Abstract
Rac or rac-like prophage harbors many genes with important physiological functions, while it remains excision-proficient in several bacterial strains including Escherichia coli, Salmonella spp. and Shigella spp. Here, we found that rac excision is induced during biofilm formation and the isogenic stain without rac is more motile and forms more biofilms in nutrient-rich medium at early stages in E. coli K-12. Additionally, the presence of rac genes increases cell lysis during biofilm development. In most E. coli strains, rac is integrated into the ttcA gene which encodes a tRNA-thioltransferase. Rac excision in E. coli K-12 leads to a functional change of TtcA, which results in reduced fitness in the presence of carbenicillin. Additionally, we demonstrate that YdaQ (renamed as XisR) is the excisionase of rac in E. coli K-12 and that rac excision is induced by the stationary sigma factor RpoS through inducing xisR expression. Taken together, our results reveal that upon rac integration, not only are new genes introduced into the host, but also there is a functional change in a host enzyme. Hence, rac excision is tightly regulated by host factors to control its stability in the host genome under different stress conditions.
Similar content being viewed by others
Introduction
Bacteriophages and bacteria are the most abundant life forms on Earth. Bacteria and phages also interact frequently and each phage infection has the potential to introduce new genetic information into the bacterial host, thereby driving the evolution of bacteria. The introduction of novel genes by phages into the bacterial host can confer beneficial phenotypes that enable the exploitation of competitive environments1,2,3,4. Prophage-encoded virulence factors make important contributions to pathogenesis, including those of Corynebacterium diphtheriae (diphtheria), Clostridium botulinum (botulism) and E. coli O157:H7 (Shiga-like toxin)5. Active prophages such as Gifsy-2 can also give the Salmonella host a competitive advantage by killing competitors and by providing immunity6. In E. coli, the genes of cryptic prophage (i.e., prophage defective in plaque formation) provide multiple benefits to the host for survival in adverse environmental conditions, such as under oxidative, acid, osmotic, antibiotic stresses and biofilm formation7. Biofilm formation is arguably the dominant lifestyle for most bacteria and prophage genes are among the most highly upregulated genes during biofilm development in E. coli and Pseudomonas aeruginosa8,9. In these cases, the reproductive success of the lysogenic bacterium carrying these new genes translates directly into an evolutionary success for the prophage resident in the chromosome.
In addition to contributing to the diversification of the bacterial genome architecture by introducing novel genes, another way for phages to affect the bacterial host is to disrupt or restore host genes upon prophage integration or excision. Prophage integration is commonly found in tRNA and tmRNA10. Integration into the coding region of genes has also been described and the prophage attachment site frequently complements the coding sequence of the protein11. In some cases, prophage integration or excision does not disrupt the gene serving as the attachment site. For example, the Gifsy-1 prophage of Salmonella integrates within the coding sequence of lepA and does not change lepA function12. Similarly, prophage e14 in E. coli K-12 inserts within the isocitrate dehydrogenase gene13 and Φ297 in E. coli O157:H7 integrates at the yecE site14 without changing the gene function. However, in other cases, inactivation of protein function by prophage integration or excision has also been described. For example, a lipase gene undergoes negative lysogenic conversion by a Staphylococcus aureus phage15,16. In the human pathogen Listeria monocytogenes, integration of prophage A118 disrupts the host gene comK encoding the major competence transcription factor17 and excision of A118 restores the function of comK18.
Prophage rac was the first defective prophage discovered in E. coli K-1219,20 and it has been regarded as a phage fossil that was acquired over 4.5 million years ago21. The rac prophage in the laboratory E. coli strain K-12 and two rac-like prophages (Sp10 and CP-933R) in two strains of enterohemorrhagic E. coli O157:H7 (Sakai and EDL933) all integrate into the ttcA gene, which encodes a tRNA-thioltransferase in these three genomes22,23. The leftmost 21 kb are greater than 99.9% identical in Sp10 and CP-933R and the leftmost 8 kb are 99.0% identical to the K-12 rac prophage23. Rac harbors 27 genes and 5 pseudogenes in E. coli K-12 and deletion of the entire rac prophage reduces resistance to acid stress, oxidative stress and antibiotic stress7. The prophage proteins in rac responsible for inhibiting cell division, including KilR, appear important for resistance to nalidixic acid and azlocillin7,24. YdaC in rac was identified in a screen for antibiotic resistance using pooled plasmids from the ASKA library that showed increased resistance to erythromycin25. The type I toxin-antitoxin pair RalR-RalA in rac increases resistance to fosfomycin26. In addition, RecE and RecT in rac are involved with RecA-dependent recombination27. Although it harbors important genes, rac remains excision proficient in E. coli K-12 and in the Sakai strain7,22. In E. coli K-12, rac has the second highest excision rate under normal growth conditions. Excision of the rac prophage would necessarily lead to the loss of rac genes due to the lack of capacity to replicate once excised28. Although rac and Sp10 are both lambda-like prophages, unlike lambda prophages, their excision is not induced upon mitomycin C treatment, which is known to trigger the host SOS responses22,29. Hence, two major questions remain for rac prophage excision: whether rac excises under stressed conditions and whether rac excision affects host gene function. In this study, we addressed both questions and found that rac excision in E. coli K-12 is induced during biofilm formation and we found that the rac deletion strain formed more biofilm in nutrient rich medium at early stages and showed high swimming motility. The function of the host gene ttcA was assessed before and after rac excision under carbenicillin stress and the results showed that rac integration caused a functional change in TtcA by creating a variant of the protein while rac excision restored it to the ancient copy. In addition, we identified the excisionase gene for rac and propose to name it xisR (excisionase gene for rac). XisR induced rac excision and bound to the rac attachment site. Furthermore, in addition to the previously identified host factor H-NS, we found that the sigma factor RpoS also increased rac excision by regulating the transcription of xisR at the stationary phase.
