Abstract
Phenylketonuria (PKU) is an inherited autosomal recessive disorder of phenylalanine metabolism, mainly caused by a deficiency of phenylalanine hydroxylase (PAH). The incidence of various PAH mutations differs among race and ethnicity. Here we report a spectrum of PAH mutations complied from 796 PKU patients from mainland China. The all 13 exons and adjacent intronic regions of the PAH gene were determined by next-generation sequencing. We identified 194 different mutations, of which 41 are not reported before. Several mutations reoccurred with high frequency including p.R243Q, p.EX6-96A > G, p.V399V, p.R241C, p.R111*, p.Y356*, p.R413P and IVS4-1G > A. 76.33% of mutations were localized in exons 3, 6, 7, 11, 12. We further compared the frequency of each mutation between populations in northern and southern China and found significant differences in 19 mutations. Furthermore, we identified 101 mutations that are not reported before in Chinese population, our study thus broadens the mutational spectrum of Chinese PKU patients. Additionally, 41 novel mutations will expand and improve PAH mutation database. Finally, our study offers proof that NGS is effective, reduces screening times and costs and facilitates the provision of appropriate genetic counseling for PKU patients.
Similar content being viewed by others
Introduction
Phenylketonuria (PKU, OMIM #261600) is an inherited disorder characterized by increased level of phenylalanine in the blood. PKU is frequently caused by functional deficiency of phenylalanine hydroxylase (PAH), an enzyme that converts phenylalanine to other compounds in the body. More than 900 mutations have been identified in PAH and recorded in the locus-specific database (LSD) known as PAHvdb (http://www.biopku.org/pah/). The occurrence of PKU varies among ethic and geographic regions, reaching approximately 1 in 15,000 newborns1. In mainland China, the average incidence is 1in 11,6142 and in Taiwan 1in 55,0573.
The frequencies and distributions of PAH mutations also differs among different populations4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32. The wide variability of common mutations between ethnic groups and geographical areas makes PAH deficiency a genetic disease of great allelic heterogeneity. Comprehensive catalog of PAH mutation spectrum will be of value to evaluate genotype-phenotype correlations, to provide genetic consultations to patients’ families as well as prenatal diagnosis and to refine diagnoses in and anticipate the dietary requirements of affected patients33,34,35.
A large-scale, unbiased comprehensive survey of PAH mutations in Chinese population was not available. Most previous analyses concerning the Chinese population were limited to a few common mutations, or were confined to a certain region of PAH, resulting in a selective bias3,12,36,37,38,39,40,41. One report carried out survey of entire PAH in a small-scale survey including 212 patients in mainland China12.
In the present study, we report a spectrum of PAH mutations complied from a large cohort of 796 PKU patients in mainland China. We determined the sequence of entire PAH gene using next-generation sequencing (NGS). Among 194 mutations identified, 41 were not reported in literature and 101 not reported in Chinese population. We believe that these results will facilitate the development of appropriate genetic counseling for PKU patients in China.
Results
Mutation spectrum
In the included cohort of Chinese patients, potential disease-causing mutations were identified on 1516 of 1592 independent alleles, corresponding to a mutation detection rate of 95.23%.A total of 720 patients were completely genotyped, whereas in the remaining 76 individuals only one causative mutation was identified; the other mutation site could not be identified by this platform. Among the fully genotyped patients, two mutations were detected in 683 of the patients, who had either compound heterozygous (n = 622) or homozygous (n = 61) genotypes, three mutations were found in 35 of the patients and four mutations were revealed in 2 of the patients. The gene analysis results were summarised in Table 1.
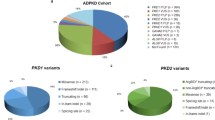
A total of 194 different types of mutations were identified, including 134 missense mutations (69.07%), 25 splice-site mutations (12.89%), 18 deletions (9.28%), 14 nonsense mutations (7.22%), 2 insertions (1.03%) and 1 silence/splice (0.52%). 76.33% of the total mutations are found in exons. Most mutations was localized in exon7 (33.44%), exon11 (13.18%), exon6 (10.48%), exon12 (10.29%), exon3 (8.94%). Interestingly, no mutations were identified in exon13.
