqsR/mqsA, by 3.7-fold ± 1.6/3.5-fold ± 1.4 fold in the ∆yeaG mutant strain following 24 h of N starvation compared to N+ conditions (Fig. 5A). Further, in the ∆yeaG mutant strain another TA module, dinJ/yafQ, was upregulated by 2.8 ± 0.9/2.7 ± 1.3 fold, following 24 h of N starvation compared to N+ conditions. In contrast, dinJ/yafQ was not upregulated in the wild-type strain (Fig. 5A). We next focused on the biology of mqsR/mqsA and dinJ/yafQ to decipher how both of these TA modules could contribute to the properties of the ∆yeaG mutant strain: The mqsR/mqsA TA module is linked to regulation of the general stress response because it directly represses the transcription of rpoS, which encodes for σ38, the RNA polymerase associated σ factor that is responsible for executing the general bacterial stress response and cspD, which encodes for the cold-shock protein D, a DNA replication inhibitor18,19. Consistent with the increased levels of mqsR/mqsA mRNA levels in 24 h N-starved ∆yeaG mutant compared to wild-type strain (Fig. 5A), mRNA levels of both rpoS and cspD genes were reduced by 2.8 ± 0.8 fold and 3.92 ± 0.77 fold respectively in 24 h N-starved ∆yeaG mutant compared to wild-type strain (Fig. 5B,C). Similarly, DinJ, the toxin component of the dinJ/yafQ TA module, has been implicated in reducing σ38 levels through directly repressing the transcription of cspE, which in turn positively affects σ38 translation through stabilisation of rpoS mRNA20. Consistent with the increased levels of dinJ/yafQ mRNA in the ∆yeaG mutant compared to the wild-type strain following 24 h of N starvation (Fig. 5A), cspE mRNA levels were reduced by 4.26 ± 1.82 fold in the ∆yeaG mutant compared to the wild-type strain (Fig. 5D). To further corroborate these results, we compared σ38 protein levels and activity, which we expected to be reduced in ∆yeaG mutant relative to the wild-type strain following 24 h N starvation: As shown in Fig. 5E, σ38 levels were 1.7 ± 0.3 fold reduced in the ∆yeaG compared to the wild-type strain. Since the transcription of katG and katE, the genes encoding two catalases capable of converting harmful hydrogen peroxide (H2O2) into water and oxygen is σ38-dependent and thus serves as a surrogate for σ38 activity, we compared catalase activity in the wild-type, ∆yeaG and ∆katE mutant bacteria. Levels of H2O2 remaining in solution were quantified following incubation with whole-cell lysates from the three strains following 24 h of N starvation. Although the effect was not as accentuated as with the ∆katE mutant strain, the catalase activity of the ∆yeaG mutant strain was significantly reduced compared to that of the wild-type strain (Fig. 5F), consistent with the reduced levels of σ38 in the ∆yeaG mutant strain (Fig. 5E). Next, to directly link the property of the ∆yeaG mutant strain is mediated by the differences in the levels of expression of mqsR/mqsA and dinJ/yafQ, we constructed ∆dinJ∆yafQ∆yeaG and ∆mqsR∆mqsA∆yeaG triple deletion mutants as well as a ∆dinJ∆yafQ∆mqsR∆mqsA∆yeaG quintuple deletion mutant. As shown in Fig. 5G and as expected, the ‘recovery’ growth curve of the ∆dinJ∆yafQ∆mqsR∆mqsA∆yeaG mutant strain almost fully resembled that of the wild-type strain, whilst those of the ∆dinJ∆yafQ∆yeaG and ∆mqsR∆mqsA∆yeaG mutant strains partly resembled that of the mutant strain. Overall, we conclude that the yeaG-dependent adaptive response to sustained N starvation is mediated by the transcriptional repression of both mqsR/mqsA and dinJ/yafQ in E. coli cells that experience sustained N starvation. The absence of yeaG results in the transcriptional de-repression of mqsR/mqsA and dinJ/yafQ transcription, which, as a consequence, has a detrimental effect on σ38-dependent transcriptional programme in the adaptive response to sustained N starvation. To further substantiate this view, we compared the expression levels of selected genes that are either positively (rpsV, katE, pdhR, bolA and poxB) or negatively (sdhB) regulated by the σ38 containing RNA polymerase by qRT-PCR in the ∆yeaG mutant and wild-type strains following 24 h of N starvation. Consistent with the results obtained so far, four out of five of the genes (katE, pdhR, bolA and poxB) that are subjected to positive regulation by σ38 showed reduced levels of expression in the ∆yeaG mutant strain; the expression level of rpsV did not significantly differ in the ∆yeaG mutant and wild-type strains following 24 h of N starvation (Fig. 5H). Whereas sdhB is negatively regulated by the σ38 containing RNA polymerase in the stationary phase of growth in rich medium through promoter occlusion21, it seems that reduced σ38 levels in the ∆yeaG strain does not detectably affect the expression levels of sdhB in 24 h N-starved E. coli.