Results
Rac excision is induced during biofilm formation
Biofilms are the preferred lifestyle of bacteria and prophages are involved in biofilm formation8,30; therefore, we tested whether prophage excision is regulated during biofilm formation in E. coli K-12 BW25113. To test the frequency of excision of the nine prophages, the fraction of cells that undergoes excision was quantified by quantitative PCR (qPCR) in a total of ~109 cells using the method described previously7. When compared to planktonic cells, the frequency of rac excision increased 6.4 ± 0.1-fold (3 per 105 cells vs 0.5 per 105 cells) in 48 h biofilm cells and the frequencies of excision of cryptic prophages CP4-6 (1.5 per 105 cells vs 0.3 per 105 cells) and CPZ-55 (0.5 per 106 cells vs 0.8 per 107 cells) were also increased but with lower overall excision (Fig. 1A). Similar to what has been reported previously, the frequency of CP4-57 excision increased 21.2 ± 0.2-fold in the biofilm cells (1.6 per 106 cells) when compared to the planktonic cells (7.5 per 108 cells)8. Prophage e14 had the highest frequency of excision (5.5 per 104 cells) in BW25113; however, e14 excision was not induced in biofilm cells (Fig. 1A). Excision of prophage Qin or CP4-44 was not detected (<1 per 108 cells), which is likely due to the lack of an active integrase for Qin and CP4-447,31. As oxidative stress is involved in biofilm formation, we tested whether oxidative stress (using 2 mM hydrogen peroxide) affected prophage excision in BW25113 cells32. Prophage e14 was the only prophage induced to excise with the addition of hydrogen peroxide (61.4 ± 0.1-fold) and rac was the only prophage repressed with the addition of hydrogen peroxide (1.9 ± 0.1-fold) (Fig. 1B). Taken together, these results show that rac excision is induced in biofilm cells but not under oxidative stress condition.
Fold change of the frequencies of prophage excision in E. coli K-12.
(A) Fold change of excision of seven prophages in biofilm cells versus planktonic cells at 48 h in E. coli K-12. (B) Fold change of excision of seven prophages with oxidative stress (2 mM H2O2) versus without oxidative stress in E. coli K-12. H2O2 was added to cells (OD600 ~ 1.0) for 90 min. Data are from two independent cultures and one standard deviation is shown in A and B.
Rac genes affect biofilm formation and motility
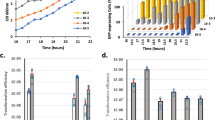
As rac is excised during biofilm formation, we tested how rac genes affect biofilm formation using the previously obtained prophage deletion strain Δrac28, constructed by us through overexpressing an engineered hns which specifically induces rac excision. When tested in rich LB medium at 37 °C, the Δrac strain formed 7.9 ± 0.2-fold more biofilm compared to the BW25113 wild-type cells at an 8 h early stage and showed the second highest fold change among the nine prophage deletion strains (Fig. 2A). However, when measured at 24 h, the biofilm formed by Δrac strain was comparable to the wild-type strain (1.3 ± 0.3-fold) (Fig. 2B). Additionally, when using M9C minimal medium supplemented with 0.4% glucose (M9C-glu) at 37 °C, the Δrac strain formed the largest amount of biofilm among the nine prophage deletion strains compared to the wild-type strain (9.5 ± 0.8-fold) at 8 h (Fig. 2C) but was comparable to the wild-type at 24 h (1.5 ± 0.2-fold) (Fig. 2D). In our previous study, we found that the Δrac strain in M9 casamino acids (M9C) medium and at 30 °C, as well as seven of the prophage deletion strains (except for ΔCP4-57 strain), formed less biofilm than the wild-type strain at 8 h7. To further test whether biofilm formation in the Δrac strain is nutrient-dependent, we tested biofilm formation at the same temperature but with the addition of 0.4% glucose (M9C-glu). Consistent with the results in LB and M9C-glu at 37 °C, the Δrac strain formed more biofilm than the wild-type, with a 27.9 ± 0.3-fold increase at 8 h (Fig. 2E) and a 4.6 ± 0.3-fold increase at 24 h (Fig. 2F). Prophage CP4-57 also undergoes induced excision during biofilm formation; we found that the ΔCP4-57 strain formed more biofilm in M9C-glu medium (Fig. 2E,F).
Biofilm formation in nine prophages deletion strains.
Early (A,C,E) and late (B,D,F) biofilm formation of E. coli K-12 BW25113 and nine isogenic prophage deletion mutants were tested in two different media (LB or M9C-glu) at two different temperatures (37 °C or 30 °C), respectively. Each data point is the average of at least ten replicate wells from two independent cultures and one standard deviation is shown here from (A–F).
As prophage excision has been associated with cell lysis during biofilm formation33, we assessed whether prophage genes contribute to cell lysis by comparing lysis in the prophage deletion strains to the BW25113 strain. Among the nine prophages, deletion of rac and DLP12 resulted in the most significant reduction in cell lysis with 5.8% ± 0.6% and 5.6% ± 0.9% of the cells lysed, respectively, compared to 11.8% ± 1.0% of the cells lysed in the wild-type biofilm cells (Fig. 3A). Furthermore, we tested the change in swimming motility in the Δrac strain. As shown in Fig. 3B,C, the Δrac strain showed much higher motility than the wild-type strain (7.3 ± 0.1-fold). Additionally, ΔCP4-57 and ΔCPS-53 strains also showed much higher motility than the wild-type strain (Fig. 3B). We and other groups have reported that the increased motility of BW25113 isogenic strains could also be caused by transposition of insertion elements (e.g., IS5 or IS1) into the master flagella operon flhDC34,35, which leads to a high and constitutive expression of flagella genes. Therefore, all of the prophage deletion strains used here were screened for the presence of insertion elements in flhDC by PCR and qPCR and we verified that all of the isogenic strains constructed did not contain any insertion elements in flhDC.