In terms of mutation frequency, p.R243Q was the most prevalent mutation (frequency 17.53%). Other mutations with relatively high frequencies were p.EX6-96A > G, p.V399V, p.R241C, p.R111*, p.Y356*, p.R413P and IVS4-1G > A (7.66%, 5.84%, 5.40%, 4.77%, 4.46%, 4.33%, 3.77%, respectively). Eight mutations, including p.R53H, p.A434D, p.R408Q, IVS7 + 2T > A, p.G247V, p.S70del, p.R261Q and p.Y166* were found with relatively lower frequencies (ranging from 1% to 3%). The remaining 178 mutations were found at relative frequencies of less than 1%.
Comparison between northern and southern Chinese populations
The frequencies of each PAH mutations were compared among geographical regions in China. We used sixteen mutations with frequency more than 1% for this comparison. Six mutations showed obvious local mutation clustering: p.V399V, p.R413P and IVS7 + 2T > A were found to be clustered in northern Chinese populations and p.R241C, p.R408Q and p.Y166* were clustered in southern Chinese populations. Among the remaining 178 mutations that exhibited a relative frequency of <1%, thirteen exhibited significant differential distribution in northern and southern China.
Novel sequence variants
Forty-one novel nucleotide lesions that have not been registered in the BIOPKU database (http://www.biopku.org/pah/) were identified in this study: p.V5Sfs*34, IVS1 + 5_ + 6delGC, IVS1-3T > C, p.F39del, p.E43*, p.N58*, p.L62Pfs*7, p.E78Q, p.E78V, p.I95del, p.D101N, p.G103D, IVS3-2A > G, p.F121V, p.E127K, p.E127G, p.G148R, p.R169S, p.R169G, p.H170P, p.E178K, p.N223I, p.F260I, p.E280Nfs*61, p.V291M, p.V291L, p.P292S, p.D296H, p.Q304K, p.T328N, p.K335E, p.A342Hfs*59, p.Y343*, IVS10-12delT, IVS10-1G > C, IVS10-1G > T, p.Y356D, IVS11-1G > C, p.T372R, p.I406V, p.Q429K.
Among these mutations, the majority were missense mutations (n = 25), followed by small deletions (n = 8), nonsense (n = 2), splice (n = 5) and insertions (n = 1). Thirty-four of the novel mutations were detected in coding regions and the remaining were located in introns.
The prediction results of novel mutations are listed in Table 2. A total of 25 novel mutations were predicted; of these, 20 mutations were predicted to be probably damaging, 5 mutations were tolerated and the remainder of the 16 mutations could not be predicted using this tool. The frequencies of the novel mutations were relatively low, which indicates that they are rare mutations.
Discussion
A comprehensive survey of the mutation spectrum of the protein of interest in a given population not only can provide insight into the structural and functional aspects of the protein as well as genotype-phenotype correlations14,15,20,22,23,26,28,42, but also facilitate genetic counseling in patients’ families. In this study, we described the molecular basis of PKU in a mainland Chinese population by analysing mutations in the PAH gene using NGS. Among a cohort of 796 patients, mutations were detected on 1516 of the 1592 independent alleles, representing a mutation detection rate of 95.23% (Table 1). A total of 194 distinct mutations were found, demonstrating the high genetic heterogeneity that is inherent in PKU.
The number of different mutations in a given population is usually high and is typically comprised of a few prevalent mutations and a large number of private mutations43. In comparing our study with previous reports, 83 of the mutations that we identified have been previously reported; however, the remaining 101 mutations (including 41 novel mutations) were reported for the first time in a Chinese mainland population12,36,37,38,39,41. As shown in Table 3, eight mutations including p.R243Q, p.EX6-96A > G, p.V399V, p.R241C, p.R111*, p.Y356*, p.R413P and IVS4-1G > A are common mutations in the mainland Chinese population, although the rank order of these mutations was different. Among them, R241C, p.EX6-96A > G, p.V399V, R413P, R243Q and R111* are also considered to be prevalent in the Chinese Taiwanese population40. The epidemiology of phenylketonuria in China is complicated. The prevalence of PKU in northern China (1/11,000) is close to what has been documented in Caucasian populations, but the prevalence in southern China is much lower44. A comparison between northern and southern China indicated marked differences in the relative frequencies of mutations. Among eight common mutations, V399V, R241C and R413P gave significant p values between two regions, with respective p values of 0.037, 0.007 and 0.002. The result that p.R413P clustered in northern China is consistent with what has been reported by Gu. et al.12. Furthermore, p.V399V has been previously detected primarily in populations of Xinjiang and northern China36,37, whereas p.R241C has been primarily detected in populations in southern China and in Taiwan patients3,45. The majority of the population in Taiwan has descended from southeast China. Based on our data, we hypothesize that the uniform distribution of V399V, R241C and R413P is a result of migration and the founder effect.
