Abstract
Marine planktonic cyanobacteria contributed to the widespread oxygenation of the oceans towards the end of the Pre-Cambrian and their evolutionary origin represents a key transition in the geochemical evolution of the Earth surface. Little is known, however, about the evolutionary events that led to the appearance of marine planktonic cyanobacteria. I present here phylogenomic (135 proteins and two ribosomal RNAs), Bayesian relaxed molecular clock (18 proteins, SSU and LSU) and Bayesian stochastic character mapping analyses from 131 cyanobacteria genomes with the aim to unravel key evolutionary steps involved in the origin of marine planktonic cyanobacteria. While filamentous cell types evolved early on at around 2,600–2,300 Mya and likely dominated microbial mats in benthic environments for most of the Proterozoic (2,500–542 Mya), marine planktonic cyanobacteria evolved towards the end of the Proterozoic and early Phanerozoic. Crown groups of modern terrestrial and/or benthic coastal cyanobacteria appeared during the late Paleoproterozoic to early Mesoproterozoic. Decrease in cell diameter and loss of filamentous forms contributed to the evolution of unicellular planktonic lineages during the middle of the Mesoproterozoic (1,600–1,000 Mya) in freshwater environments. This study shows that marine planktonic cyanobacteria evolved from benthic marine and some diverged from freshwater ancestors during the Neoproterozoic (1,000–542 Mya).
Similar content being viewed by others
Introduction
Cyanobacteria have fundamentally transformed the geochemistry1,2 of our planet. Multiple lines of geochemical evidence support the occurrence of intervals of profound global environmental change at the beginning and end of the Proterozoic (2,500–542 Mya)3,4,5. While it is widely accepted that the presence of molecular oxygen in the early fossil record was the result of cyanobacteria activity, little is known about how cyanobacteria evolution (e.g., habitat preference) may have contributed to changes in biogeochemical cycles through Earth history. Geochemical evidence has indicated that there was a first step-increase in the oxygenation of the Earth’s surface, which is known as the Great Oxidation Event (GOE), in the early Paleoproterozoic (2,500–1,600 Mya)1,2. A second but much steeper increase in oxygen levels, known as the Neoproterozoic Oxygenation Event (NOE)4,6,7, occurred at around 800 to 500 Mya5,8. Recent chromium (Cr) isotope data point to low levels of atmospheric oxygen in the Earth’s surface during the mid-Proterozoic3, which is consistent with the late evolution of marine planktonic cyanobacteria during the Cryogenian9; both types of evidence help explain the late emergence and diversification of metazoans10.
Understanding the evolution of planktonic cyanobacteria is an essential question because their origin fundamentally transformed the nitrogen and carbon cycles towards the end of the Pre-Cambrian9. It remains unclear, however, what evolutionary events led to the emergence of open-ocean planktonic forms within cyanobacteria and how these events relate to geochemical evidence during the Pre-Cambrian4. So far, it seems that ocean geochemistry (e.g., euxinic conditions during the early- to mid-Proterozoic)4,7,11 and nutrient availability12 likely contributed to the apparent delay in diversification and widespread colonization of open ocean environments by planktonic cyanobacteria during the Neoproterozoic9.
Marine phytoplankton today contribute to almost half of the Earth’s total primary production13. Within the cyanobacteria, only a few lineages colonized the open-ocean (i.e., Crocosphaera and relatives, cyanobacterium UCYN-A, Trichodesmium, as well as Prochlorococcus and Synechococcus)14,15,16,17. From these lineages, N-fixing cyanobacteria are particularly important because they exert a control on primary productivity and the export of organic carbon to the deep ocean14, by converting nitrogen gas (N2) into ammonium (NH4+), which is later used to make amino acids and proteins. Marine picocyanobacteria (i.e., Prochlorococcus and Synechococcus) numerically dominate most phytoplankton assemblages in modern oceans contributing importantly to primary productivity16,17,18. While some planktonic cyanobacteria are unicellular and free living cells (e.g., Crocosphaera, Prochlorococcus, Synechococcus), others have established symbiotic relationships with prymnesiophyte algae15. Amongst the filamentous forms, Trichodesmium are free-living and form aggregates. However, filamentous heterocyst-forming cyanobacteria (e.g., Richelia, Calothrix) are found in association with diatoms such as Hemiaulus, Rhizosolenia and Chaetoceros19,20,21.
While environmental conditions might have prevented the widespread diversification of planktonic forms during most of the Pre-Cambrian9, the evolutionary history of marine planktonic cyanobacteria (e.g., habitat preferences, morphology) likely played an important role in the events surrounding the emergence of complex life in the oceans. Data from 131 cyanobacterial genomes was used to carry out large-scale multi-gene analyses of cyanobacteria; these analyses provide robust evidence for the early evolution of filamentous forms and mat-forming/benthic cyanobacteria and a delay in the emergence of marine planktonic cyanobacteria towards the end of the Pre-Cambrian. Two separate data sets (protein and nucleotide sequence data) and five different types of substitution models (including the CAT-GTR model) were used to explore the timing of key evolutionary events that led to the late emergence of planktonic cyanobacteria. Bayesian stochastic character mapping analyses were performed to study the evolutionary traits involved in the emergence of marine planktonic cyanobacteria such as loss of filamentous forms (and presumably intracellular communication), decrease in cell diameter and shifts in habitat preference within cyanobacteria. This study also shows that marine planktonic cyanobacteria evolved from benthic marine and freshwater ancestors.