Rac genes affect cells lysis and cell motility.
(A) Cell lysis in nine prophage deletion strains as measured in 48 h biofilm cells. (B) Motility of nine prophage deletion strains in low salt motility agar plates. Data are from two independent cultures and one standard deviation is shown in A and B. (C) Motility of Δrac and wild-type strains on high salt motility plates. Photographic image was taken by one of the first authors XL. All of the isogenic strains constructed did not contain any insertion elements in the promoter region of flhDC operon.
Rac excision causes amino acid changes in TtcA
The prophage rac resides between ttcA and the pseudogene ttcC’ in E. coli K-12 BW25113 (Fig. 4A). By comparing the DNA sequences near the attachment region before rac excision (in the wild-type strain) and after excision (in the Δrac strain), we found that excision of rac results in a change of four TtcA amino acids near the N-terminus (Fig. 4B) due to the imperfect match of the left and the right attachment sites (Fig. 4C). The same change was obtained when the rac excisionase is overproduced, which confirms that this change is indeed caused by the natural excision of rac. Two previously identified rac-like prophages, CP-933R and SP10, are both inserted into the same ttcA site of E. coli O157:H7 Str. EDL933 and E. coli O157:H7 Str. Sakai, respectively23. Through a bioinformatics search, we found that although there are differences (one nucleotide) among the right attachment sites of these three strains, the same changes in the TtcA protein should occur when the rac and rac-like prophages are excised from the host genome of these two strains (Fig. 4D).
Changes in TtcA before and after rac excision.
(A) Excision of rac was induced by overproduction of excisionase and integrase proteins in E. coli K-12 BW25113. (B) Rac excision caused a change of four amino acids of TtcA at the N-terminus. Numbers on the top indicate the amino acid position in TtcA. (C) The nucleotide sequence of the left and the right attachments of rac before excision (attL and attR) as well as the attachment site after rac excision (attB) in E. coli K-12 BW25113. Letters labeled under the attB sequence indicate the partial TtcA’ amino acid sequence. Numbers on the bottom indicate the amino acid position in TtcA’. (D) The nucleotide sequence of the two attachments sites (attL and attR) of rac-like prophage, CP-933R in E. coli O157:H7 Str. EDL933 strain and Sp10 in E. coli O157:H7 Str. Sakai strain. Arrows indicate the translational start of TtcA.
To explore the possibility that rac affects ttcA upon its integration, we analyzed the TtcA sequences and the upstream and downstream regions of ttcA in all of the E. coli strains available in the EcoGene-RefSeq database (http://www.ecogene.org/refseq). We also included closely related strains of Shigella and Salmonella that harbor rac or rac-like prophages in our analysis. The alignment results show that although the N-terminus of TtcA is rather variable, the protein can be categorized into two distinctive groups depending on whether or not they harbor rac-like prophages (Fig. 5). For the 10 strains of E. coli and 4 strains of Shigella flexneri without a rac-like prophage inserted in ttcA, such as E. coli O103:H2 Str. 12009, E. coli UMN026 and Shigella flexneri 5 Str. 8401 (Fig. 5, above black line), the N-terminus of TtcA is the same as the TtcA’ variant created by rac excision in E. coli K-12 strains. While in the other 13 strains harboring rac or rac-like prophages in ttcA, such as E. coli E22, Shigella sonnei 53G and Salmonella enterica subsp. serovar Enteritidis Str. P125109, the N-terminus of TtcA is the same as the E. coli K-12 strain with rac (Fig. 5, below black line). These results suggest that the TtcA’ variant formed after rac excision in E. coli K-12 is the ancient copy of this protein and that rac integration changed the ancient TtcA. One exception is the uropathogenic E. coli strain CFT073, which does not harbor a rac-like prophage in ttcA but does possess a Φ-CTF073-potB prophage inserted into the potB gene. Prophage Φ-CTF073-potB carries similar structural genes (e.g., capsid and tail protein) and a recombination gene (e.g., recE) to those found in rac-like prophages36.
Sequence alignment of TtcA in bacteria without or with rac integrated.
Alignment of TtcA sequences was performed using the ClustalX program. Sequences from 14 strains without rac integrated at ttcA (above the black line) and from 13 bacteria strains with rac integrated at ttcA (below the black line) as well as the E. coli CFT073 strain (box indicated) with rac integrated elsewhere other than ttcA gene. Numbers on the top indicate the amino acid position in TtcA. Portion with identical amino acids in the 28 species are excluded (dot indicated) and only non-identical amino acids are highlighted. Species abbreviations: 12009, E. coli O103:H2 Str.12009; HS, E. coli HS; IAI1, E. coli IAI1; UMN026, E. coli UMN026; E24377A, E. coli E24377A; EC4115, E. coli EC4115; TW14359, E. coli O157:H7 Str. TW14359; O78, E. coli APEC O78; 8739, E. coli ATCC 8739; P12B, E. coli P12B; 8401, Shigella flexneri 5 Str. 8401; 2002017, Shigella flexneri 2002017; 2457T, Shigella flexneri 2A Str. 2457T; 301, Shigella flexneri 2A Str. 301, MG1655, E. coli K-12 MG1655; DH10B, E. coli K-12 DH10B; W3110, E. coli K-12 W3110; EDL933, E. coli O157:H7 Str. EDL933; Sakai, E. coli O157:H7 Str. Sakai; BW2952, E. coli BW2952; SE11, E. coli SE11; BL21, E. coli BL21 (DE3); E22, E. coli E22; 53G, Shigella sonnei 53G; P125109, Salmonella enterica subsp. serovar Enteritidis Str. P125109; 11128, E. coli O111:H- Str. 11128; DH1, E. coli DH1; CFT073, E. coli CFT073.