It is well known that different ethnic groups have their own distinctive and diverse PAH mutant allele series that include either one or a few prevalent founder alleles46. When comparing PAH mutational data between different ethnic groups, correlations between the mutations in and the genetic histories of the investigated populations were found. Marked differences were identified when comparing PAH mutations with ≥ 3% frequency (totaling 34) between Asian and European countries (Table 4). Five mutations, including p.R243Q, p.EX6-96A > G, p.R241C, p.R413P and IVS4-1G > A, were found to be common mutations in East Asian countries such as China, Japan14 and Korea8, accounting for 53.76%, 69.70% and 62.10% of the total mutations respectively. Three mutations, including p.R111*, p.Y356* and p.T278I, were frequently detected in China and Japan14, China and Korea8, Japan14 and Korea8 respectively. The remaining mutations, including p.V399V, p.A259T, p.R252W, p.Y325* and p.V388M, were found to be common in only one country. In sharp contrast, these mutations except for p.R252W, the above mentioned mutations were either rarely detected or undetected in West Asia and Europe countries. For example in Iran17 and Turkey18, three common mutations including p.R261Q, p.P281L and IVS10-11G, A were shared, However these mutations were either rare or did not occur in East Asia. Instead, they were prevalent in select Europe countries. In Europe, p.R408W was found to be the common mutation, ranking first in the Czech Republic28 (East Europe) and Germany5 (West Europe) and second in Danemark4 (North Europe) and Serbia32(South Europe). These results suggest that p.R408W was the most prevalent founder allele in the European population46. In contrast, p.R408W was either rarely detected or undetected in populations of East Asia, whereas it was the most common mutation in Turkish populations18. The remaining mutations were only found to be prevalent in subsets of the four countries. For example, p.L48S was the most prevalent mutation in Serbian populations32 and it was also common in Turkish populations18. The p.R158Q, p.A403V, p.Y414C and IVS12 + 1G > A mutations were relatively common in only two countries. Based on the above comparison, we identified that there were several overlaps of mutant allele distributions between West Asia and Europe and that the mutations that were common in East Asia were different from these.
In the present study, the high mutation detection rate of 95.23% was similar to previous studies in which sequencing analysis of the PAH gene was conducted12,15, but it was relatively lower than the results from studies that employed exon analysis combined with multiplex ligation-dependent probe amplification (MLPA)14,20,24,28. This is because NGS is able to detect small deletions and insertions, whereas it is not able to detect large deletions or duplications. Despite scanning the entirety of the PAH coding region and its exon–intron boundaries, no mutations were detected in 76 alleles. The most likely explanation behind this is that the mutations are located in regions that were not detectable in this study (for example, in the promoter regions, the 5′ and 3′ UTRs, in non-coding RNA binding sites, or in the intronic sequences far away from exon–intron boundaries). Alternatively, the mutations may have been large deletions or duplications.
Using NGS as a routine genetic diagnostic tool enables thousands of DNA sequences to be simultaneously obtained in notably reduced turnaround times and at a significantly reduced cost. Furthermore, this technique provides high sensitivity, specificity and coverage (including all coding regions of the involved exons and adjacent intronic regions). However, the biggest limitation to using NGS is the need to analyse and interpret complicated data.
We believe our study will provide guidance for future medical practice such as prenatal screening and early diagnosis of PKU. Diagnostic methods can be developed based on the known characteristics of a population. Currently, PKU screening is performed in newborn babies as a part of the tertiary prevention in birth defects preventive network in China. Developing a new method for screening might enhance primary prevention. Based on the mutational spectrum presented in this study, our hope is that carrier screening can be conducted preceding gestation, which would offer timely guidance with respect to prenatal diagnosis for couples who are both carriers.
