Abstract
It is an urgent need to develop new drugs for Mycobacterium tuberculosis (Mtb) and the enzyme, dihydrofolate reductase (DHFR) is a recognised drug target. The crystal structures of methotrexate binding to mt- and h-DHFR separately indicate that the glycerol (GOL) binding site is likely to be critical for the function of mt-DHFR selective inhibitors. We have used in silico methods to screen NCI small molecule database and a group of related compounds were obtained that inhibit mt-DHFR activity and showed bactericidal effects against a test Mtb strain. The binding poses were then analysed and the influence of GOL binding site was studied by using molecular modelling. By comparing the chemical structures, 4 compounds that might be able to occupy the GOL binding site were identified. However, these compounds contain large hydrophobic side chains. As the GOL binding site is more hydrophilic, molecular modelling indicated that these compounds were failed to occupy the GOL site. The most potent inhibitor (compound 6) demonstrated limited selectivity for mt-DHFR, but did contain a novel central core (7H-pyrrolo[3,2-f]quinazoline-1,3-diamine), which may significantly expand the chemical space of novel mt-DHFR inhibitors. Collectively, these observations will inform future medicinal chemistry efforts to improve the selectivity of compounds against mt-DHFR.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is a deadly infectious disease caused by Mycobacterium tuberculosis (Mtb) that is particularly prevalent in South-East Asia and Africa. In 2013, it is estimated that 9 million people developed TB and 1.5 million died from the disease1. Despite the fact that death from TB is often preventable, the rapid increase of multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis has resulted in an urgent need to develop new drug targets for Mtb2,3.
The enzyme, dihydrofolate reductase (DHFR), catalyzes NADPH-dependent reduction of dihydrofolate to tetrahydrofolate, which is a precursor of cofactors necessary for the synthesis of thymidylate, purine nucleotides, methionine, serine and glycine that are required for DNA, RNA and protein synthesis4,5. Specific inhibitors of mycobacterial DHFR (mt-DHFR) that are active against live Mtb cells have been developed, suggesting that such inhibitors may be useful for treating TB6. Highly potent inhibition of DHFR has been achieved with analogues of the substrate, dihydrofolate and one of the most well-known inhibitors is Methotrexate (MTX, Fig. 1), which binds to both human DHFR (h-DHFR) and mt-DHFR without any significant selectivity7,8. Such inhibitors contain 2,4-Diaminopteridin as the central core and are referred to as classical inhibitors. However, the full pteridinediamine structure is not required and non-classical inhibitors were subsequently developed that contain pyrimidine-2,4-diamines or analogues as the central cores, such as Trimethoprim (TMP)9, Pyrimethamine10,11, Methylbenzoprim12 and Bromo-WR9921013,14 (Fig. 1).
Analysis of the crystal structures of MTX binding to mt- and h-DHFR indicate that mt-DHFR binds with MTX, glycerol (GOL) and NADPH (PDB ID: 1DF7) (Fig. 2)15, but the GOL binding site is essentially absent in h-DHFR (PDB ID: 1OHJ, h-DHFR complexed with NADPH and PT523)16. The mt-DHFR active site can be roughly divided into 2 parts, the dihydrofolate and GOL binding sites. Most DHFR inhibitors target the dihydrofolate binding site. The GOL binding site is a relative small pocket and is very close to the dihydrofolate binding site, such that it might be treated as an extension of dihydrofolate binding site. It has been assumed that the GOL binding site is critical for the function of mt-DHFR selective inhibitors15,17. For example, El-Hamamsy et al., designed and synthesised a group of compounds containing GOL-like side chains and among these compounds, compound El-7a (Fig. 1) showed notable selectivity for inhibition effects on mt-DHFR against h-DHFR17. However, the assay they used to evaluate the abilities of El-7a was based on TB5 Saccharomyces cerevisiae carrying the DHFR genes from M. tuberculosis and human. Therefore, there is no direct evidence to show El-7a can inhibit the growth of M. tuberculosis.