The yeaG-dependent adaptive response to sustained N starvation involves the transcriptional repression of mqsR/mqsA and dinJ/yafQ toxin-antitoxin genes.
(A) Expression levels of ten E. coli TA gene pairs in wild-type and ∆yeaG mutant strain calculated as fold change in mRNA between N+ and N-24 h N-starved conditions and normalised to 16S expression. Error bars represent sd (n = 2; except mqsR/mqsA, where n = 4). (B–D) Expression levels of cspD, rpoS and cspE in wild-type and ∆yeaG mutant strain calculated as fold change in mRNA between N+ and N-24 h N starved conditions and normalised to 16S expression. Error bars represent sd (n = 4). (E) Levels of RpoS are reduced in ∆yeaG mutant strain following 24 h N starvation. Representative immunoblot of whole-cell extracts of bacterial cells sampled after 20 min and 24 h of N starvation probed with anti-RpoS and anti-DnaK (loading control) antibody. Graph shows quantification of bands by densitometry (n = 3), calculated as the ratio in Optical Density between the bands corresponding to RpoS and DnaK. Error bars represent sd (n = 4). (F) RpoS-dependent catalase activity is impaired in the ∆yeaG mutant strain following 24 h of N starvation. Catalase activity was assessed as a function of concentration of H2O2 remaining in solution after incubation with 24 h N-starved wild-type, ∆yeaG or ∆katE bacterial lysates. Error bars represent sd (n = 3). (G) Deletion of dinJ/yafQ and mqsR/mqsA TA modules in the ∆yeaG strain leads to complementation of the mutant phenotype, whilst deletion of only one TA modules in the ∆yeaG strain leads to only partial complementation. The ‘recovery’ growth curves for wild-type, ∆yeaG, ∆dinJ∆yafQ∆yeaG, ∆mqsR∆mqsA∆yeaG and ∆dinJ∆yafQ∆mqsR∆mqsA∆yeaG mutant strains were obtained by OD600nm readings at hourly time points. (H.) Expression levels of sdhB, rpsV, katE, pdhR, bolA and poxB at N-24 calculated as fold change in mRNA between the wild-type and ∆yeaG mutant strain and normalised to 16S expression. Error bars represent sd (n = 3). Statistical analyses for all data-sets were performed by Student’s t test (*P < 0.05; **P < 0.01) relative to the wild-type strain.