Rac excision causes a functional change in TtcA
Large-scale phenotypic screening of all Keio mutants from PortEco showed that the ΔttcA knockout strain has an altered fitness score when exposed to carbenicillin37. As shown in Fig. 6A, the ΔttcA strain showed increased metabolic activity compared to that of the wild-type strain without carbenicillin. In contrast, in the presence of 4 μg/mL carbenicillin, the ΔttcA strain showed reduced metabolic activity compared to that of the wild-type strain at later times (after 14 h) (Fig. 6B). Since the rac left attachment site (attL) is inside the coding region of ttcA, in the ΔttcA strain, the removal of the whole ttcA coding region would completely abolish rac excision. In order to investigate whether rac excision causes a functional change in TtcA, a native TtcA (before rac is excised) and the TtcA’ variant (after rac is excised) were produced in the same ΔttcA host strain, thereby ruling out the potential effect caused by differences in the frequency of rac excision in the wild-type strain and in the ΔttcA strain. The results show that the cells producing native TtcA have a much higher metabolic rate than the TtcA’ variant in the presence of carbenicillin, which suggests that rac excision causes a reduction in the ability of TtcA to cope with carbenicillin stress (Fig. 6C). As a negative control, the metabolic rate of cells overexpressing the native TtcA was comparable to cells overexpressing TtcA’ in the absence of carbenicillin (Fig. 6D). Taken together, these results show that rac excision causes a change in the host gene ttcA that leads to reduced fitness in the presence of carbenicillin stress.
Rac excision causes a functional change in TtcA.
Metabolic rate of the wild-type and ΔttcA strains in the absence of carbenicillin (A) or in the presence of 4 μg/mL carbenicillin (B). Metabolic rate of the ΔttcA strain overexpressing a wild-type TtcA (before rac excision) or overexpressing TtcA’ (TtcA variant after rac excision) with 4 μg/mL carbenicillin (C) and in the absence of carbenicillin (D). Data are from two independent cultures and one standard deviation is shown.
YdaQ is the rac excisionase
Excisionase enzymes are required for lambdoid prophage excision by interacting with integrase38 and the integrase gene is usually adjacent to the attachment site on the chromosome in order to interact with its DNA target39. In rac, an integrase intR is found next to the ttcA gene in which the 5′-end serves as the left attachment site in E. coli K-1223. Next to intR lies ydaQ, which encodes a small protein of 71 amino acids (Fig. 7A). Here, we tested the ability of YdaQ to induce rac excision. When yadQ is deleted, rac excision was reduced 9.4 ± 0.8-fold during the exponential phase and 13.6 ± 0.3-fold during the stationary phase (Fig. 7B), while overexpressing ydaQ increases rac excision 9.7 ± 0.2-fold (Fig. 7C). Deletion of other genes nearby intR, such as ralR and ralA, did not affect rac excision (data not shown). To determine whether YadQ can bind the attachment site like other excisionases, we performed electrophoresis mobility shift assays (EMSA) using purified YdaQ with an N-terminal hexahistidine tag (His-tagged) (Fig. 7D) which is verified by mass spectrometry (Supplementary Table S1). Two DNA fragments, one containing the right attachment site of rac (AttR) and one that does not contain the right attachment site of rac (AttM) were used as probes (Fig. 7E). The EMSA results show that YdaQ bound and shifted the right attachment site of rac. In addition, the binding increased with the increasing concentrations of YdaQ (Fig. 7F, AttR panel). As a negative control, YadQ did not bind to the biotin-labeled AttM fragment (Fig. 7F, AttM panel). In addition, possible dimers were observed when YadQ was purified (Fig. 7D, lane 3) and the addition of the reducing agent dithiothreitol (DTT) greatly reduced the dimerization of YdaQ (Fig. 7D, lane 4). Taken together, these results suggest that YdaQ controls rac excision and thus, we propose to rename ydaQ as xisR (excisionase for rac prophage).
YdaQ (XisR) controls rac excision.
(A) Schematic representation of the intR and xisR genes as well as the neighboring genes of prophage rac. (B) Excision rates of rac in the ΔxisR and in BW25113 strains. (C) Excision rates of rac in the wild-type strain overexpressing xisR via pCA24N-xisR. IPTG (1 mM) was added to BW25113/pCA24N-xisR cells (OD600 ~ 1.0) for 4 h. Error bars represent standard deviations for triplicate cultures in B and C. (D) XisR was purified via pET28b-xisR. IPTG (1 mM) was added to induce xisR expression for 4 h (lane 2) and cells without IPTG were included as a negative control (lane 1). Possible dimers of XisR were observed during purification (lane 3) and the addition of the reducing agent dithiothreitol (DTT) reduced the dimerization of XisR (lane 4). The triangle indicates the dimers of XisR and the asterisk indicates the XisR monomers. (E) Schematic representation of the PCR amplified fragments, AttR (294 bp) and AttM (291 bp), which are used as probes in EMSA. The AttR fragment contains the rac attR region and attR upstream and downstream regions, while AttM only contains the upstream region of attR. (F) EMSA results show that the purified XisR binds to the biotin-labeled AttR fragment and that, the binding increases with increasing concentrations of XisR. As a negative control, XisR does not bind to the biotin-labeled AttM fragment. Equal amounts of labeled probe was added in each lane.