In the present study, pharmacophore models were built using in silico methods that considered the influence of the GOL binding site. A group of related compounds were obtained and tested for their ability to inhibit mt-DHFR activity and for the bactericidal effects against a test Mtb strain. Of these, a group of compounds that significantly inhibited the growth of Mtb (without strong selectivity against h-DHFR) were obtained. In order to understand the relatively low selectivity (5 times vs h-DHFR for the most potent compound), the influence of GOL binding site was studied using molecular dynamic simulations and free energy calculations, as it was believed that the GOL binding site could be important for the selectivity.
Methods
Selection of Docking Libraries
The National Cancer Institute compounds library (NCI Release 4, 265,242 compounds, published in May 2012) was chosen for computer screening. The 3D conformations of all the molecules were generated using the Sybyl (v 7.3; Tripos) virtual screening tools.
System Setup
The crystal structure of mt-DHFR in complex with NADPH, GOL and MTX (PDB ID: 1DF7)15 was taken as the receptor. The bound ligand, MTX, was extracted and used as a reference to indicate the binding site. The receptor was treated with Amber 1218, not only for virtual screening and molecular docking, but also for subsequent molecular dynamic simulations. The parameters of NADPH were obtained from AMBER parameter database (http://www.pharmacy.manchester.ac.uk/bryce/amber/, uploaded by U. Ryde) and the ab initio QM calculations were performed on GOL using the B3LYP 6-31G* basis set within Gaussian0919. The molecular geometries were optimised and the atom-centered point charges were calculated to fit the electrostatic potential using RESP20. The receptor was treated by leap module in Amber 12 with amber ff12SB (for the protein part) and Generalized Amber Force Field (GAFF, for the GOL part)21 force field and then the receptor was saved in the pdb format for virtual screening and molecular docking.
Virtual screening
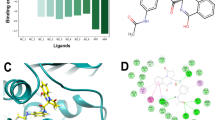
A total of 78 inhibitors of mt-DHFR were collected from the PDB (ligands complexed with protein) and various literature sources17,22,23,24,25. All these compounds were docked into mt-DHFR to understand their interactions with the protein. A 3D-pharmacophore model was then built and used to search the NCI database (National Cancer Institute small molecule database release 4) using the Unity module within Sybyl, as shown in Fig. 3A. As compound El-7a17 was predicted to occupy the GOL binding site which may cause the selectivity against h-DHFR, a decoy compound template was designed and used as a reference to perform a similarity search using vROCS (version 3.1.2, Openeye)26 with default settings (Fig. 3B). In the molecule template, the acylamino side chain was used to mimic the hydrogen bond acceptors and donors on GOL.
Molecular Docking
FRED Docking
FRED (v 3.0.1; Fast Rigid Exhaustive Docking)27 is a protein-ligand docking program released by Openeye. The hits obtained from previous virtual screening were treated with the Omega2 (v 2.5.1.4)28 module in the Openeye package using the default settings to generate a multi-conformers molecular library and saved at most 200 conformations per compound. The hits library was then docked to mt-DHFR using FRED with default settings and ranked by the Chemgauss 4 scoring function. The top 500 compounds were recorded for further consideration.
GOLD Docking
GOLD (v 5.2.2 Genetic Optimization for Ligand Docking)29 was used to dock each ligand 10 times, starting each time from a different random population of ligand orientations and using the default automatic genetic algorithm parameter settings. All torsion angles in each compound were allowed to rotate freely and the results of the different docking runs were ranked using Gold Score. The top 500 molecules were considered for experimental testing.
Generation of recombinant enzymes and in vitro assays for Mtb and human DHFR
Recom-binant DHFR enzymes were expressed and purified as described previously30. Enzyme assays were performed in 100 mM HEPES, 50 mM KCl, pH 7.0 at 25 °C. The absorbance decrease at 340 nm representing the oxidation of NADPH was monitored with a spectrophotometer (Cary 50, Varian, Palo Alto, CA). Inhibitors were added to a 1 ml cuvette at various concentrations, with the individual DHFRs (20 nM) and 40 μM of NADPH and the reaction was initiated by the addition of 40 μM of dihydrofolate. For IC50 determinations, 20 nM of DHFRs was incubated with 40 μM of cofactor NADPH and seven serially diluted concentrations of the inhibitors for 1 minute. The reaction was initiated by the addition of 40 μM dihydrofolate. The reaction progress was measured for 2 minutes and the linear region was used to determine the initial velocity parameters. Percent inhibition values from different concentration points were analysed by the curve fitting program supported by the Collaborative Drug Discovery.