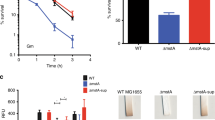
The absence of yeaG reduces bacterial cell viability during sustained N starvation
Since σ38-dependent gene expression is central to full adaptation to diverse stress conditions, including N starvation and thus ultimately to the survival of the bacterial cell, we next investigated how the deletion of yeaG impacts bacterial survival following sustained N starvation. Wild-type and ∆yeaG mutant bacterial numbers were assessed by plate count of CFU during N starvation over 5 days. As shown in Fig. 6A, following an initial period of little change, bacterial numbers began to drop after 72 h of N starvation for both strains. For the wild-type strain there is a slow but steady decrease in bacterial numbers. However, in comparison, the viability of the ∆yeaG mutant strain declines significantly faster. This faster rate of death was not observed in the ∆yeaG mutant strain that was complemented with a plasmid-borne wild-type copy of yeaG, whose bacterial numbers were comparable to the wild-type strain throughout sustained N starvation (Fig. 6A). However, as expected, a similar decrease in the number of viable bacterial cells was observed after 72 h of N starvation when the ∆yeaG mutant strain was complemented with catalytic mutant variants of yeaG (Fig. 6B). This observation further corroborates previous results (Fig. 3), which suggests that the catalytic activity of the AAA+ and STK domain of YeaG is important for its function. We thus conclude that absence of yeaG impairs the ability of E. coli to survive sustained periods of N starvation, most likely because the σ38-dependent adaptive response cannot be fully executed. Consistent with a role for YeaG in allowing bacteria to adapt to adverse growth environments, a comparative analysis of virulence carried out with a S. Typhimurium ∆yeaGH and wild-type strains indicated that mutant bacteria displayed lower proliferation in the spleen (Fig. 6C), thus implying that yeaG could be a determinant the overall fitness of S. Typhimurium.
The absence of yeaG reduces bacterial cell viability during sustained N starvation.
(A) The viability of wild-type, ∆yeaG mutant and ∆yeaG mutant complemented with plasmid-borne YeaG (∆yeaG pBAD18-yeaG) during sustained N starvation was determined by colony-forming unit (CFU) count over 5 days. Error bars represent s.e.m. (n = 3). Statistical analyses were performed by one-way ANOVA (*P < 0.05). (B) The viability of wild-type, ∆yeaG, ∆yeaG complemented with plasmid-borne YeaG (∆yeaG pBAD18-yeaG) or ∆yeaG mutant strain expressing variants of YeaG with deleterious mutations in the eSTK catalytic domain (pBAD18-yeaG K426A; pBAD18-yeaG Y382Stop) was determined by colony-forming unit (CFU) count after 20 min, 72 h & 120 h in N starvation. Error bars represent sd (n = 3). Statistical analyses were performed by one-way ANOVA (*P < 0.05). (C) Competitive Index (CI) analysis of Salmonella Typhimurium ∆yeaGH. C57 BL/6 mice (n = 5) were inoculated intragastrically with equal numbers of wild-type and mutant strain. Bacteria were recovered from infected spleens after 5 days post-inoculation and strains distinguished based on antibiotic resistance by replica plating. A CI value of 0.64 (p = 0.037), indicating moderate attenuation of the ∆yeaGH mutant strain in vivo, was obtained by calculating the ratio of wild-type to ∆yeaGH bacteria recovered (output), divided by the ratio of wild-type to ∆yeaGH mutant bacteria present in the inoculum (input). The scatter plot displays values obtained for individual mice and the mean is indicated. (D) Metabolically-inactive bacteria are more likely to withstand long-term stress. Representative histogram of GFP fluorescence shows wild-type population distribution according to metabolic activity at selected time points. The insert diagram shows the levels of GFP fluorescence measured at 3.5 h following induction of GFP expression during ‘recovery’ growth. Bacteria were subsequently transferred to media without inducer or N and GFP fluorescence was assessed again after 24 h in starvation conditions (see text for details).