Methods
Subjects
A total of 796 unrelated patients from 29 separate newborn screening centres of China were enrolled. These patients were diagnosed at birth either through a neonatal screening program or based on clinical presentation. Demographic data, including age, consanguinity, family history and geographical origin and biochemical testing data, including plasma phenylalanine (Phe) levels, dihydropteridine reductase activity, urinary biopterin and neopterin ratio and tetrahydrobiopterin loading, were collected. The ages of the included patients ranged from 6- months to 5-years old. In families with more than one patient, only one member of each sibling pair was included in the study of mutation frequency. The numbers of patients in northern China and southern China that were divided by the Qinling Mountains and the Huaihe River were 557 and 239, respectively. Both parents of the included patients were native.
These patients were classified into one of three separate phenotype categories according to their pretreatment plasma Phe levels, including mild hyperphenylalaninaemia (Phe 120–600 umol/L), mild PKU(Phe 600-1200 μmol/L) and classic PKU (Phe >1200 μmol/L)47. Among the 796 patients, 145 (18.22%) were mild hyperphenylalaninemia(MHP), 215 (27.01%) were mild PKU(mPKU) and 418 (52.51%) were classic PKU(cPKU), the remaining 18 patients (2.26%) could not be classified because the pretreatment Phe levels were not available(Table 5). The entire list of patients’ phenotype and blood Phe levels can be found as Supplementary Table S1. The parental permissions and informed consents were obtained from the parents of all patients. The research was approved by the Ethics Committee of West China Second University Hospital, Sichuan University (No: 2014018) and followed the tenets of the Declaration of Helsinki.
DNA extraction
Blood samples (4 ml) were collected from each patient and their parents by venipuncture in EDTA. Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp® DNA Blood Mini Kit (Qiagen, Cat.No.51106, Germany) according to the recommended protocol.
Next-generation sequencing
A series of unique primers were designed to amplify all 13 exons and their surrounding introns, which covered 200bp upstream and 200bp downstream of the exons of the PAH gene. A pair of primers that were designed to amplify approximately 150 bp of the human β-globin gene (HBB) (accession number AY260740) was used as an internal quality control to identify false negatives caused by inadequate DNA or failed PCR.
To improve the throughput of the assay, the PCR primers were designed to not only amplify the target DNA but also to provide a unique primer index for each of the 96 samples in each of the plates (multiple index PCR). This strategy resulted in 96 sets of 10-bp-long nucleic acid tags that were individually included at the 5′ ends of each of the PAH and HBB primers. The second index that was used for each sample was the 8-bp-long nucleic acid tag from the library adapter sequence, which identified the specific 96-well plate that each sample was included in. This index was attached to the amplicons of the samples through an adapter preparation process. Using this “double index system,” hundreds of samples can be mixed together and detected in one sequencing chip at the same time.
PCR was performed on a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA), with a cycling protocol that consisted of denaturation at 94 °C for 30 s, 56° C for 30 s and 72 °C for 1 min. After 35 cycles, gel electrophoresis was used to verify the quality of the amplified DNA, only eligible DNAs was included in the library preparation. The PCR amplification products were prepared for DNA pooling and the diverse adapter library was added onto the amplification products. After a concentration of DNA was obtained that could satisfy the requirements of the library preparation method, gene mutations were sequenced using an Illumina Hiseq 2000 (Illumina Inc, San Diego, CA, USA) sequencing instrument.
After sequencing the samples, the raw sequence data were analysed using in-house software. First, all sequence data were traced back to the specimens from which they arose according to the sequences of the primer and adapter indices. Second, the amplicon sequences of each of the samples were aligned with standard reference PAH sequences from the database PAHvdb (http://www.biopku.org/pah/); SNPs were found in target areas and relevant information was noted.
All of the PAH gene sequencing reactions and analyses were performed in the Centre of BGI Health clinical laboratory, Shenzhen, China.