Results
Phylogenetic relationships
An increase in genome sequencing and taxon-sampling have allowed for broad coverage of a range of morphologies, lifestyles and metabolisms within cyanobacteria22. The analyses performed here included a large phylogenetic data set consisting of 131 genome taxa with a total of 56,251 amino acids (aa) and 4,555 base pairs (bp). Whilst analyses have recovered well-supported monophyletic groups previously reported9,22,23,24,25,26, new genomic data have revealed novel deep-branching relationships of major cyanobacteria lineages22,24 (Fig. 1 and Supplementary Fig. S1). In this study Pseudanabaena appears as an early divergent lineage within cyanobacteria (Fig. 1 and Supplementary Fig. S1) occurring in 88% of the Maximum Likelihood trees generated for each gene alignment (137 genes) generated in SATé 2.2.327. A basal position for Pseudanabaena is consistent with recent large-scale multi gene studies22,28. Previous studies suggesting that Pseudanabaena is a derived lineage were based on SSU rRNA datasets9,25.
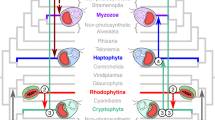
The origin and diversification of cyanobacteria as inferred from geologic time.
The phylogenetic tree was estimated based on 135 proteins and two ribosomal RNAs (SSU and LSU) from 131 taxa implementing Maximum Likelihood in RAxML GUI v.1.161. Bayesian relaxed molecular clock analyses were carried out in MCMCtree31. For the tree shown age estimates were estimated under the independent rates model33 for the RNA data set. Four calibrations (brown circles) were used2,67,68,69 for the tree shown and were treated as soft bounds. The root of the tree was set with a maximum age of 2,700 Myr63 and a minimum age of 2,320 Myr2. Numbered nodes 1–10 indicate divergence times for clades and key evolutionary events in the evolution of cyanobacteria including: the first origin of filamentous cells, ancestors of the Microcyanobacteria and Macrocyanobacteria, unicellular N-fixers and the marine Synechococcus and Prochlorococcus clades. Age estimates are given in Table 1, which includes the corresponding values for the posterior 95% confidence intervals.
Genomic data have also clarified problematic phylogenetic relationships such as the positioning of the filamentous LPP group (Fig. 1 and Supplementary Fig. S1). New data strongly support sister relationships between the LPP clade (Supplementary Fig. S1) and Prochlorothrix, Synechoccocous elongatus and the SynPro clade (Synechococcus, Prochlorococcus, Cyanobium). While the inclusion of recently sequenced genomes22,24,29,30 suggest a new placement for Trichodesmium (Fig. 1 and Supplementary Fig. S1), more Oscillatoria-like genomes are needed to fully understand the placement of this important lineage. Modern marine planktonic cyanobacteria evolved within two major groups of cyanobacteria, here referred to as the Microcyanobacteria and the Macrocyanobacteria since they are well-supported monophyletic clades (Supplementary Fig. S1). Whilst the Microcyanobacteria contain lineages with smaller cell diameters (<3 μm), the Macrocyanobacteria contain lineages with larger cell diameters (>3 μm; Supplementary Fig. S3). The Macrocyanobacteria are the most taxonomically and ecologically diverse clade including lineages such as Synechocystis, Pleurocapsa, Microcystis, Trichodesmium and the Nostocales, amongst others (Fig. 1).
Relaxed molecular clock analyses
Age divergences were estimated using two independent data sets, RNA (SSU and LSU: 4,555 bp) and proteins (18 genes: 4,980 aa) and applying a Bayesian approach31,32. Four calibration points were implemented, three of which have been previously used9,25. Relaxed molecular clock analyses were performed under the independent-rates model33, which has been shown to be the best fitting molecular clock model for cyanobacteria based on Bayes Factors9. Four different models of molecular evolution were implemented for proteins and RNA in MCMCtree and the CAT- GTR model for proteins and RNA in Phylobayes (Table 1). The implementation of two different maximum ages for the origin of oxygenic photosynthesis (i.e., 3,000 and 2,700 Myr) resulted in different age estimates for the origin of filamentous forms (node 2). While an older maximum age (3,000 Myr) predicts the origin of filamentous forms (node 2) before the GOE with estimates ranging between 2,665 and 2,559 Mya, a younger maximum age (2,700 Myr) predicts filamentous forms appearing around the time of the GOE between 2,460 and 2,351 Mya (Fig. 1, Table 1). Overall an older maximum age (3,000 Myr) tends to make ages older across all analyses.
Results were consistent across models of molecular evolution within each data set. There is strong evidence for a Neoproterozoic or early Cambrian origin for marine unicellular N-fixers (i.e., the Crocosphaera clade) and the filamentous Nodularia spumigena CCY9414. Age estimates appear to be younger for Prochlorococcus (nodes 9) and Synechococcus (node 10) based on the nucleotide data set, in contrast to the protein data set (Table 1). All analyses however provide robust evidence for the relatively late evolution of marine planktonic cyanobacteria. Other marine N-fixers evolved during the Phanerozoic such as Richelia (a diatom symbiont) and the cyanobacterium UCYN-A clade (a coccolithophore symbiont; Fig. 1). Age estimates across all analyses are summarized in Table 1 and Fig. 1 and are mostly in broad agreement.
Bayesian trait evolution analyses
The earliest cyanobacteria were likely unicellular (node 1) and inhabited low salinity environments (Fig. 2 and Supplementary Fig. S4). Living relatives of these early divergent lineages have been isolated from terrestrial/freshwater environments (e.g., Pseudanabaena PCC6802 Cyanothece PCC7425, Fig. 1), hot-springs (e.g., Thermosynechococcus elongatus BP-1) and coastal marine habitats (e.g., Acaryochloris and Synechococcus PCC7336). Bayesian stochastic character mapping analyses revealed that filamentous cyanobacteria evolved early on and different molecular clock analyses indicate filamentous forms evolved around 2,665 to 2,351 Mya and the GOE (node 2; Figs 1 and Supplementary Fig. S2). Ancestors of early filamentous forms likely resembled modern relatives of Pseudanabaena and the LPP clade (nodes 2 and 3; Fig. 2). All Basal Lineages and the Microcyanobacteria have retained small cell diameters exhibiting cells that are less than 3 μm, with most lineages exhibiting diameters that are less than 2 μm (Supplementary Fig. S3 and Supplementary Table S2). Interestingly, further decrease in cell diameter characterizes the evolution of the marine Prochlorococcus within the SynPro clade26. Also a switch from filamentous to unicellular cell types occurred (node 5; Figs 1 and 2) around 1,994 to 1,421 Mya (Table 1).