The population of wild-type E. coli with increased metabolic activity following sustained N starvation displays impaired viability
The absence of yeaG clearly has a detrimental effect on the viability of E. coli during sustained period of N starvation (Fig. 6). Results also suggest that, unlike the wild-type strain, the ∆yeaG strain exists in a single population with increased metabolic activity following sustained N starvation (Fig. 4). To determine if the differences in population metabolic heterogeneity result in reduced viability of the ∆yeaG mutant compared to the wild-type strain following sustained periods of N starvation, we took advantage of the heterogeneous population profile of the wild-type bacterial population during ‘recovery’ growth (Fig. 4). After 3.5 h of ‘recovery’ growth, where metabolic heterogeneity in the wild-type population was evident (Fig. 4), bacteria were transferred to fresh growth media without arabinose (the inducer for GFP expression) and N, thereby stopping both GFP production and bacterial growth. The aim here was to ‘freeze’ the existing population profile that was present at 3.5 h during ‘recovery’ growth. After a further period of 72 h in N starvation, the decrease in CFU in the bacterial populations seen in Fig. 6A was accompanied by the disappearance of the wild-type population displaying increased metabolic activity (i.e. increased GFP production, representing 34% of the total bacteria analysed) (Fig. 6D). Because pre-formed matured GFP molecules are stable for at least 96 h inside live bacterial cells22, the loss of bright GFP bacteria can be attributed to bacterial death. We conclude that the population of wild-type E. coli with increased metabolic activity following sustained N starvation displays impaired viability. Therefore, the ∆yeaG mutant strain is less viable than the wild-type strain, because it, unlike the latter, exists in a single and metabolically active population following sustained N starvation (Figs 4 and 6A).
Discussion
The operon containing the yeaG and yeaH genes is highly conserved in E. coli and related bacteria. Results from several transcriptomics studies have shown that this operon is expressed in response to a diverse range of stresses. Whilst yeaH encodes a protein of little sequence or structural similarity to any known proteins, the product of yeaG is an eSTK. The impetus for the present study was the lack of a biological role for yeaG in bacterial stress responses. We have now assigned a role for yeaG in the adaptation of E. coli to sustained N starvation.
It is well established that rpoS is not expressed in exponentially growing bacteria and the regulation of its expression is tightly controlled at the transcription, translation and protein stability and activity levels as it impacts multiple physiological properties of the cell that affect growth. Therefore, managing transcription and translation of rpoS is an essential aspect of the bacterial stress response. Our results suggest that yeaG acts upstream of rpoS in the regulatory cascade that allows adaptation to sustained N starvation. In the scenario proposed in Fig. 7, in response to sustained N starvation, YeaG represses the transcription of mqsR/mqsA and dinJ/yafQ TA genes. This positively impacts rpoS transcription and translation, which as a consequence will result in the execution of the rpoS-dependent gene expression programme, needed to fully implement the adaptive response to sustained N stress. Conversely, in the absence of yeaG, our results show that the transcription of mqsR/mqsA and dinJ/yafQ TA genes become de-repressed in response to sustained N starvation. This negatively impacts rpoS at the transcriptional and translational levels and collectively results in an incomplete execution of the rpoS-dependent adaptive response to sustained N stress. Interestingly, a ΔrpoS mutant grows poorly compared to the wild-type and ∆yeaG strains in the recovery growth experiment (Figure S2), We thus propose that the correct intracellular levels of rpoS is important to instigate the appropriate adaptive response to sustained N starvation, whereas the absence of rpoS is likely to have significant pleiotropic effects on how E. coli adapts to N stress. This observation further underscores the view that yeaG, acting upstream of rpoS, ensures that intracellular levels of rpoS in E. coli exposed to sustained N starvation are maintained to allow the appropriate adaptive response to be executed. The mechanistic basis by which the YeaG mediated repression of mqsR/mqsA and dinJ/yafQ transcription occurs remains elusive and will be the subject of future studies. However, since MqsR, MqsA, DinJ or YafQ are not phosphorylated in a global phosphoproteome analysis of E. coli cells from stationary phase batch cultures grown in minimal media23, it is unlikely that they serve as substrates for YeaG. It is well established that a variety of regulatory mechanisms (anti-adaptor proteins, small RNAs) contribute to increasing σ38 levels (via either increased rpoS translation and decreased σ38 degradation) in response to suboptimal growth conditions24,25,26,27 including nutrient deprivation and thus we cannot exclude an effect of YeaG on these mechanisms. Nevertheless, we previously reported that NtrC couples the Ntr stress response and the stringent response via the activation of relA transcription1,4. The new results now indicate that the full coupling of these two major bacterial stress responses to N stress adaptation may also involve YeaG. Further, the results also highlight the requirement for the three major E. coli σ factors for implementing the full Ntr response: σ54 (for the NtrC-mediated activation of Ntr genes, including yeaG), σ70 (for nitrogen assimilation control protein (Nac)-mediated regulation of Ntr response genes) and σ38 (for the general stress response).