Microbroth culture for bacteriostatic and bactericidal activity
The anti-mycobacterial activity of the compounds was tested by the measurement of OD readings and agar plate cultures. The test strain used was a stock culture of M. tuberculosis H37Ra (ATCC 25177) stored at −80 °C. Prior to testing, the stock culture was thawed and grown on Middlebrook 7H10 agar (Difco, USA) to check for viability and purity. To prepare the inoculum, a suspension of the culture in Middlebrook 7H9 broth (Difco, USA) was adjusted to a turbidity equivalent to McFarland standard no.1 and further diluted to a final concentration of approximately 105 cfu/ml. The compounds were dissolved in DMSO and diluted in 1% DMSO in 7H9 broth to obtain 100 μg/ml, 50 μg/ml, 10 μg/ml, 5 μg/ml 1 μg/ml and 0.1 μg/ml of compound. Each dilution was pipetted in 150 μl duplicates into a sterile 96-well microtitre plate. Three controls were set up with (a) 1% DMSO in 7H9 broth (b) 7H9 broth without DMSO and (c) p-amino salicylic acid (PAS) at 4 μg/ml (26.12 μM) in 7H9 broth with 1% DMSO. All wells were then inoculated with 10 μl of the mycobacterial culture at 105 cfu/ml, sealed with parafilm and incubated at 36 °C for 28 days. OD readings (at 630 nm wavelength) were taken daily using TECAN spectrophotometer with Magellan software version 7.1. In addition, on day 14 and day 28, 10 μl was removed from each well for subculturing on compound-free 7H10 agar plates which were incubated up to six weeks at 36 °C. The number, size and time of appearance of colonies in each subculture was recorded. A random sample of colonies was stained for acid-fastness to exclude non-mycobacterial contamination. Bactericidal activity was indicated by no growth of the test strain in the subculture, up to six weeks of incubation. Compounds showing less increase in OD readings than the no-compound controls but positive growth in the subcultures were deemed to have bacteriostatic activity.
Molecular Dynamic Simulation
Each selected compound and mt-DHFR complex was explicitly solvated in a truncated octahedral box of TIP3P model water (at least 12 Å from the complex to avoid periodic artifacts from occurring) by using Amber 12 with the amber ff12SB force field. Enough Na+ ions were added to neutralize the charges of the system by using the “addions” command line within the tleap module (AmberTools 12, which adds counterions around the complex using a Coulombic potential on a grid). The whole system was first optimised by energy minimisation, followed by 40 ns molecular dynamic simulation. To obtain the parameters for the ligands and GOL, QM calculations were performed by using the B3LYP 6-31G* basis set within Gaussian09 to optimise molecular geometries and the atom-centred point charges were calculated to fit the electrostatic potential using RESP. The parameters of NADPH were obtained from the AMBER parameter database, as described previously. GOL and each ligand were treated by GAFF for parameters. We used the Principal Component Analysis (PCA) approach to check for equilibration and sampling. At least the last 20 ns stable, equilibrated trajectories of each simulation were taken for energy analysis.
The standard equilibration protocol used was as follows:
-
1
The solvent was energy minimised by 50 steps of the steepest descent method and then followed by 10000 steps of the conjugate gradient method with the default nonbonded cutoff of 8 Å, while the solute was held fixed.
-
2
The entire system was energy minimised with the same settings as the previous step.