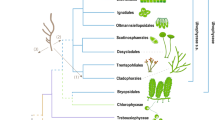
Timing and trends in cell diameter, loss of filamentous forms and habitat preference within cyanobacteria.
Nodes shown (1–10) correspond to Fig. 1 and Table 1. Stars represent common ancestors that appear in Fig. 1 but ages are not given in Table 1. The timing of the Great Oxidation Event (GOE) is after ref. 2, the Lomagundi-Jatuli Excursion after ref. 55 and Gunflint formation after ref. 73. Green lines represent freshwater lineages and blue lines represent marine lineages based on Bayesian inference of character evolution (stochastic character mapping analyses; Supplementary Fig. S4). Cartoons are not drawn according to scale; taxa with smaller cell diameter are shown at the bottom and larger cell diameter at the top.
All analyses suggest that the Macrocyanobacteria clade, exhibiting larger cell diameters (>3 up to 50 μm), may have evolved just after the GOE with age estimates ranging between 2,386 and 1,894 Mya (node 4; Fig. 1 and Table 1 and Supplementary Fig. S3). Within this clade two opposite evolutionary trends were found: 1) an increase in cell diameter (e.g., Fischerella and Mastigocladopsis) within the Nostocales and 2) a decrease in cell diameter (node 7) within the clade containing Microcystis and Crocosphaera relatives (Supplementary Table S2, Supplementary Fig. S3). A switch from filamentous to unicellular forms also occurred (node 7, Fig. 1 and Supplementary Fig. S2) around 1,437 to 1,047 Mya in freshwater habitats (Supplementary Fig. S4). Whilst unicellular marine N-fixing cyanobacteria (e.g., Crocosphaera and relatives) and Nodularia spumigena CCY9414 diverged from freshwater ancestors, Trichodesmium evolved from filamentous coastal marine lineages (Fig. 1 and Supplementary Fig. S4).
Discussion
Stem vs crown groups
Recent genomic data have improved the resolution of the cyanobacteria tree of life helping with the interpretation of the geological record9,25,28. Cyanobacteria fossils with a cell diameter bigger than 3 μm appeared in the Belcher Subgroup with fossils such as colonial coccoids (Eoentophysalis) and colonial ellipsoids (Eosynechococcus)34,35. Oscillatoria-like filamentous fossils (e.g., Gunflintia) appeared in the Gunflint iron formation36 and Halythrix sp. in the Belcher subgroup34. At approximately 1,900 Myr, microfossils with increased cell diameters as well as sheaths became common35. These findings are consistent with the evolutionary studies shown here in which ancestors with inferred cell diameters larger than 3 μm (node 4), the Macrocyanobacteria, postdate the GOE (Table 1, Fig. 2)25. It is therefore likely that the first appearance of reliable cyanobacteria fossils observed at around 2,000 Myr35,37 is indicative of an ancient cyanobacteria radiation23. Interestingly, age estimates based on molecular clock studies show a lag between the early origin of oxygenic photosynthesis25,28,38,39 and the first reliable evidence of cyanobacteria in the fossil record at around 2,000 Myr35,37,40. The Macrocyanobacteria clade (node 4) also evolved traits necessary for establishing thick laminated mats23,25,26 and have shown a significant shift in diversification rates39. It is not surprising that this clade contains the highest taxonomic and ecological diversity of modern cyanobacteria.
Age estimates shown here suggests that there was a delay between the appearance of the first reliable cyanobacteria fossils and the ancestors of the crown groups containing marine planktonic cyanobacteria (e.g., Nostocales/Gloeocapsa, Arthrospira/Trichodesmium and Pleurocapsa/Microcystis/Crocosphaera clades; Table 1; Fig. 1). The great majority of modern cyanobacteria can be traced back to the late Paleoproterozoic and the Mesoproterozoic, implying that older cyanobacteria fossils (e.g., Gunflintia) belong to stem groups with no living relatives. Crown groups with morphologies that required cell differentiation and division of labor (e.g., Nostocales) evolved during the Mesoproterozoic (Fig. 1). Cell differentiation is particularly important for some marine N-fixing planktonic cyanobacteria that evolved during the Neoproterozoic (e.g., Nodularia spumigena) and Cretaceous (e.g., Richelia). The evolution of cell differentiation mechanisms would have involved a specific program of gene expression including the induction of regulatory genes and of genes encoding the proteins for the morphological and biochemical differentiation for specialized cells (i.e., heterocyst)26. Comparative genomic studies have shown that more complex morphologies characteristic of crown groups (e.g., Nostocales) required the evolution of additional genes involved in signal transduction and transcription-related functional categories22. Genomic and trait evolution studies have also revealed that more complex morphologies within cyanobacteria exhibit bigger genome sizes26 presumably as a result of the more elaborate metabolic processes involved in these lineages41.