By generating phenotypically diverse subpopulations, bacteria employ so called ‘bet-hedging’ strategies to maximise survival by increasing the chance that a small percentage of the population, called persisters, will be better adapted to any sustained adverse change in the growth environment28. We have demonstrated that sustained N starvation results in the generation of a metabolically heterogeneous population in a yeaG-dependent manner. This seems to clearly confer a physiological advantage by promoting bacterial survival during sustained periods of N starvation. Our results imply that yeaG might potentially influence for the formation of persisters during sustained N starvation that can be less susceptible to killing by antibiotics. Notably and in support of this view, the transcription of cspD, a gene required for persister formation29, becomes de-repressed during sustained N starvation through the yeaG-mediated repression of mqsR/mqsA transcription.
In summary, this study provides new insights into mechanisms used by E. coli to adapt to sustained N starvation. Further, since E. coli is still one of the most widely used systems for the production of recombinant proteins in industrial bioprocessing, managing changes in metabolic performance, such as augmenting the metabolic activity and population uniformity in the ∆yeaG strain, could represent a novel route for optimising such processes. This is especially relevant since phenotypic heterogeneity is a major obstacle in the context of improving the robustness of bioprocessing procedures involving bacteria30. Further, since the metabolic state clearly influences the susceptibility of bacteria to antibiotics31, these findings could potentially be exploited as an innovative way to combat antibiotic-recalcitrant bacteria that cause disease. Importantly, this study has uncovered a novel aspect of the regulatory basis that underpins the Ntr response in E. coli and highlights the dynamic and layered complexity of bacterial stress response networks.
Methods
Bacterial strains, plasmids and growth conditions
The strains used in this study were derivatives from Escherichia coli K-12 or Salmonella Typhimurium strain NTCC 12023 and are listed in Table S2 in the supplementary material. The E. coli ∆yeaGH and S. Typhimurium ∆yeaGH double mutant strains were constructed using the λ red recombinase method for gene deletion32; briefly, a kanamycin resistance cassette was introduced in the bacterial chromosome at the location of the two adjacent genes, thereby replacing them. The E. coli ∆dinJ∆yafQ∆yeaG triple mutant strain was obtained by transduction of the knockout yeaG allele from ∆yeaG (donor strain) into the ∆dinJ∆yafQ double mutant (recipient strain) using the P1 bacteriophage. Similarly the E. coli ∆dinJ∆yafQ∆mqsR∆mqsA∆yeaG quintuple mutant was obtained by transduction of the knockout mqsRA allele from ∆mqsR∆mqsA (donor strain) in to the triple mutant ∆dinJ∆yafQ∆yeaG (recipient strain) cured of its kanamycin cassette using pCP20, using the P1 bacteriophage. Bacteria were grown at 37 °C, 180 rpm, in Gutnick minimal media (33.8 mM KH2PO4, 77.5 mM K2HPO4, 5.74 mM K2SO4, 0.41 mM MgSO4), supplemented with Ho-LE trace elements33 and 0.4% (w/v) glucose and containing either 10 mM NH4Cl (for precultures) or 3 mM NH4Cl (for ‘pre-starvation’ and ‘recovery’ growth). In all growth assays, bacteria were subcultured in 50 ml of growth media for a starting OD600 of 0.05. Where appropriate, growth medium was supplemented with 50 μg/ml ampicillin. For strains carrying complementation (pBAD18) or dual-fluorescence (pFCcGi) plasmids, arabinose was added at 0.1% and 0.2% (w/v), respectively, for induction of gene expression. Plasmids used in this study are listed in Table S2 in the supplementary material. Ammonium concentration in the media was calculated using the Aquaquant ammonium quantification kit (Merck Millipore), according to the manufacturer’s instructions.