Validation tests of Sanger sequencing
When a given patient’s mutation locus was detected, it was amplified by polymerase chain reaction (PCR) from a parent’s sample and then sequenced bidirectionally in an ABI-3730 DNA analyser. This not only validated the locus but also confirmed the carrier status of the parents. The PCR cycling protocol consisted of an initial denaturation at 95 °C for 3 min, followed by 35 cycles, of 95 °C for 40 s, 55 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 10 min.
Pathologic analysis of novel mutations
A SIFT prediction was performed (http://sift.jcvi.org/) using the “SIFT Human SNPs” tool to obtain predictions for nonsynonymous SNPs. The annotation version was Homo sapiens GRCh37 Ensembl 63. A list of the chromosomal positions and alleles corresponding to the 41 novel mutations were uploaded into the import the web-site.
Statistical analysis
Statistical analysis was performed using statistical package for social science software (SPSS version 16.0). Mutational frequencies were calculated by the counting method. An x2 analysis was performed to test for differences between two geographic populations. A p value <0.05 was considered statistically significant.
Additional Information
How to cite this article: Li, N. et al. Molecular characterisation of phenylketonuria in a Chinese mainland population using next-generation sequencing. Sci. Rep. 5, 15769; doi: 10.1038/srep15769 (2015).
References
Mitchell, J. J., Trakadis, Y. J. & Scriver, C. R. Phenylalanine hydroxylase deficiency. Genet Med 13, 697–707 (2011).
Shi, X. T. et al. Newborn screening for inborn errors of metabolism in mainland china: 30 years of experience. JIMD Rep 6, 79–83 (2012).
Chien, Y. H. et al. Mutation spectrum in Taiwanese patients with phenylalanine hydroxylase deficiency and a founder effect for the R241C mutation. Hum Mutat 23, 206 (2004).
Guldberg, P., Henriksen, K. F. & Guttler, F. Molecular analysis of phenylketonuria in Denmark: 99% of the mutations detected by denaturing gradient gel electrophoresis. Genomics 17, 141–146 (1993).
Aulehla-Scholz, C. & Heilbronner, H. Mutational spectrum in German patients with phenylalanine hydroxylase deficiency. Hum Mutat 21, 399–400 (2003).
Kasnauskiene, J., Giannattasio, S., Lattanzio, P., Cimbalistiene, L. & Kucinskas, V. The molecular basis of phenylketonuria in Lithuania. Hum Mutat 21, 398 (2003).
Pronina, N. et al. The molecular basis of phenylketonuria in Latvia. Hum Mutat 21, 398–399 (2003).
Lee, D. H. et al. The molecular basis of phenylketonuria in Koreans. J Hum Genet 49, 617–621 (2004).
Bercovich, D. et al. Genotype-phenotype correlations analysis of mutations in the phenylalanine hydroxylase (PAH) gene. J Hum Genet 53, 407–418 (2008).
Karacic, I. et al. Genotype-predicted tetrahydrobiopterin (BH4)-responsiveness and molecular genetics in Croatian patients with phenylalanine hydroxylase (PAH) deficiency. Mol Genet Metab 97, 165–171 (2009).
Dahri, S. et al. Mutation analysis of phenylketonuria patients from Morocco: high prevalence of mutation G352fsdelG and detection of a novel mutation p.K85X. Clin Biochem 43, 76–81 (2010).
Zhu, T. et al. Mutational spectrum of phenylketonuria in the Chinese Han population: a novel insight into the geographic distribution of the common mutations. Pediatr Res 67, 280–285 (2010).
Bashyam, M. D. et al. Phenylalanine hydroxylase gene mutations in phenylketonuria patients from India: identification of novel mutations that affect PAH RNA. Mol Genet Metab 100, 96–99 (2010).
Okano, Y., Kudo, S., Nishi, Y., Sakaguchi, T. & Aso, K. Molecular characterization of phenylketonuria and tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency in Japan. J Hum Genet 56, 306–12 (2011).
Rivera, I. et al. Phenylalanine hydroxylase deficiency: molecular epidemiology and predictable BH4-responsiveness in South Portugal PKU patients. Mol Genet Metab 104 Suppl, S86–92 (2011).
Kostandyan, N. et al. The spectrum of phenylketonuria genotypes in the Armenian population: identification of three novel mutant PAH alleles. Mol Genet Metab 104 Suppl, S93–96 (2011).