A historical perspective
Previous broad taxonomic and phylogenomic studies of prokaryotes studies have inferred a terrestrial/freshwater ancestry of cyanobacteria42,43. Trait evolution studies of cyanobacteria, implementing large genomic data sets studies, have come to similar conclusions23,25,29. The Bayesian stochastic character mapping analyses presented here confirm that cyanobacteria first evolved in freshwaters. Moreover, most cyanobacterial lineages inhabited benthic, terrestrial and/or shelf environments for most of the Proterozoic. Interestingly, in modern habitats, benthic cyanobacteria are much more taxonomically diverse. This is perhaps due to the great variety of available niches in coastal environments (e.g., intertidal or infralittoral areas)44. The early establishment of mat-forming filamentous cyanobacteria (Fig. 1)25,28 and subsequent dominance of benthic microbial communities would have restricted primary productivity to terrestrial and ocean margins (Fig. 2). The small area of fresh waters and ocean margins imply that the global biogeochemical impact of oxygenic photosynthesis would have been minimal until cyanobacteria started colonizing the open ocean9,25, which currently covers approximately two-thirds of the Earth’s surface41. Some marine planktonic lineages had a freshwater ancestor (Fig. 2). This is illustrated by the unicellular marine clades such as Crocosphaera and SynPro. Marine lineages adapted to marine environments by acquiring the machinery that enables them to osmoregulate in marine environments such as the set of genes responsible for the synthesis of compatible solutes: glucosylglycerol (GG), glucosylglycerate (GGA) and glycine betaine (GB)45,46.
This study has revealed that decrease in cell size and a switch from filamentous to unicellular forms or loss of filamentous forms, in the lead up to the origin of the Crocosphaera and the SynPro clades, likely played a key role in the emergence of a planktonic life style. Convergent evolution with regard to the emergence of unicellular phytoplankton forms suggests similar selective pressures taking place (e.g., nutrient starvation) on the evolutionary history of these lineages. Other strategies such as gas vesicles evolved to cope with buoyancy regulation amongst some marine filamentous lineages such as Trichodesmium and Nodularia spumigena.
Marine planktonic habitats likely provided a challenging environment for cyanobacteria to proliferate into since the ocean remained anoxic for most of the Proterozoic4,47,48. Under anoxic conditions, including episodes of euxinia (anoxic environments with the presence of hydrogen sulphide) and ferruginous conditions (see recent reviews on ocean geochemistry of the Pre-Cambrian4,11), key trace metals essential for N-fixation would have been depleted. This is particularly the case for micronutrients such as molybdenum (Mo) an essential constituent of the nitrogenase enzyme involved in N-fixation49. While marine mat-forming cyanobacteria such as Microcoleus chthonoplastes are capable of performing sulphide-dependent anoxygenic photosynthesis, this biogeochemical process appears to serve as a detoxification mechanism50 in response to an inherent active sulphur cycle found in microbial mats. In microbial mats, the establishment of vertically stratified layers (known as laminated structures) allows for the spatial separation of oxygenic photosynthesis and N-fixation (an anaerobic process inhibited by oxygen)50. The segregation of biogeochemical processes could have persisted through most of the Pre-Cambrian, allowing the coexistence of N-fixers and oxygenic phototrophs. Interestingly, Trichodesmium is the only modern planktonic filamentous N-fixer that diverged from mat-forming relatives51.
Marine planktonic cyanobacteria
Only a few lineages within cyanobacteria adapted to the lack of nutrients (oligotrophy) characteristic of the Earth’s open ocean. Age estimates suggest that there was an interval of more than a billion years between the timing of origin of the common ancestor of the Macrocyanobacteria and the origin of modern marine planktonic N-fixers (Fig. 1, Table 1). Nitrogen-fixing cyanobacteria evolved different morphologies and physiological strategies within the Macrocyanobacteria. At least three lineages evolved independently towards the end of the Pre-Cambrian: Crocosphaera and relatives, Trichodesmium and Nodularia spumigena. Ancestors of the unicellular N-fixers, Crocosphaera clade, underwent decrease in cell diameters and a switch from filamentous to unicellular forms. While unicellular marine N-fixers, the Crocosphaera clade, diverged from freshwater relatives, Trichodesmium evolved from filamentous mat-forming cyanobacteria which have modern relatives found in benthic and in marine littorals9,51. Within the Nostocales, Nodularia spumigena CCY9414, a lineage currently found in the Baltic Sea in salty or brackish waters, diverged from freshwater relatives towards the end of the Pre-Cambrian (Fig. 1). Recent planktonic lineages such as Richelia and the cyanobacterium UCYN-A clade evolved as symbionts during the Cretaceous.
Decrease in cell diameter was part of the evolutionary history of the major primary producers, Prochlorococcus and Synechococcus within the Microcyanobacteria (Fig. 1). The abundant marine Prochlorococcus and Synechococcus shared a common ancestor and evolved within the Microcyanobacteria (Fig. 1). The SynPro clade is sister to the filamentous Prochlorothrix and nested within the LPP clade (Fig. 1 and Supplementary Fig. S1). This phylogenetic relationship indicates that there was a switch from filamentous to unicellular forms39,52 in which cell adhesion and intracellular communication were likely lost. Within the evolution of the marine SynPro clade, there is also a trend in the decrease of genome and cell size17,26. Convergent evolution with regard to a decrease in cell diameter highlights the advantage of smaller cells when inhabiting oligotrophic environments. In modern oceans small phytoplankton cells usually dominate phytoplankton communities under oligotrophic conditions, such as the oceanic gyres, whereas larger phytoplankton cells are more abundant along continental margins and in upwelling zones, where nutrient concentrations tend to be higher and more variable53. Divergence times (Table 1) mostly agree with a Neoproterozoic or early Phanerozoic origin for Prochlorococcus (node 9) and the marine Synechococcus (node 10).