Rac excision is regulated by RpoS through excisionase
We found that rac has a higher excision rate in stationary phase growth compared to exponential phase growth (Fig. 7B). Our previous results showed that rac excision was regulated by the host factor H-NS during the stationary phase28. In this study, we found that cells lacking H-NS still exhibit a higher excision rate in the stationary phase compared to exponential phase (5.7 ± 0.2-fold) (Fig. 8A), which suggests that other host factors besides H-NS may also control rac excision. We next tested whether rac excision is regulated by the stationary phase sigma factor RpoS. After deleting rpoS, there was no significant difference in rac excision between the cells in the exponential growth phase and in the stationary growth phase (1.7 ± 0.5-fold) (Fig. 8A), which suggests that RpoS is involved with rac excision. The qRT-PCR results show that the expression level of xisR was highly induced (110.3 ± 1.0-fold) in the wild-type strain but was only slightly induced in the rpoS deletion strain from the exponential phase to stationary phase (3.4 ± 0.5-fold) (Fig. 8B). Taken together, these results show that rac excision is induced in the stationary phase and that RpoS plays a role in regulating xisR expression.
RpoS regulates rac excision by inducing xisR expression.
(A) Excision rates of rac in BW25113, Δhns and ΔrpoS strains were measured during two different growth phases. Fold change indicates the change of rac excision rates in the stationary phase versus in the exponential phase. (B) Transcripts of xisR in the in the BW25113 and in ΔrpoS strains at two different growth phases. Fold change indicates the change of gene expression in the stationary phase versus in the exponential phase. Expression of rrsG was used to normalize the total RNA in different samples. Data are from two independent cultures and one standard deviation is shown in A and B.
Discussion
Collectively, our results support the hypothesis that rac integration or excision affects host function, especially under stress conditions. In this study, we tested whether rac integration or excision affects the function of the protein encoded by the host gene ttcA, which serves as one of the attachment sites for rac in E. coli K-12. We found that the amino acid sequence and function of TtcA were altered after rac excised. Previously, we have shown that the excision of rac decreased cell survival in the presence of nalidixic acid and azlocillin primarily through the KilR protein, which inhibits cell division7,40. Recently, we also found that the deletion of the type I toxin/antitoxin pair RalR/RalA in rac reduced metabolic activity in the presence of fosfomycin26. Conversely, studies have also shown that deletion of the complete rac prophage greatly increased resistance to bicyclomycin, which functions through inhibition of the termination factor Rho and the resistance is conferred by deletion of kilR and the remaining downstream operon but not DNA downstream of kilR41. Taken together, these results reveal that the integration or excision of the rac prophage can lead to a change in host fitness in the presence of different antibiotics. Thus, prophages can increase host fitness in the presence of antibiotics not only by introducing new genes (e.g., kilR, ralRA) into the host but also by affecting the function of the host protein by altering the coding sequence of the protein upon integration.
In this study, we found that rac excision is induced during biofilm formation (Fig. 1A), which is supported by the increased excision seen in the stationary phase (Fig. 7B), as biofilm cells are most similar to stationary phase cells42. Two proteins from rac, KilR and RalR, can both lead to cell death when overproduced24,26, but whether RalR or KilR can trigger cell death or cell lysis in biofilms requires further investigation. Conversely, recent studies have also shown that prophage excision under a specific condition, such as biofilm formation, will create genetic or potential phenotypic variation at the population level by generating a sub-population of prophage-free cells, which may benefit the population as a whole43,44. Here, we found that the rac deletion strain showed high swimming motility and formed more biofilm at an early stage in nutrient-rich medium. Excision of prophage CP4-57 in E. coli K-12 is induced during biofilm formation at early stages in both nutrient-rich and nutrient-poor medium, which led to a formation of slow-growing cells with increased motility8. In Pseudomonas aeruginosa, the conversion of prophage Pf4 into the super infective form occurred in dispersed biofilms, which is correlated with the appearance of small colony variants in the dispersal population44. In Streptococcus pneumoniae, a small population of cells undergoes prophage induction and contributes to the eDNA release during biofilm formation45. The excision of prophage A118 is specifically induced during intracellular growth primarily within phagosomes, which restores a functional competence gene that is required for the efficient phagosomal escape of L. monocytogenes18. These results suggest that bacteria can use various ways to respond to stress that involve editing their genomes through prophage excision. Hence, prophages not only contribute to the diversification of genome architecture among different bacterial strains but also contribute to intra-population diversification in a biofilm or in association with a eukaryotic host.
Previous attempts to identify prophage excision under normal growth conditions have shown very low excision frequencies7,46,47, which indicates stable residence of prophages under these conditions. To date, several host proteins have been described to control the expression of foreign DNA in bacteria. H-NS plays a key role in selectively silencing the transcription of horizontally acquired genes in Gram-negative bacteria48,49,50. In the nine prophages in E. coli K-12, H-NS only controls the excision of rac, which suggests a selective repression of H-NS on different prophages28. Similarly, the DNA-binding protein Hha only induces cryptic prophage CP4-57 and DLP12 excision8 and the transcription terminator factor Rho only represses cryptic prophage CPS-53 excision51. For the prophages CP4-57 and DLP12, Hha was found to bind to the attachment sites of these two prophages via nickel-enrichment DNA microarrays, thus facilitating homologous recombination during prophage excision52. In addition, the alternative sigma factor RpoH (σH) in Staphylococcus aureus is recruited by a temperate phage to regulate prophage integration and excision46. Here, we found that the stationary sigma factor RpoS regulates rac excision. Taken together, these results collectively reveal that recruitment of host factors by phages for integration/excision modulation may provide a novel strategy to sense host conditions and further influence prophage fitness and the correlative bacterial fitness. In addition, unlike rac in E. coli K-12, two rac-like prophages in E. coli O157:H7 retain replication machinery and can further infect other strains, including E. coli K-12, once excised20. Therefore, further study on the excision and transmissibility of rac or rac-like prophages under different stress conditions in other pathogenic strains is necessary.
Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The experiments were conducted in Luria-Bertani (LB) medium53 at 37 °C unless specified. Kanamycin (50 μg/mL) was used for culturing strains with single-gene knockouts from the Keio Collection54, chloramphenicol (30 μg/mL) was used for maintaining the pCA24N-based plasmids and kanamycin (50 μg/mL) was used to maintain the pET28b-based plasmids.
Plasmid construction
The primers used for the construction of the plasmids are listed in Table S2. For the construction of pCA24N-ttcA and pCA24N-ttcA’, the coding region of the target gene was PCR amplified from genomic DNA isolated from the BW25113 and from the Δrac strains, respectively. The PCR products were phosphorylated, purified and ligated into vector pCA24N as previously described26,55. The ligation mixture was transformed into BW25113 strains. The constructs were confirmed by PCR followed by DNA sequencing using primers pCA24N-F and pCA24N-R. For the construction of pET28b-xisR, the coding region of xisR was PCR amplified using the primer pair pET28b-xisR-F/R, the PCR product was digested with NcoI and HindIII and the digested PCR product was ligated into digested pET28b. The construct was confirmed by PCR followed by DNA sequencing using primers T7-F and T7-R.
Biofilm cell collection
Biofilm cells were collected as previously described with minor modifications56. Overnight cultures of the BW25113 and isogenic prophage deletion strains were inoculated into a 2 L shake flask (to reach OD600 ~ 0.05) containing 250 mL LB and 10 g glass wool (Corning Glass Works, Corning, NY), during which time the cells formed a biofilm on the surface of the glass wool. After incubation with shaking at 180 rpm at 37 °C for 48 h, the planktonic cells in contact with biofilms were taken directly from the culture. At the same time, the glass wool was taken from the culture and quickly washed twice in 100 mL ice-cold 0.85% NaCl. The biofilm cells were removed from the glass wool by sonication at 22 Watts for 2 min in a FS3 water bath sonicator (Fisher Scientific, Pittsburgh, PA, USA) submerged in 200 mL of ice-cold 0.85% NaCl. The biofilm cells were collected by centrifugation at 4 °C for 30 s to remove the NaCl. The cell pellets from the planktonic state growth and biofilm growth were saved at -80 °C until used.
Biofilm assay
Biofilm formation assays were carried out in 96-well polystyrene plates (Corning Costar, Cambridge, MA, USA) using 0.1% crystal violet staining57. Overnight E. coli K-12 BW25113 and the isogenic prophage deletion strain cultures were diluted to an OD600 of 0.05 in LB or M9C-glucose (M9 minimal medium with 0.4% casamino acids and 0.4% glucose) medium without shaking for 8 h or 24 h. The cell growth was measured at 620 nm. The biofilm cells were stained with 0.1% crystal violet and measured at 540 nm. To remove the growth effects, biofilm formation was normalized by bacterial growth for each strain. Two independent cultures were used for each strain.
Biofilm lysis assay
As E. coli K-12 BW25113 and the isogenic prophage deletion mutants do not have a complete lacZ gene, the plasmid pLP170 was introduced to provide a constitutive expression of lacZ58,59. Thus, BW25113 cells harboring pLP170 cells will release β-galactosidase into the supernatant only when the cells are lysed. The biofilm cells were collected using the method described above with the exception that 100 μg/mL carbenicillin was added to maintain the pLP170 plasmid. After 48 h of incubation, the supernatant was collected after removing the cells in the culture and the biofilm cells were collected after removal from the glass wool by sonication. The β-galactosidase activity was measured using the supernatant and the biofilm cells as described previously58. The percentage of lysis was calculated by quantifying the β-galactosidase activity in the supernatant compared to the total β-galactosidase activity in the biofilm cells and in the supernatant.
Protein expression and purification
For protein purification, E. coli BL21 (DE3) cells containing plasmid pET28b-xisR were grown overnight in LB supplemented with kanamycin. The overnight cultures were diluted 1:100 in LB and 1 mM IPTG was added when the OD600 reached ~1.0. The cells were collected by centrifugation after IPTG induction for 4 h, resuspended in lysis buffer (50 mM NaH2PO4·2H2O, pH 8.0, 300 mM NaCl) and were disrupted using a Fast-Prep 24 instrument (MP Biomedicals, Santa Ana, CA). The cell debris was removed by centrifugation and the His-tagged protein was purified using Ni-NTA agarose (Qiagen) following the manufacturer’s instructions. The eluted His-tagged proteins were collected and dialyzed against buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.4) and were stored at 4 °C.
Electrophoretic mobility shift assay (EMSA)
The DNA fragment AttR covering the attR site was PCR amplified using primer pair rac attR-F/R (Table S2). As a negative control, fragment AttM that only contains the upstream region of attR was also PCR amplified using primer pair rac-attM-F/R. DNA fragments were purified and labeled using the Biotin 3′ End DNA Labeling Kit (Thermo scientific, Rockford, USA). The purified protein and DNA fragments were mixed following the protocol as described in the LightShift Chemiluminescent EMSA kit (Thermo scientific, Rockford, USA). The binding reaction mixtures were incubated at 25 °C for 1 h. The protein-DNA complexes were separated by electrophoresis in 0.5 × Tris-borate-EDTA (TBE) polyacrylamide gels and were then transferred to nylon membranes. The DNA fragments were visualized using the Chemiluminescence Nucleic Acid Detection Module Kit (Thermo scientific, Rockford, USA).