-
3
The solvent was subjected to a short (20 ps) MD simulation at a temperature of 100 K, under constant pressure conditions. For this and all following simulations, MD simulations were performed with explicit solvent models and in the NPT ensemble (T = 300 K; P = 1 atm). Periodic boundary conditions (PBC) and particle-mesh-Ewald method (PME)31 were used to model long-range electrostatic effects, with the temperature coupled to an external bath using a weak coupling algorithm32. The cutoff non-bonded interaction was set as 8 Å. The bond interactions involving H-atoms were constrained by using the SHAKE algorithm.
-
4
Over 20 ps, the solvent temperature was raised to 300 K. During both this phase and the last, position restraints on every solute atom (force constant 100 kcal/mol/Å2) maintained them in their energy-minimised conformation.
-
5
Over a series of 20 ps constant–pressure simulations at 300 K, the restraints on the solute were gradually relaxed (50, 25, 10, 5, 2 and then 1 kcal/mol/Å2).
-
6
The final 200 ps MD simulations were performed as the equilibration runs without any restraints.
After the energy minimisation and equilibrations, production MD was run for 40 ns in an NPT ensemble at 1 atm and 300 K. The time step necessary to solve the Newton’s equations was chosen to be equal to 2 fs and the trajectory files were collected every 10 ps for the subsequent analysis. All trajectory analysis was performed with the Ptraj module in the AmberTools 12 and examined visually using VMD software33.
In the present work, in total 9 series of molecular simulations were performed and they are: El-7a in complex with mt-DHFR in the absence (Pose 1 and 2) and in the presence (Pose 3) of GOL; compound 2 in complex with mt-DHFR in the absence (Pose 1, 2 and 3) and in the presence (Pose 4) of GOL; compound 6 in complex with mt-DHFR and in complex with h-DHFR.
Binding Free Energy Calculations
The binding free energy can be calculated using equation 1 from a well-equilibrated molecular dynamics simulation:

Where G is the average free energy calculated from a set of structures taken from the equilibrated simulation (snapshots). Two popular choices to calculate the binding free energy of the snapshots are the Poisson–Boltzman (PBSA) and generalised Born (GBSA) models34,35,36,37,38, where SA corresponds to an estimation of the non-polar solvation free energy based on a simple surface area term.
In the present study, one thousand snapshots collected from the last 20 ns stable simulations at 20 ps intervals were used and MM-PBSA was chosen to calculate the binding free energies with the Python script, MMPBSA.py, included in AMBERTOOLS 13. The nonpolar solvation free energy (ΔGnp) was determined by the solvent accessible surface area (SASA) according to equation 2.

Where the surface tension γ and the offset β were set to the standard values of 0.00542 kcal/mol/Å2 and 0.92 kcal/mol, respectively. Other options were set to default settings. The entropy was estimated by using the Normal Mode program within the AMBER suite. Because these calculations are computationally intensive, only 100 snapshots for each MD trajectories were used for the normal-mode analysis of the equilibrated structure.
In the present work, in total 8 series of binding free energy calculations were performed and they are: El-7a in complex with mt-DHFR in the absence (Pose 1 and 2) of GOL; compound 2 in complex with mt-DHFR in the absence (Pose 1, 2 and 3) and in the presence (Pose 4) of GOL; and compound 6 in complex with mt-DHFR and in complex with h-DHFR.
Results and Discussion
Virtual screening and Molecular Docking
The docking results were clustered by using Discovery Studio and then carefully analysed manually. After removal of duplicates and those molecules that have already been previously reported as mt-DHFR inhibitors, 213 compounds (158 molecules selected from the hit list based on vRocs searching and 55 from Sybyl searching) were chosen from the final hit list. Following manual inspection with the combined aims of covering the maximum amount of chemical space while avoiding non drug-like function groups, 40 compounds (26 molecules based on vRocs searching and 14 on Sybyl searching) were subsequently selected and ordered from NCI to test for their ability to inhibit mt-DHFR and h-DHFR.
Inhibition of DHFR activities in vitro
The 40 compounds were tested for their ability to inhibit recombinant mt-DHFR and h-DHFR activity in in vitro enzyme assays. Eight compounds showed strong inhibition effects on mt-DHFR and three of these compounds (6, 7 and 8) were more potent inhibitors of mt-DHFR compared to h-DHFR ( Table 1).