Lack of evidence for early marine planktonic cyanobacteria
After the GOE, it has been assumed that marine cyanobacteria were responsible for the increase in primary productivity (Lomagundi–Jatuli excursion) and carbon burial around 2,200 to 2,060 Mya2,54,55. Yet if marine planktonic cyanobacteria were involved in the carbon burial (e.g., Lomagundi-Jatuli excursion), evolutionary studies have provided no evidence so far supporting the survival of early marine planktonic lineages (Fig. 1 and Supplementary Fig. S4) or perhaps these lineages have not yet been discovered. While marine living planktonic cyanobacteria cannot be traced back to the early Paleoproterozoic (Figs 1 and 2), we cannot discard the possibility that early marine cyanobacteria lineages went extinct after the GOE due to changes in marine water chemistry resulting from freely available oxygen (e.g., euxinic conditions)4,11,12. It is clear that more efforts are needed studying early divergent lineages of cyanobacteria in order to unravel the transition from a terrestrial to marine biosphere.
Concluding remarks
This study sheds light on the evolutionary steps that led to the recent evolution of marine planktonic cyanobacteria. Loss of filamentous forms, decrease in cell diameter and shifts in habitat preference from freshwater to marine were involved in the emergence of planktonic forms. Most groups of contemporary cyanobacteria can be traced back to the Mid-Proterozoic. Early cyanobacteria likely inhabited benthic, terrestrial and/or coastal marine environments. The early restriction of cyanobacteria to terrestrial and costal environments, in contrast to the vast oceans, helps explain the delay in the oxidation of the Earth’s surface during the Pre-Cambrian. Primary productivity would have significantly increased once planktonic phytoplankton became prominent towards the end of the Pre-Cambrian and early Phanerozoic.
Materials and Methods
Alignment and taxon sampling
Alignments including 135 protein-coding genes (56,251 aa) and two ribosomal RNAs (4,555 bp) were analysed for 131 genome taxa. All sequence data for the 131 cyanobacterial genomes were obtained from GenBank (http://www.ncbi.nlm.nih.gov) and using Geneious R6. The chosen genes (135 proteins and two ribosomal RNA: SSU and LSU) were universally present in cyanobacterial taxa, evolutionarily conserved and had a minimum number of gene duplications23,56. Principal coordinates analyses were used to identify ortholog genes that belong to a conserved ‘core’ gene set in cyanobacteria56. For the protein data set, genes chosen represent a wide diversity of cellular functions. These functions range from Information Processing (IP; transcription, translation, DNA replication and repair), Metabolism (Met), Cellular Processes (CP; including cell division, cell envelope biogenesis, motility and secretion) and General Function Prediction (GFP). For a detailed list with names and description of the genes included in this study see Blank and Sánchez-Baracaldo25. Each gene was aligned independently using SATé 2.2.327, a multiple sequence alignment and phylogenetic reconstruction program. Single gene alignments generated in SATé were imported into Mesquite v. 2.7557 to obtain ‘nexus’ and ‘phylip’ format files for subsequent analyses. Single alignments were later concatenated into a single nexus format file using Sequence Matrix v 100.058. Two concatenated matrices were obtained: one for protein-coding genes and a second one for ribosomal RNAs; both of these matrices were used to estimate tree topologies as described below.
Phylogenetic analyses
Maximum likelihood analyses were performed in RAxML 7.2.659 and Phylobayes32. A multiple gene approach was implemented to establish the deep-branching relationships in the cyanobacterial tree. All sequence data for 131 cyanobacterial genomes were obtained from GenBank (http://www.ncbi.nlm.nih.gov) and using Geneious 6.1.4. A total of 135 protein-coding genes (with 56,251 aa) and two ribosomal RNAs (with a total of 4,555 bp) were used to establish phylogenetic relationships of cyanobacteria. ProTest v.2.460 was used to estimate the best model of evolution for the protein set. To analyse the protein sequences I implemented the LG model and G (gamma-distribution with 4 rate categories). Matrices containing the protein and RNA data set were imported into RAxML GUI v.1.161 and up to 50 maximum likelihood trees were generated. To obtain statistical support analyses were performed in RAxML 7.0.359 (Supplementary Fig. S1). RAxML analyses recovered the same well-supported monophyletic groups previous reported by recent phylogenomic studies22,23,25,28,62.
Bayesian divergence time estimation
Divergence times were estimated implementing a Bayesian relaxed molecular clock approach in MCMCTree31 and Phylobayes32 (Table 1). Two independent data sets were assembled to estimate age divergences: 1) eighteen proteins and 2) two RNA genes (SSU and LSU). The independent-rates model was implemented as previous studies of cyanobacteria have shown that this model is favored over the auto-correlated rates model9. Since cyanobacteria have an ancient origin, independent rates more likely represent greater variation in inherited factors in contrast to auto-correlated rates9. Because the current implementation of MCMCtree and Phylobayes do not allow the use of mixed (nucleotides and amino acids) data sets. Ages were estimated using both type of data sets separately as follows: 1) Protein data set with a total of 18 proteins (AtpA, AtpB, AtpH, L1, L4, L5, NdhH, PetB, PetD, PsaA, PsbE, RbcL, S10, S13, S19, SecY, TufA and Ycf3) and 2) nucleotide data set including two RNAs (SSU and LSU). Supplementary Table S1 contains a list of gene names and substitution rates for the analyses performed in this study.
In MCMCtree, I implemented four models of nucleotide and protein evolution (See Supplementary Information on line). For all age calibrations, soft bounds were specified with 2.5% tail probabilities above/below these limits, allowing for molecular data to correct for conflicting fossil information. In Phylobayes, I implemented the CAT-GTR replacement model for both nucleotides and amino acids32. For all non-calibrated nodes, I used a birth-death prior on divergence times I also performed two separate experiments two independent experiments with permissive gamma distributed root priors that allowed a 95% credibility interval of the root node to range between 2,320–2,700 Mya and 2,320–3,000 Mya. See supplementary information for a more detailed description of the relaxed molecular clock analyses performed.