Western Blotting
For immunoblotting, the following commercial antibodies were used: mouse monoclonal anti-RpoS (WP009), at a dilution of 1 : 5 000 (NeoClone) and mouse monoclonal anti-DnaK (8E2/2), at a dilution of 1 : 10 000 (Enzo Life Sciences), in conjunction with the anti-mouse ECL horseradish peroxidase (HRP)-linked secondary antibody, at a dilution of 1 : 10 000 and mouse monoclonal anti-polyHistidine-Peroxidase (A7058), at a dilution of 1 : 5 000 (Sigma-Aldrich).
Quantitative real-time PCR (qRT-PCR)
RNA samples were obtained from bacteria at specific time points by stabilisation with Qiagen RNA Protect reagent (Qiagen) and extracted using Qiagen RNeasy Mini kit (Qiagen) and PureLink DNase Set (Invitrogen). Purified RNA was stored at −80 °C in nuclease-free water. cDNA was amplified from 100 ng of RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). qRT-PCR reactions were as follows: 1 μl cDNA, 5 μl TaqMan Fast Universal PCR Master Mix (Applied Biosystems), 0.5 μl TaqMan probe (Applied Biosystems), in a total volume of 10 μl. Amplification was performed on an Applied Biosystems StepOnePlus Real-Time PCR system using the following conditions: 50 °C (2 min), 95 °C (10 min), followed by 40 cycles of 95 °C (15 s) and 60 °C (1 min). Primer and probe mixtures were custom designed (Custom TaqMan Gene Expression Assays, Applied Biosystems) and sequences are provided in Table S3 in the supplementary material. For analysis of TA mRNA, qRT-PCR reactions were performed using SybrGreen PCR master mix (Applied Biosystems) with 400 ng cDNA and 2 μM specific primers (listed on Table S3). Amplification was done on Rotor-Gene 3000 (Corbett Life Science) with the following cycling parameters: 95 °C (10 min), followed by 45 cycles of 95 °C (10 s), 55 °C (30 s), 68 °C (15 s) and 72 °C (40 s). All real-time analysis was performed in triplicate and quantification of 16S expression served as internal control.
Measurement of bacterial growth and viability
To establish growth curves, bacterial growth was measured over time by optical density (OD600nm). In ‘recovery’ growth, similar numbers of viable cells were confirmed at the starting point. To measure bacterial replication by Fluorescence Dilution, dilution of GFP fluorescence was assessed in ‘recovery’ growth, in the absence of arabinose, with pre-induced strains carrying pFCcGi. At selected times, bacteria were fixed using 2% (w/v) paraformaldehyde in PBS for 10 min at room temperature, resuspended in PBS solution and kept at 4 °C until analysis by flow cytometry. Bacterial replication was measured as the fold change in the geometric mean values of GFP fluorescence intensity between a given time point and 0 h. Number of viable cells was determined by colony forming unit (CFU) ml−1 of bacteria at selected time points, by plating serial dilutions onto LB agar. Bacterial growth was also measured as the fold change in CFU ml−1 between selected time points and 0 h.
Flow cytometry
For each sample, 10,000 bacterial events were acquired on a LSR Fortessa flow cytometer (BD) using FACSDiva software. GFP and mCherry fluorescence intensities were detected at 525/50 nm and 610/20 nm, respectively. Data analysis was done using the FlowJo software version 10 (TreeStar). To analyse GFP fluorescence, bacteria were previously gated on mCherry-positive signal.
Catalase Activity
Catalase activity of bacterial cells was measured using Catalase Assay kit (Sigma-Aldrich), according to the manufacturer’s instructions. Bacteria were pelleted from 1 ml aliquot of culture, washed with Assay Buffer and lysed by sonication, prior to colorimetric assay for calculation of hydrogen peroxide substrate remaining in solution.