Antibacterial activity
In the microbroth cultures for the demonstration of mycobacterial inhibition, the 7H9 broth and 1% DMSO controls showed a steady increase in OD readings and yielded abundant growth of M. tuberculosis H37Ra colonies within 14 days of incubation on 7H10 agar. In the presence of para-aminosalicylic acid (PAS), one of the standard anti-TB drugs, there was only scanty growth plus a two-week delay in the appearance of colonies. These results are consistent with uninhibited mycobacterial multiplication in 1% DMSO and a more than two-log reduction in growth in the presence of PAS. All eight compounds showed varying degrees of growth inhibition on the test strain, seen as total inhibition of growth, reduction in colony counts, delayed appearance of colonies or slower increase in colony size. The final colony counts after four weeks of exposure to the compounds are shown in Table 2. At 100μg/ml, three compounds (4, 6 and 8) showed bactericidal activity. At 50 μg/ml, only one (compound 6) was still bactericidal, while at 10 μg/ml, only compound 6 (at 25.37 μM) showed a notable reduction in colony count. Compound 7 showed the least growth inhibition, affecting the colony size but not the colony count. After exposure to 50 μg/ml of compound 8, the test strain did not grow in the 14-day subculture but formed abundant colonies in the 28-day subculture. This unexpected finding raised the possibility of induced resistance in which small numbers of mutants, when given a selective advantage in the presence of the compound, multiplied to become a predominantly resistant population. Overall, at 100 μg/ml, compounds 4, 6 and 8 showed better activity than PAS; at 50 μg/ml, compounds 2, 3 and 4 showed similar activity as PAS; but at 10 μg/ml, only compound 6 (25.37 μM) retained the same activity as PAS (4 μg/ml, 26.12 μM).
Molecular Dynamic Simulation
The influence of GOL on the binding of compound El-7a
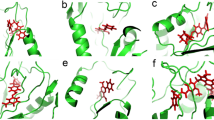
We were interested to examine the influence of GOL in the binding of potential mt-DHFR inhibitors because the GOL binding site is essentially absent in h-DHFR (PDB ID: 1OHJ, h-DHFR complexed with NADPH and PT523)10. Therefore, the binding poses of compound El-7a were analysed in the absence or presence of GOL. In the absence of GOL, compound El-7a can be docked with two binding poses (Pose 1, the propanetriol group occupied the GOL site; Pose 2, the propanetriol group pointed to the opposite direction of GOL site, Fig. 4). In the presence of GOL, compound El-7a could only be docked with one binding pose (pose 3) (both the propanetriol group and the benzene ring pointed to the opposite direction of GOL site). Molecular dynamic simulations, MM-PBSA and Normal Mode calculations were performed on these three binding poses to generate more detailed conformational and energy information.
In the absence of GOL, both pose 1 and pose 2 were stable. The average structures from the trajectories shown in Fig. 5 indicate that in pose 1 compound El-7a formed hydrogen bonds with Ile5, Asp27, Gln28, Leu24 and Tyr100, while in pose 2, it formed hydrogen bonds with Ile5 and Ile94. MM-PBSA and Normal Mode calculations were performed on these two simulations and the results showed the binding free energy of pose 1 was lower than that of pose 2, which indicated that pose 1 is a more stable binding pose (Table 3). This result suggests the propanetriol group of compound El-7a may occupy the GOL binding site. The free energy decomposition of MM-PBSA calculation was performed to analyze the interaction of El-7a with the active site in Pose 1. We observed that 6 residues showed strong contributions to the binding free energy (lower than −1.0 kcal/mol). They are Ile5 (−1.61 Kcal/mol), Trp6 (−1.89 Kcal/mol), Ala7 (−1.36 Kcal/mol), Ile20 (−1.66 Kcal/mol), Trp22 (−2.43 Kcal/mol) and Phe31 (−1.83 Kcal/mol). Among these residues, Ile5, Trp6, Ala7 and Phe31 show strong interactions with the pyrimidine-2,4-diamines group and Ile20 and Trp22 form interactions with the propanetriol group (GOL binding site). The major free energy contribution of Ile20 comes from the VDW interaction, whilst the major contributions of Trp22 are from both VDW (−0.95 Kcal/mol) and electrostatic (−2.41 Kcal/mol) interactions.