Fossil constraints
Experiments reported here (Table 1) were performed implementing two maximum ages for the cyanobacterial root: 2,700 Myr63 and 3,000 Myr64. The minimum age for the cyanobacterial root was set at 2,320 Myr (the rise in atmospheric oxygen)2. Based on geochemical evidence a younger (2,500 Myr)65 age is also reported for the origin of oxygenic photosynthesis. I also calibrated the trees with fossils exhibiting unique morphological features that could be assigned to well-supported groups such as the Nostocales and the Pleurocapsales. In the Nostocales, akinetes are thick-walled dormant cells to enable a response to extreme cold and desiccation resistant environmental conditions66. These specialized cells are present in most species amongst the Nostocales and likely evolved once in this group. Akinetes have been shown to have a single ancestor based on trait evolution studies23. A maximum age of 2,100 Myr was used for this monophyletic group for which it has been hypothesised that specialised cells such as the heterocyst evolved in response to free available oxygen in the atmosphere67. The Pleurocapsales are characterised by having multiple fission, a unique phenotypic property that distinguishes members of this group from other cyanobacteria. The Pleurocapsales also belong to a well-supported monophyletic group (Supplementary Fig. S1) including two strains of Pleurocapsa (PCC 7319 and PCC 7327). A minimum age of 1,700 Myr68 and a maximum age of 1,900 Myr (Gunflint iron formation and the first appearance of reliable cyanobacteria fossils observed in the fossil record35,37 were implemented. Finally, a maximum age of 110 Myr was implemented for Hemiaulus69 as these organisms host the symbiont Richelia19,20,21.
Bayesian inference of character evolution
To infer the evolution of cell type, cell diameter and habitat, I used Bayesian stochastic character mapping70. Analyses were implemented in SIMMAP v1.571 and used relative time calibrated trees generated in MCMCtree31 for the protein set as described above. No prior on the rate parameter was used, as I wanted to use the branch lengths as a direct estimate of rate of evolution. A β prior is implemented in SIMMAP on the symmetry of the transition rate matrix. Cell type or morphology were coded as 0 = unicellular and 1 = filamentous. Cell diameter data were coded as discrete characters, where 0 = average cell diameter raging from 1 to 2 μm, 1 = average cell diameter raging 2 to 3 μm, 2 = average cell diameter raging 3 to 5 μm and 3 = average cell diameter equal to or greater than 5 μm. Habitat were coded as 0 = freshwater and 1 = brackish, marine or hypersaline. Character states were obtained from the Bergey’s Manual of Systematic Bacteriology72, previous studies of trait evolution of cyanobacteria25 and other cyanobacteria studies22. For binary characters (habitat and cell type) the bias parameter was drawn from a symmetrical β prior. Since SIMMAP uses a symmetrical β prior on the symmetry of the transition rate matrix, this influences the degree to which transitions favor state 0 over 1. The shape of the β distribution is described by the α parameter and discretized into κ categories. I performed sensitivity experimenting using three different α distributions, where α used the following values, 0.1, 1 and 10. Analyses for habitat and cell type used α = 1. For multi-state characters (cell diameter) the bias parameter between states is specified as simply 1/κ, where k is the number of states. The overall rate of substitution for both of these classes is a branch length multiplier drawn from a prior gamma distribution. Supplementary Table S2 contains the characters states used for the three characters studied here.
Additional Information
How to cite this article: Sánchez-Baracaldo, P. Origin of marine planktonic cyanobacteria. Sci. Rep. 5, 17418; doi: 10.1038/srep17418 (2015).
References
Holland, H. D. The oxygenation of the atmosphere and oceans. Philos. T. R. Soc. B. 361, 903–915, doi: 10.1098/rstb.2006.1838 (2006).
Bekker, A. et al. Dating the rise of atmospheric oxygen. Nature 427, 117–120, doi: 10.1038/nature02260 (2004).
Planavsky, N. J. et al. Low Mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science 346, 635–638 (2014).
Lyons, T. W., Reinhard, C. T. & Planavsky, N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014).
Sahoo, S. K. et al. Ocean oxygenation in the wake of the Marinoan glaciation. Nature. 489, 546–549, doi: 10.1038/nature11445 (2012).
Och, L. M. & Shields-Zhou, G. A. The Neoproterozoic oxygenation event: Environmental perturbations and biogeochemical cycling. Earth-Sci Rev. 110, 26–57, doi: 10.1016/j.earscirev.2011.09.004 (2012).
Canfield, D. E., Poulton, S. W. & Narbonne, G. M. Late-Neoproterozoic Deep-Ocean Oxygenation and the Rise of Animal Life. Science 315, 92–95 (2007).
Fike, D. A., Grotzinger, J. P., Pratt, L. M. & Summons, R. E. Oxidation of the Ediacaran Ocean. Nature 444, 744 747
Sánchez-Baracaldo, P., Ridgwell, A. & Raven, J. A. A Neoproterozoic transition in the marine nitrogen cycle. Curr. Biol. 24, 652–657, doi: 10.1016/j.cub.2014.01.041 (2014).
Erwin, D. H. et al. The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals. Science 334, 1091–1097, doi: 10.1126/science.1206375 (2011).
Poulton, S. W. & Canfield, D. E. Ferruginous Conditions: A Dominant Feature of the Ocean through Earth’s History. Elements 7, 107–112, doi: 10.2113/gselements.7.2.107 (2011).
Anbar, A. D. & Knoll, A. H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142, doi: 10.1126/science.1069651 (2002).
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 281, 237–240 (1998).