Competitive infections
All animal experiments were performed in accordance with the Animals Scientific Procedures Act 1986 and were approved by the Imperial College London Ethical Review Committee. Infection studies were carried out with 6–8 week old female C57BL/6mice (Charles River). Groups of 5 mice were inoculated intragastrically with 0.2 ml of bacteria resuspended in PBS solution. The bacteria inoculum was 5 × 108 CFU of combined wild-type and mutant strains (2.5 × 108 of each strain, at a ratio of 1:1). Bacterial input was quantified by plating serial dilutions of the inoculum and strains were differentiated based on antibiotic resistance. Mice were sacrificed after 5 days and dilution series of spleen lysates were plated for CFU count (output) and strains were differentiated based on antibiotic resistance. Values for Competitive Index (CI) were calculated as the ratio of mutant to wild-type in the output divided by that of the input.
Bioinformatics
Alignments were performed using the Molecular Evolutionary Genetics Analysis (MEGA) software version 634. Phylogenetic dendrogram was obtained from multiple sequence alignments using the maximum likelihood method. The full amino acid sequence of E. coli YeaG was used in the analysis.
Statistical analysis
Unless specified otherwise, data is represented as the mean average of 3 independent experiments or more and variation is shown by standard error of mean (s.e.m.). Statistically significant relationships were determined using Student’s t-test or one-way analysis of variance (ANOVA), with a probability (p) value < 0.05 being the statistical significance considered.
Additional Information
How to cite this article: Figueira, R. et al. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci. Rep. 5, 17524; doi: 10.1038/srep17524 (2015).
References
Brown, D. R., Barton, G., Pan, Z., Buck, M. & Wigneshweraraj, S. Combinatorial stress responses: direct coupling of two major stress responses in Escherichia coli. Microbial Cell 1, 315–317, 10.15698/mic2014.09.168 (2014).
Zimmer, D. P. et al. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97, 14674–14679, 10.1073/pnas.97.26.14674 (2000).
Srivatsan, A. & Wang, J. D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105, 10.1016/j.mib.2008.02.001 (2008).
Brown, D. R., Barton, G., Pan, Z., Buck, M. & Wigneshweraraj, S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nature communications 5, 4115, 10.1038/ncomms5115 (2014).
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F. & Hengge, R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters and sigma factor selectivity. J. Bacteriol. 187, 1591–1603, 10.1128/JB.187.5.1591-1603.2005 (2005).
Gyaneshwar, P. et al. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187, 1074–1090, 10.1128/JB.187.3.1074-1090.2005 (2005).
Erickson, K. D. & Detweiler, C. S. The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62, 883–894, 10.1111/j.1365-2958.2006.05420.x (2006).
Perez, J., Castaneda-Garcia, A., Jenke-Kodama, H., Muller, R. & Munoz-Dorado, J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc. Natl. Acad. Sci. USA 105, 15950–15955, 10.1073/pnas.0806851105 (2008).
Pereira, S. F., Goss, L. & Dworkin, J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212, 10.1128/MMBR.00042-10 (2011).
Tyagi, N., Anamika, K. & Srinivasan, N. A framework for classification of prokaryotic protein kinases. PLoS One 5, e10608, 10.1371/journal.pone.0010608 (2010).
Krupa, A. & Srinivasan, N. Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes. BMC Genomics 6, 129, 10.1186/1471-2164-6-129 (2005).
Wendler, P., Ciniawsky, S., Kock, M. & Kube, S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823, 2–14, 10.1016/j.bbamcr.2011.06.014 (2012).
Tagourti, J., Landoulsi, A. & Richarme, G. Cloning, expression, purification and characterization of the stress kinase YeaG from Escherichia coli. Protein Expr. Purif. 59, 79–85, 10.1016/j.pep.2008.01.005 (2008).
Lowder, M. & Oliver, J. D. The Use of Modified GFP as a Reporter for Metabolic Activity in Pseudomonas putida. Microb. Ecol. 41, 310–313, 10.1007/s002480000094 (2001).