A simulation was performed on the system of mt-DHFR in complex with NADPH, GOL and El-7a. As shown in Fig. 6A, at the beginning of the simulation, GOL had strong interactions with its binding pocket (Trp22, Leu24, Asp27 and Glu28). With the simulations, compound El-7a slowly changed its orientation; the propanetriol group of compound El-7a moved towards the GOL binding site. Asp27 gradually moved close to the propanetriol group of compound El-7a and formed hydrogen bonds between them. During this process, GOL was slowly expelled from the binding site and left the protein complex (Fig. 6B). At the end of simulation, GOL moved out of the protein completely and the propanetriol group of compound El-7a moved to the GOL binding site and formed hydrogen bonds with Asp27 and Leu24. The results of these simulations suggest that the GOL binding site could be a potential drug target. It must be addressed at here that the GOL is purely a crystallisation artifact, as mt-DHFR is usually stored with glycerol to maintain its activity. Therefore, GOL may affect the in vitro enzyme assays, but it unlikely can be a necessary co-factor for the mt-DHFR in a biologically relevant situation. The simulation with GOL was performed only for the reason to confirm El-7a has a strong ability to bind to GOL binding site, even when the binding site has been occupied by GOL.
Binding pose analyses of compounds 1–8
Based on previous simulation of compound El-7a, we asked whether compounds 1-8 identified in the present study could bind into the GOL binding site. By comparing the chemical structures of compounds 1–8 with compound El-7a, it was observed that only compounds 1, 2, 3 and 4 might cause a similar effect as compound El-7a because they contain side chains at 6 position on the pyrimidine-2,4-diamines. However, the side chain of compound 4 is much larger than the GOL binding site, which significantly reduces such a possibility. All eight compounds were docked again by GOLD in the absence of GOL and ten binding poses were saved for analysis. As we assumed, only compounds 1-3, were observed docking with dual binding poses. Among these three compounds, compound 2 showed the strongest inhibition effects on mt-DHFR, so it was selected for molecular dynamic simulations for more accurate binding pose analysis. In absence of GOL, compound 2 can be docked with three binding poses: Pose 1 (Fig. 7A), no side group binding to GOL binding site; Pose 2 (Fig. 7B), naphthalene ring binding to GOL binding site; and Pose3 (Fig. 7C), benzene ring binding to GOL binding site. In the presence of GOL, only one binding pose is available, which is similar to the pose 1 and we refer to this as Pose 4 (Fig. 7D). Molecular dynamic simulations, MM-PBSA and Normal Mode calculations were performed on these four binding poses for more detailed conformational and energy analysis.
Through the conformational analysis, we anticipated compound 2 cannot bind into the GOL binding site, because either the naphthalene ring or benzene ring is hydrophobic and cannot have strong interactions with those hydrophilic residues at the GOL binding site. As shown in Table 4, in the absence of GOL, the binding pose 1 showed the lowest binding free energy in comparison with Pose 2 and Pose 3, which may indict compound 2 cannot occupy the GOL binding site, although the energy difference is not very significant. The free energy decomposition showed that in poses 2 and 3, although the naphthalene ring or benzene ring occupy the GOL binding site respectively, none of the key residues (Trp22, Leu24, Asp27 and Glu28) in the GOL binding site have significant free energy contributions (greater than −1 Kcal/mol) for ligand binding. In the presence of GOL, the binding pose 4 shows a similar binding free energy with the consideration of entropy in comparison with the binding pose 1 in the absence of GOL. In binding pose 4, there are strong VDW interactions between GOL and compound 2 and the total free energy contribution is −1.37 Kcal/mol, which is the 4th strongest contributions among all the residues and GOL itself has a strong interaction with Trp22 (free energy contribution −2.07 Kcal/mol). The results suggest that GOL functions as a bridge to link compound 2 and mt-DHFR and by this way GOL enhances the enthalpy term of the binding free energy of the compound 2 in in vitro enzyme assays, but it also slightly increases the entropy penalties, so that the final binding free energies of compound 2 in pose 1 and pose 4 are almost same. However, in the real biological situation as per the antibacterial activity assays, GOL is unlikely to be a cofactor binding in the GOL binding site, so that binding Pose 1 may be the favourable binding pose in this case. Most DHFR inhibitors target the dihydrofolate binding site. The GOL binding site is a relatively small pocket that is very close to the dihydrofolate binding site and may be treated as an extension of the dihydrofolate binding site. The dihydrofolate binding site is relative large, so although those compounds with large hydrophobic side chains, such as compound 2, cannot occupy the GOL binding site, it can still fit into the dihydrofolate binding site. Once they bind into this site, the reduction of dihydrofolate to tetrahydrofolate can be prevented leading to the inhibition of Mtb growth.