Zehr, J. P. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 19, 162–173 doi: 10.1016/j.tim.2010.12.004 (2011).
Thompson, A. W. et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550, doi: 10.1126/science.1222700 (2012).
Johnson, Z. I. et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311, 1737–1740 (2006).
Scanlan, D. J. et al. Ecological Genomics of Marine Picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299 (2009).
Flombaum, P. et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 110, 9824–9829, doi: 10.1073/pnas.1307701110 (2013).
Foster, R. A. et al. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. The ISME J. 5, 1484–1493, doi: 10.1038/ismej.2011.26 (2011).
Villareal, T. Laboratory culture and preliminary characterization of the nitrogen-fixing Rhizosolenia-Richelia symbiosis. Mar. Ecol. 11, 117–132 (1990).
Janson, S., Wouters, J., Bergman, B. & Carpenter, E. J. Host specificity in the Richelia-diatom symbiosis revealed by hetR gene sequence analysis. Environ. Microbiol. 1, 431–438 (1999).
Shih, P. M. et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. P. roc Natl. Acad. Sci. USA 110, 1053–1058, doi: 10.1073/pnas.1217107110 (2013).
Sánchez-Baracaldo, P., Hayes, P. K. & Blank, C. E. Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology 3, 145–165 (2005).
Bombar, D., Heller, P., Sánchez-Baracaldo, P., Carter, B. J. & Zehr, J. P. Comparative genomics reveals surprising divergence of two closely related strains of uncultivated UCYN-A cyanobacteria. The ISME J. 8, 2530–2542, doi: 10.1038/ismej.2014.167 (2014).
Blank, C. E. & Sánchez-Baracaldo, P. Timing of morphological and ecological innovations in the cyanobacteria–a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23, doi: 10.1111/j.1472-4669.2009.00220.x (2010).
Larsson, J., Nylander, J. A. & Bergman, B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol. Biol. 11, 187 (2011).
Liu, K., Raghavan, S., Nelesen, S., Linder, C. R. & Warnow, T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science 324, 1561–1564, doi: 10.1126/science.1171243 (2009).
Schirrmeister, B. E., Gugger, M. & Donoghue, P. C. J. Cyanobacteria and the Great Oxidation Event: evidence from genes and fossils. Palaeontology, doi: 10.1111/pala.12178 (2015).
Dagan, T. et al. Genomes of Stigonematalean Cyanobacteria (Subsection V) and the Evolution of Oxygenic Photosynthesis from Prokaryotes to Plastids. Genome Biol. Evol. 5, 31–44 (2013).
Donia M. S. et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc. Natl. Acad. Sci. USA 108, E1423–E1432 (2011).
Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Lartillot N., Lepage T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286 2288
Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. Plos Biol. 4, e88 (2006).
Hofmann, H. J. Precambrian microflora, Belcher Islands, Canada: significance and systematics. J. Paleontol. 50, 1040–1073 (1976).
Golubic, S. & Seong-Joo, L. Early cyanobacterial fossil record: preservation, palaeoenvironments and identification. Eur. J. Phycol. 34, 339–348 (1999).
Wacey, D. et al. Nanoscale analysis of pyritized microfossils reveals differential heterotrophic consumption in the ~1.9-Ga Gunflint chert. Proc. Natl. Acad. Sci. USA 110, 8020–8024, doi: 10.1073/pnas.1221965110 (2013).
Sergeev, V. N., Gerasimenko, L. M. & Zavarzin, G. A. Proterozoic history and present state of cyanobacteria. Mikrobiologiia 71, 725–740 (2002).
Falcón, L. I., Magallón, S. & Castillo, A. Dating the cyanobacterial ancestor of the chloroplast. The ISME J., 4, 777–783, doi: 10.1038/ismej.2010.2 (2010).
Schirrmeister, B. E., de Vos J. M., Antonelli A. & C., B. H. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc. Natl. Acad. Sci. USA 110, 1791–1796 (2013).
Brasier, M. D. Green algae (Chlorophyta) and the question of freshwater symbiogenesis in the early proterozoic. J. Phycol. 49, 1036–1039 (2013).
Raven, J. A., Beardall, J., Larkum, A. W. & Sánchez-Baracaldo, P. Interactions of photosynthesis with genome size and function. Philos. T. R. Soc. B. 368, 20120264, doi: 10.1098/rstb.2012.0264 (2013).
Battistuzzi, F. U., Feijao, A. & Hedges, S. B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy and the colonization of land. BMC Evol. Biol. 4, 44, doi: 10.1186/1471-2148-4-44 (2004).
Sleep, N. H. & Bird, D. K. Evolutionary ecology during the rise of dioxygen in the Earth’s atmosphere. Philos. T. R. Soc. B. 363, 2651–2664, doi: 10.1098/rstb.2008.0018 (2008).
Coelho, S. M., Simon, N., Ahmed, S., Cock, J. M. & Partensky, F. Ecological and evolutionary genomics of marine photosynthetic organisms. Mol. Ecol. 22, 867–907, doi: 10.1111/mec.12000 (2012).
Hagemann, M. Genomics of Salt Acclimation: Synthesis of Compatible Solutes among Cyanobacteria. Adv. Bot. Res. 65, 27–55, doi: 10.1016/B978-0-12-394313-2.00002-0 (2013).
Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 35, 87–123, doi: 10.1111/j.1574-6976.2010.00234.x (2011).
Scott, C. et al. Tracing the stepwise oxygenation of the Proterozoic ocean. Nature 452, 456–U455, doi: 10.1038/nature06811 (2008).
Planavsky, N. J. et al. Widespread iron-rich conditions in the mid-Proterozoic ocean. Nature 477, 448–U495, doi: 10.1038/nature10327 (2011).