Quantification of frequency of prophage excision
The frequency of prophage excision under different conditions was quantified by quantitative PCR (qPCR). The number of chromosomes that are devoid of rac or other prophages was quantified using qPCR primers (Table S2) as previously described7. The number of total chromosomes was quantified by a reference gene, adenylosuccinate synthase (purA). A quantification method based on the relative amount of a target gene versus a reference gene was used7,60. Total DNA was isolated using a TIANamp Bacteria DNA kit (Tiangen, China) and was used as the template for the qPCR reaction using the Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix (Thermo scientific, Rockford, USA). The reaction and analysis was performed using the StepOne Real-Time PCR System (Applied Biosystems).
Quantitative real-time reverse-transcription PCR (qRT-PCR)
For conducting qRT-PCR, total RNA was purified using a RNAprep Pure Cell/Bacteria kit (Tiangen, China). A total of 1 μg RNA was used for reverse transcription using the Reverse Transcription system kit (Promega, Madison, USA). The housekeeping gene rrsG (16S rRNA gene) was used to normalize the gene expression data. The remaining procedures followed those described above for the qPCR assay.
Metabolic assay
Metabolic activities in the presence of carbenicillin were measured using the Biolog kit (Hayward, CA, USA) as previously described26. Briefly, overnight cultures of BW25113 and the isogenic deletion strains were diluted in LB medium to an OD600 of 0.05 and allowed to grow until the OD600 reached ~1.0. The cells were diluted to an OD600 0.07 in 1 × IF-10a (Cat. No. 72264) and were further diluted 200-fold into a solution containing 1 × IF-10a, 1 × BioLog Redox Dye D (Cat. No. 74224) and 1 × Rich medium (2.0 g/L tryptone, 1.0 g/L yeast extract and 1.0 g/L NaCl) to a final OD600 of 0.0003. A volume of 100 μL of this cell suspension containing of 4 μg/mL carbenicillin was transferred into 96-well microtiter plates and was incubated at 37 °C without shaking. The metabolic activity was measured via the absorbance at 590 nm, which indicates the intracellular reduced state due to formazan (purple) formed from a tetrazolium dye.
Additional Information
How to cite this article: Liu, X. et al. Physiological Function of Rac Prophage During Biofilm Formation and Regulation of Rac Excision in Escherichia coli K-12. Sci. Rep. 5, 16074; doi: 10.1038/srep16074 (2015).
References
Canchaya, C., Proux, C., Fournous, G., Bruttin, A. & Brüssow, H. Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238–276 (2003).
Yu, Z. C. et al. Filamentous phages prevalent in Pseudoalteromonas spp. confer properties advantageous to host survival in Arctic sea ice. ISME J 9, 871–881 (2015).
Matos, R. C. et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet 9, e1003539 (2013).
Chen, J. & Novick, R. P. Phage-mediated intergeneric transfer of toxin genes. Science 323, 139–141 (2009).
Brüssow, H., Canchaya, C. & Hardt, W.-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602 (2004).
Bossi, L., Fuentes, J. A., Mora, G. & Figueroa-Bossi, N. Prophage contribution to bacterial population dynamics. J. Bacteriol. 185, 6467–6471 (2003).
Wang, X. et al. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147 (2010).
Wang, X., Kim, Y. & Wood, T. K. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J 3, 1164–1179 (2009).
Whiteley, M. et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001).
Williams, K. P. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucl. Acids Res. 30, 866–875 (2002).
Bobay, L. M., Rocha, E. P. & Touchon, M. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 30, 737–751 (2013).
Figueroa-Bossi, N. & Bossi, L. Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33, 167–176 (1999).
Campbell, A. M. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174, 7495–7499 (1992).
De Greve, H., Cao, Q. Z., Deboeck, F. & Hernalsteens, J. P. The Shiga-toxin VT2-encoding bacteriophage Φ297 integrates at a distinct position in the Escherichia coli genome. Biochim. Biophys. Acta 1579, 196–202 (2002).
Lee, C. Y. & Iandolo, J. J. Lysogenic conversion of Staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J. Bacteriol. 166, 385–391 (1986).
Carroll, J. D., Cafferkey, M. T. & Coleman, D. C. Serotype F double- and triple-converting phage insertionally inactivate the Staphylococcus aureus β-toxin determinant by a common molecular mechanism. FEMS Microbiol. Lett. 106, 147–155 (1993).
Loessner, M. J., Inman, R. B., Lauer, P. & Calendar, R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes : implications for phage evolution. Mol. Microbiol. 35, 324–340 (2000).
Rabinovich, L., Sigal, N., Borovok, I., Nir-Paz, R. & Herskovits, A. A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150, 792–802 (2012).
Kaiser, K. & Murray, N. E. Physical characterisation of the “Rac prophage” in E. coli K12. Mol. Gen. Genet. 175, 159–174 (1979).
Low, B. Restoration by the rac locus of recombinant forming ability in recB- and recC- merozygotes of Escherichia coli K-12. Mol. Gen. Genet. 122, 119–130 (1973).
Perna, N. T. et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533 (2001).
Asadulghani, M. et al. The defective prophage pool of Escherichia coli O157: Prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog 5, e1000408 (2009).
Casjens, S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49, 277–300 (2003).
Conter, A., Bouche, J. P. & Dassain, M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J. Bacteriol. 178, 5100–5104 (1996).
Soo, V. W., Hanson-Manful, P. & Patrick, W. M. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc. Natl Acad. Sci. USA 108, 1484–1489 (2011).
Guo, Y. et al. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucl. Acids Res. 42, 6448–6462 (2014).