Zare-Karizi, S. et al. Mutation spectrum of phenylketonuria in Iranian population. Mol Genet Metab 102, 29–32 (2011).
Dobrowolski, S. F. et al. Molecular genetics and impact of residual in vitro phenylalanine hydroxylase activity on tetrahydrobiopterin responsiveness in Turkish PKU population. Mol Genet Metab 102, 116–121 (2011).
Georgiou, T. et al. The spectrum of mutations identified in Cypriot patients with phenylalanine hydroxylase deficiency detected through neonatal screening. Clin Biochem 45, 588–592 (2012).
Groselj, U. et al. Five novel mutations and two large deletions in a population analysis of the phenylalanine hydroxylase gene. Mol Genet Metab 106, 142–148 (2012).
Baturina, O. A., Bondar, A. A., Tupikin, A. E., Zhabin, S. G. & Morozov, I. V. Mutation analysis of the phenylalanine hydroxylase gene of phenylketonuria patients of Kemerovskaya Oblast’ and Saha Republic. Tsitol Genet 46, 40–47 (2012).
Djordjevic, M. et al. Molecular Genetics and Genotype-Based Estimation of BH4-Responsiveness in Serbian PKU Patients: Spotlight on Phenotypic Implications of p.L48S. JIMD Rep 9, 49–58 (2012).
Bueno, M. A. et al. Molecular epidemiology and genotype-phenotype correlation in phenylketonuria patients from South Spain. J Hum Genet 58, 279–284 (2013).
Polak, E. et al. Phenylalanine hydroxylase deficiency in the Slovak population: genotype-phenotype correlations and genotype-based predictions of BH4-responsiveness. Gene 526, 347–355 (2013).
Trunzo, R. et al. Mutation analysis in hyperphenylalaninemia patients from South Italy. Clin Biochem 46, 1896–1898 (2013).
Murad, H., Dabboul, A., Moassas, F., Alasmar, D. & Al-Achkar, W. Mutation spectrum of phenylketonuria in Syrian population: genotype-phenotype correlation. Gene 528, 241–247 (2013).
Karam, P. E., Alhamra, R. S., Nemer, G. & Usta, J. Spectrum of mutations in Lebanese patients with phenylalanine hydroxylase deficiency. Gene 515, 117–122 (2013).
Reblova, K. et al. Hyperphenylalaninemia in the Czech Republic: genotype-phenotype correlations and in silico analysis of novel missense mutations. Clin Chim Acta 419, 1–10 (2013).
Bik-Multanowski, M. et al. Molecular genetics of PKU in Poland and potential impact of mutations on BH4 responsiveness. Acta Biochim Pol 60, 613–616 (2013).
Leuders, S. et al. Influence of PAH Genotype on Sapropterin Response in PKU: Results of a Single-Center Cohort Study. JIMD Rep 13, 101–109 (2014).
Ho, G. et al. The Molecular Bases of Phenylketonuria (PKU) in New South Wales, Australia: Mutation Profile and Correlation with Tetrahydrobiopterin (BH4) Responsiveness. JIMD Rep 14, 55–65 (2014).
Djordjevic, M. et al. Molecular Genetics and Genotype-Based Estimation of BH4-Responsiveness in Serbian PKU Patients: Spotlight on Phenotypic Implications of p.L48S. JIMD Rep 9, 49–58 (2013).
Guttler, F. & Guldberg, P. Mutation analysis anticipates dietary requirements in phenylketonuria. Eur J Pediatr 159 Suppl 2, S150–153 (2000).
Dobrowolski, S. F. et al. Mutations in the phenylalanine hydroxylase gene identified in 95 patients with phenylketonuria using novel systems of mutation scanning and specific genotyping based upon thermal melt profiles. Mol Genet Metab 91, 218–227 (2007).
Dobrowolski, S. F. et al. A limited spectrum of phenylalanine hydroxylase mutations is observed in phenylketonuria patients in western Poland and implications for treatment with 6R tetrahydrobiopterin. J Hum Genet 54, 335–339 (2009).
Song, F. et al. Phenylketonuria mutations in Northern China. Mol Genet Metab 86 Suppl 1, S107–118 (2005).