The binding pose of compound 6
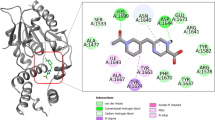
In the present study, as compound 6 was the most potent inhibitor of mt-DHFR and exhibited the strongest bactericidal activity, we analysed its binding pose in more detail. Compound 6 was docked in the presence and absence of GOL and this showed no significant differences between these two binding poses. The binding pose of compound 6 is similar to the bound MTX in the crystal structure. Figure 8 shows the results of the docking study in the absence of GOL, with the blue dotted area representing the binding site of MTX (or dihydrofolate) and the purple dotted indicating the binding site of GOL. As predicted, compound 6 occupies the MTX binding site without touching GOL binding site. In this binding pose, it can be observed from the docking results that compound 6 formed 5 hydrogen bonds with Gln28, Asp27, Ile5, Tyr100 and Ile94 and it also formed strong hydrophobic interaction with Pro51 and Phe31.
The binding poses of compound 6 bound with mt-DHFR and h-DHFR were compared. It was observed that the binding pockets of these two proteins are highly conserved and the binding poses of compound 6 are almost identical (Fig. 9), which is consistent with the fact that compound 6 only shows a very slight degree of selectivity against h-DHFR. DHFR inhibitors usually contain a central core of 2,4-diaminopteridin (classical inhibitors) or pyrimidine-2,4-diamines (non-classical inhibitors). However, compound 6 contains a novel central core (7H-pyrrolo[3,2-f]quinazoline-1,3-diamine), which will significantly expand the chemical space of novel DHFR inhibitors.
Molecular dynamic simulations were performed on compound 6 in complex with h-DHFR and mt-DHFR in the absence of bound GOL, as GOL is not a cofactor in the real biological situation. Through the conformation analyses along both trajectories, compound 6 was tightly bound to both proteins and the binding poses were stable along the entire simulations. The MM-PBSA and Normal Mode calculations were performed on both systems and it was found that the binding free energies of these two simulations were similar (Table 5), but the one with h-DHFR is slightly lower than that of mt-DHFR (only about −0.76 Kcal/mol), which does not fully match the experimental data. The reason for this is likely to be a result of system error, because such small differences in binding affinities cannot be accurately predicted by MM-PBSA and Normal Mode calculations, especially when the receptors are two different proteins. We also observed that the binding free energy of compound 2 with mt-DHFR is lower than that of compound 6, which contradicts the experimental data, most likely because the MM-PBSA and Normal Mode approaches are better at selecting poses for one compound than ranking molecules with significantly different structures (which is certainly true for compounds 2 and 6).