Handa, N. & Kobayashi, I. Type III restriction is alleviated by bacteriophage (RecE) homologous recombination function but enhanced by bacterial (RecBCD) function. J. Bacteriol. 187, 7362–7373 (2005).
Hong, S. H., Wang, X. & Wood, T. K. Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli. Microb. Biotechnol. 3, 344–356 (2010).
Roberts, J. W. & Roberts, C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc. Natl Acad Sci. USA 72, 147–151 (1975).
Watnick, P. & Kolter, R. Biofilm, City of microbes. J. Bacteriol. 182, 2675–2679 (2000).
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997).
Boles, B. R. & Singh, P. K. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl Acad Sci. USA 105, 12503–12508 (2008).
Godeke, J., Paul, K., Lassak, J. & Thormann, K. M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J 5, 613–626 (2011).
Wang, X. & Wood, T. K. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J 5, 1517–1525 (2011).
Zhang, Z. & Saier, M. H. A novel mechanism of transposon-mediated gene activation. PLoS Genet 5, e1000689 (2009).
Lloyd, A. L., Rasko, D. A. & Mobley, H. L. T. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 189, 3532–3546 (2007).
Nichols, R. J. et al. Phenotypic landscape of a bacterial cell. Cell 144, 143–156 (2011).
Cho, E. H., Gumport, R. I. & Gardner, J. F. Interactions between integrase and excisionase in the phage lambda excisive nucleoprotein complex. J. Bacteriol. 184, 5200–5203 (2002).
Fogg, P. C. M., Rigden, D. J., Saunders, J. R., McCarthy, A. J. & Allison, H. E. Characterization of the relationship between integrase, excisionase and antirepressor activities associated with a superinfecting Shiga toxin encoding bacteriophage. Nucl. Acids Res. 39, 2116–2129 (2011).
Greer, H. The kil gene of bacteriophage lambda. Virology 66, 589–604 (1975).
Cardinale, C. J. et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938 (2008).
Beloin, C. & Ghigo, J. M. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13, 16–19 (2005).
Savage, V. J., Chopra, I. & O’Neill, A. J. Population diversification in Staphylococcus aureus biofilms may promote dissemination and persistence. PloS ONE 8, e62513 (2013).
Rice, S. A. et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J 3, 271–282 (2009).
Carrolo, M., Frias, M. J. O., Pinto, F. R., Melo-Cristino, J. & Ramirez, M. r. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE 5, e15678 (2010).
Tao, L. A., Wu, X. Q. & Sun, B. L. Alternative sigma factor sigma(H) modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog 6, e1000888 (2010).
Sozhamannan, S. et al. The Bacillus anthracis chromosome contains four conserved, excision-proficient, putative prophages. BMC Microbiol. 6, 34 (2006).
Dorman, C. J. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5, 157–161 (2007).
Navarre, W. W. et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 (2006).
Lucchini, S. et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 3, e38 (2007).
Menouni, R., Champ, S., Espinosa, L., Boudvillain, M. & Ansaldi, M. Transcription termination controls prophage maintenance in Escherichia coli genomes. Proc. Natl. Acad. Sci. USA 110, 14414–14419 (2013).
Garcia-Contreras, R., Zhang, X. S., Kim, Y. & Wood, T. K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS ONE 3, e2394 (2008).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, N.Y (1989).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006 0008 (2006).
Kitagawa, M. et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 (2005).
Ren, D., Bedzyk, L. A., Thomas, S. M., Ye, R. W. & Wood, T. K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524 (2004).
Fletcher, M. The effects of culture concentration and age, time and temperature on bacterial attachment to polystyrene. Can. J. Microbiol. 23, 1–6 (1977).
Ueda, A. & Wood, T. K. Connecting quorum sensing, c-di-GMP, pel polysaccharide and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5, e1000483 (2009).
Preston, M. J. et al. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65, 3086–3090 (1997).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 29, e45 (2001).
Acknowledgements
This work was supported by the National Basic Research Program of China (Grant No. 2013CB955701), by the National Science Foundation of China (31290233 and 31270214) and China Postdoctoral Science Foundation funded project (2013M542217 and 2014T70830). XW is the 1000-Youth Elite Program recipient in China. TW was supported by the ARO (W911NF-14-1-0279) and is the Biotechnology Endowed Professor at the Pennsylvania State University. We are grateful for KEIO and ASKA collection.
Author information
Authors and Affiliations
Contributions
X.W., T.W. and X.L. conceived and designed the study. X.L., Y.L., Y.G., Z.Z., B.L. and X.C. performed the experiments, X.L., Y.L., Y.G. and X.W. analyzed the data. X.L. and X.W. wrote the paper. T.W. critically reviewed the paper. All of the authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, X., Li, Y., Guo, Y. et al. Physiological Function of Rac Prophage During Biofilm Formation and Regulation of Rac Excision in Escherichia coli K-12. Sci Rep 5, 16074 (2015). https://doi.org/10.1038/srep16074
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16074
This article is cited by
-
Environmental pH is a key modulator of Staphylococcus aureus biofilm development under predation by the virulent phage phiIPLA-RODI
The ISME Journal (2021)
-
RETRACTED ARTICLE: Cryptic prophages in a blaNDM-1-bearing plasmid increase bacterial survival against high NaCl concentration, high and low temperatures, and oxidative and immunological stressors
Journal of Microbiology (2020)
-
Prophage protein RacR activates lysozyme LysN, causing the growth defect of E. coli JM83
Scientific Reports (2019)
-
Phage or foe: an insight into the impact of viral predation on microbial communities
The ISME Journal (2018)
-
Cold adaptation regulated by cryptic prophage excision in Shewanella oneidensis
The ISME Journal (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.