Yu, W. Z. et al. Mutation characteristics of the PAH gene in four nationality groups in Xinjiang of China. J Genet 87, 293–297 (2008).
Zhou, Y. A. et al. Mutations of the phenylalanine hydroxylase gene in patients with phenylketonuria in Shanxi, China. Genet Mol Biol 35, 709–713 (2012).
Mao, X. M. et al. Analysis of mutations in exon 7 of phenylalanine hydroxylase gene among children with phenylketonuria in Ningxia, China. Zhongguo Dang Dai Er Ke Za Zhi 16, 259–262 (2014).
Liang, Y. et al. The mutation spectrum of the phenylalanine hydroxylase (PAH) gene and associated haplotypes reveal ethnic heterogeneity in the Taiwanese population. J Hum Genet 59, 145–152 (2014).
Yu, W. et al. Characterization of phenylalanine hydroxylase gene mutations in phenylketonuria in Xinjiang of China. Int J Clin Exp Med 7, 4406–4412 (2014).
Santos, L. L. et al. Variations in genotype-phenotype correlations in phenylketonuria patients. Genet Mol Res 9, 1–8 (2010).
Avigad, S. et al. Compound heterozygosity in nonphenylketonuria hyperphenylalanemia: the contribution of mutations for classical phenylketonuria. Am J Hum Genet 49, 393–399 (1991).
Liu, T. T., Chiang, S. H., Wu, S. J. & Hsiao, K. J. Tetrahydrobiopterin-deficient hyperphenylalaninemia in the Chinese. Clin Chim Acta 313, 157–169 (2001).
Zhang, M. et al. Two novel mutations in phenylalanine hydroxylase gene and in vitro expression analysis on mutation Arg252Gln. Chin Med Sci J 12, 22–25 (1997).
Zschocke J. Phenylketonuria mutations in Europe. Hum Mutat 21, 345–356 (2003).
Lindner, M. Treatment of phenylketonuria variants: European recommendations. In Blau N., ed. PKU and BH4: advances in phenylketonuria and tetrahydrobiopterin. Heilbronn: SPS Verlagsgesellschaft mbH. 180–187 (2006).
Acknowledgements
We thank all of the children and parents who participated in this study and provided personal information. We thank the staff of the newborn screening centres that were involved in the recruitment and data collection processes. This study was supported by the National “Twelfth Five-Year” Plan for Science & Technology Support of China (grant ID: 2014BAI06B01), the National Science & Technology basic work Project of the Ministry of Science and Technology of China (grant ID: 2014FY110700), the National “Twelfth Five-Year” Plan for Science & Technology Support of China (grant ID: 2012BAI09B04) and the Program for Changjiang Scholars and Innovative Research Team in University (grant ID: IRT0935).
Author information
Authors and Affiliations
Contributions
N.N.L. and J.Z. developed the study design, conducted the study and drafted the manuscript. Z.L. and J.T. assisted in organising and collecting the samples. H.T.J. and S.C. conducted genetic data acquisition and interpretation. J.P.S. and L.T.Z. analysed the data. X.H.L., Y.D., X.J. and Y.L. participated in reviewing, editing and revising the manuscript. W.W. conceived this study. All authors have read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Li, N., Jia, H., Liu, Z. et al. Molecular characterisation of phenylketonuria in a Chinese mainland population using next-generation sequencing. Sci Rep 5, 15769 (2015). https://doi.org/10.1038/srep15769
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15769
This article is cited by
-
Identification of deep intronic variants of PAH in phenylketonuria using full-length gene sequencing
Orphanet Journal of Rare Diseases (2023)
-
The spectrum of phenylalanine hydroxylase variants and genotype–phenotype correlation in phenylketonuria patients in Gansu, China
Human Genomics (2023)
-
Identification of phenylketonuria patient genotypes using single-gene full-length sequencing
Human Genomics (2022)
-
Characterization of phenylalanine hydroxylase gene variants and analysis of genotype–phenotype correlation in patients with phenylalanine hydroxylase deficiency from Fujian Province, Southeastern China
Molecular Biology Reports (2022)
-
The study of the full spectrum of variants leading to hyperphenylalaninemia have revealed 10 new variants in the PAH gene
Metabolic Brain Disease (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.