Zerkle, A. L., House, C. H., Cox, R. P. & Canfield, D. E. Metal limitation of cyanobacterial N2 fixation and implications for the Precambrian nitrogen cycle. Geobiology 4, 285–297 (2006).
Stal, L. Cyanobacterial mats and stromatolites. In: Ecology of Cyanobacteria II. Springer. (Netherlands), pp 65–126. Whitton, B. A. (Springer, 2012).
Abed, R. M. M., Palinska, K. A., Camoin, G. & Golubic, S. Common evolutionary origin of planktonic and benthic nitrogen-fixing oscillatoriacean cyanobacteria from tropical oceans. FEMS Microbiol. Lett. 260, 171–177, doi: 10.1111/j.1574-6968.2006.00321.x (2006).
Schirrmeister, B. E., Antonelli, A. & Bagheri, H. C. The origin of multicellularity in cyanobacteria. BMC Evol. Biol. 11, doi: Artn 4510.1186/1471-2148-11-45 (2011).
Flombaum, P. & et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 110, 9824–9829, doi: 10.1073/pnas.1307701110 (2013).
Papineau, D., Purohit, R., Fogel, M. L. & Shields-Zhou, G. A. High phosphate availability as a possible cause for massive cyanobacterial production of oxygen in the Paleoproterozoic atmosphere. Earth Planet Sc. Lett. 362, 225–236, doi: 10.1016/j.epsl.2012.11.050 (2013).
Kump, L. R. et al. Isotopic Evidence for Massive Oxidation of Organic Matter Following the Great Oxidation Event. Science 334, 1694–1696, doi: 10.1126/science.1213999 (2011).
Shi, T. & Falkowski, P. G. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc. Natl. Acad. Sci. USA 105, 2510–2515, doi: 10.1073/pnas.0711165105 (2008).
Maddison, W. & Maddison, D. Mesquite: a modular system for evolutionary analysis. Version 2.75. Available from http://mesquiteproject.org (2009). (Accessed: 10th January 2012).
Vaidya, G., Lohman, D. J. & Meier, R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180, doi: 10.1111/j.1096-0031.2010.00329.x (2011).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690, doi: 10.1093/bioinformatics/btl446 (2006).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105, doi: 10.1093/bioinformatics/bti263 (2005).
Silvestro, D. & Michalak, I. raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337, doi: 10.1007/s13127-011-0056-0 (2012).
Latysheva, N., Junker, V. L., Palmer, W. J., Codd, G. A. & Barker, D. The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 28, 603–606, doi: 10.1093/bioinformatics/bts008 (2012).
Brocks, J. J., Buick, R., Summons, R. E. & Logan, G. A. A reconstruction of Archean biological diversity based on molecular fossils from the 2.78 to 2.45 billion-year-old Mount Bruce Supergroup, Hamersley Basin, Western Australia. Geochim. Cosmochim. Acta 67, 4321–4335 (2003).
Crowe, S. A. et al. Atmospheric oxygenation three billion years ago. Nature 501, 535-+, doi: 10.1038/nature12426 (2013).
Rasmussen, B., Fletcher, I. R., Brocks, J. J. & Kilburn, M. R. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455, 1101–U1109, doi: 10.1038/nature07381 (2008).
van Dok, W. & Hart T. B. Akinete Differentiation in Anabaena circinalis (Cyanophyta). J. Phycol. 32, 557–565 (1996).
Tomitani, A., Knoll, A. H., Cavanaugh, C. M. & Ohno, T. The evolutionary diversification of cyanobacteria: Molecular-phylogenetic and paleontological perspectives. P. Natl. Acad. Sci. USA 103, 5442–5447, doi: 10.1073/pnas.0600999103 (2006).
Zhang, Y. & Golubic, S. Endolithic microfossils (cyanophyta) from early Proterozoic stromatolites, Hebei, China. Acta Micropaleontol. Sin. 4, 1–3 (1987).
Sims, P. A., Mann, D. G. & Medlin, L. K. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402, doi: 10.2216/05-22.1 (2006).
Huelsenbeck, J. P., Nielsen, R. & Bollback, J. P. Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158, doi: 10.1080/10635150390192780 (2003).
Bollback, J. P. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7, 88, doi: 10.1186/1471-2105-7-88 (2006).
Castenholz, R. W. et al. Phylum BX. Cyanobacteria. In: Bergey’s Manual® of Systematic Bacteriology, Springer New York. New York edn (2001).
Fralick, P., Davis, D. W. & Kissin, S. A. The age of the Gunflint Formation, Ontario, Canada: single zircon U–Pb age determinations from reworked volcanic ash. Can. J. Earth Sci. 39, 1085–1091, doi: 10.1139/e02-028 (2011).
Acknowledgements
I thank Martin Brasier for discussions on the early Earth geochemical record; Sam Price and Nathan Chrismas for assistance with the analyses; Andy Ridgwell and Emma J Stone for helpful comments on the manuscript. Funding support for this work came from a Royal Society Dorothy Hodgkin Fellowship.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sánchez-Baracaldo, P. Origin of marine planktonic cyanobacteria. Sci Rep 5, 17418 (2015). https://doi.org/10.1038/srep17418
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17418
This article is cited by
-
Genetics re-establish the utility of 2-methylhopanes as cyanobacterial biomarkers before 750 million years ago
Nature Ecology & Evolution (2023)
-
Elucidating the picocyanobacteria salinity divide through ecogenomics of new freshwater isolates
BMC Biology (2022)
-
Increased biomass and carbon burial 2 billion years ago triggered mountain building
Communications Earth & Environment (2021)
-
Evolution of the structure and impact of Earth’s biosphere
Nature Reviews Earth & Environment (2021)
-
Dating Alphaproteobacteria evolution with eukaryotic fossils
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.