Ceroni, F., Algar, R., Stan, G. B. & Ellis, T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods 12, 415–418, 10.1038/nmeth.3339 (2015).
Maisonneuve, E. & Gerdes, K. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548, 10.1016/j.cell.2014.02.050 (2014).
Gerdes, K. & Maisonneuve, E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66, 103–123, 10.1146/annurev-micro-092611-150159 (2012).
Wang, X. et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366, 10.1038/nchembio.560 (2011).
Kim, Y. et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12, 1105–1121, 10.1111/j.1462-2920.2009.02147.x (2010).
Hu, Y., Benedik, M. J. & Wood, T. K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 14, 669–679, 10.1111/j.1462-2920.2011.02618.x (2012).
Levi-Meyrueis, C. et al. Repressor activity of the RpoS/sigmaS-dependent RNA polymerase requires DNA binding. Nucleic Acids Res. 43, 1456–1468, 10.1093/nar/gku1379 (2015).
Helaine, S. et al. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. USA 107, 3746–3751, 10.1073/pnas.1000041107 (2010).
Soares, N. C., Spat, P., Krug, K. & Macek, B. Global dynamics of the Escherichia coli proteome and phosphoproteome during growth in minimal medium. J. Proteome Res. 12, 2611–2621, 10.1021/pr3011843 (2013).
Battesti, A., Majdalani, N. & Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213, 10.1146/annurev-micro-090110-102946 (2011).
Mika, F. & Hengge, R. Small RNAs in the control of RpoS, CsgD and biofilm architecture of Escherichia coli. RNA Biol. 11, 494–507, 10.4161/rna.28867 (2014).
Landini, P., Egli, T., Wolf, J. & Lacour, S. sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 6, 1–13, 10.1111/1758-2229.12112 (2014).
Battesti, A., Majdalani, N. & Gottesman, S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc. Natl. Acad. Sci. USA 112, 5159–5164, 10.1073/pnas.1504639112 (2015).
Veening, J. W., Smits, W. K. & Kuipers, O. P. Bistability, epigenetics and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210, 10.1146/annurev.micro.62.081307.163002 (2008).
Kim, Y. & Wood, T. K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391, 209–213, 10.1016/j.bbrc.2009.11.033 (2010).
Delvigne, F., Zune, Q., Lara, A. R., Al-Soud, W. & Sorensen, S. J. Metabolic variability in bioprocessing: implications of microbial phenotypic heterogeneity. Trends Biotechnol. 32, 608–616, 10.1016/j.tibtech.2014.10.002 (2014).
Peng, B. et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–261, 10.1016/j.cmet.2015.01.008 (2015).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645, 10.1073/pnas.120163297 (2000).
Atlas, R. M. Handbook of microbiological media vol 1. Fourth edition. (CRC Press, 2010).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729, 10.1093/molbev/mst197 (2013).
Acknowledgements
This work was supported by a Wellcome Trust Investigator Award and grants from the Biotechnology and Biological Sciences Research Council (BBSRC).
Author information
Authors and Affiliations
Contributions
R.F., D.F., M.J.G.E., L.B. and D.R.B. performed the experiments; Z.P. performed preliminary work; R.F., D.R.B., S.H. and S.W. performed the data analysis; R.F., D.R.B., S.H. and S.W. wrote the manuscript and S.W. provided overall direction for the project.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Figueira, R., Brown, D., Ferreira, D. et al. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci Rep 5, 17524 (2015). https://doi.org/10.1038/srep17524
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17524
This article is cited by
-
Identification of serine/threonine kinases that regulate metabolism and sporulation in Clostridium beijerinckii
Applied Microbiology and Biotechnology (2022)
-
Mutant phenotypes for thousands of bacterial genes of unknown function
Nature (2018)
-
Functional analysis of PrkA - a putative serine protein kinase from Mesorhizobium alhagi CCNWXJ12-2 - in stress resistance
BMC Microbiology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.