A free energy decomposition of MM-PBSA calculation was performed to analyze the interaction of compound 6 with both mt-DHFR and h-DHFR. We observed that 4 residues contributed strongly to the binding free energy of compound 6 with mt-DHFR (lower than −1.0 Kcal/mol). They are Ile5 (−1.56 Kcal/mol), Trp6 (−1.61 Kcal/mol), Phe31 (−2.32 Kcal/mol) and Leu50(−1.02 Kcal/mol). Among these residues, Ile5 (−1.35 Kcal/mol VDW, −1.73 Kcal/mol electrostatic interactions) and Trp6 (−1.19 Kcal/mol VDW, −0.94 Kcal/mol electrostatic interactions) show both VDW and electrostatic interactions and Phe31 and Leu50 mainly show strong VDW interaction. In the interactions between h-DHFR and compound 6, there are five residues (Ile7 (−1.74 Kcal/mol), Val8 (−1.54 Kcal/mol), Phe31 (−1.10 Kcal/mol), Phe34 (−2.34 Kcal/mol) and Ile60 (−1.50 Kcal/mol) that demonstrated strong free energy contributions (lower than −1.0 Kcal/mol). Among them, only Ile7 show free energy contributions from both VDW (−1.33 Kcal/mol) and electrostatic interactions (−1.72 Kcal/mol) and the remaining residues mainly contribute to the binding free energy via VDW interactions. The hydrophobic residue Phe31 in h-DHFR showed a strong free energy contribution, but the corresponding residue in Mt-DHFR, Gln28 (with a polar side chain, labeled in red in Fig. 9c), only showed a weak free energy contribution (−0.37 Kcal/mol). This difference could be a potential direction for future molecular design attempts to improve the selectivity of compound 6 analogues, for example by adding more polar groups at the corresponding position. Another more obvious direction is the addition of a GOL like group on compound 6 to occupy the GOL binding site.
Conclusion
Using in silico methods, we have identified a set of mt-DHFR inhibitors that inhibited recombinant mt-DHFR and h-DHFR to varying degrees and that showed bactericidal effects against M. tuberculosis H37Ra. Molecular simulations indicated that the GOL binding site can be occupied by inhibitors containing appropriate side chains. Among the compounds identified in the present study, only the compounds 1, 2, 3 and 4 have such possibility. However, they all contain large hydrophobic side chains at 6 position on the pyrimidine-2,4-diamines. As the GOL binding site is more hydrophilic and contains hydrogen bond acceptors and donors, these compounds failed to occupy the GOL site, which could be one of the reasons that the present inhibitors did not show strong selectivity against h-DHFR. Compound 6 was the most potent inhibitor obtained in the present study. It contains a novel central core (7H-pyrrolo[3,2-f]quinazoline-1,3-diamine), which will significantly expand the chemical space of novel DHFR inhibitors. This compound did show a small degree of selectivity against h-DHFR and can be used as a lead compound for further optimization and will inform future medicinal chemistry efforts to improve the selectivity of compounds against mt-DHFR.
Whilst the GOL binding site is not essential for mt-DHFR enzyme activity, it is thought to be important for the selectivity for mt-DHFR against h-DHFR. Our modelling study suggests hydrophilic side groups may be able to occupy this binding site. This approach may lead to new inhibitors being discovered that specifically bind to mt-DHFR. However, it is cautionary to note that as Mtb has a thick cell wall that only allows highly hydrophobic compounds to pass through, adding a hydrophilic side group to compounds may prevent them from crossing the mycobacterial cell wall. Consequently, although such compounds may be able to specifically inhibit mt-DHFR, they may not be able to act on Mtb whole cells. Therefore, finding a balance between selectivity and activity on Mtb remains a major challenge in anti-TB drug discovery.
Additional Information
How to cite this article: Hong, W. et al. The identification of novel Mycobacterium tuberculosis DHFR inhibitors and the investigation of their binding preferences by using molecular modelling. Sci. Rep. 5, 15328; doi: 10.1038/srep15328 (2015).
References
World Health Organization (WHO). Global tuberculosis report (2014). Available at: http://www.who.int/tb/publications/global_report/en/.
Nachega, J. B. & Chaisson R. E. Tuberculosis drug resistance: a global threat. Clin. Infect. Dis. 36, 24–30 (2003).
Raviglione, M. & Smith I. XDR tuberculosis-implications for global public health. N. Engl. J. Med. 356, 656–659 (2007).
Blakley, R.L. Dihydrofolate reductase. In Folates and Pterines [ Blakley, R. L. & Benkovic, S. J. (eds)] [191–253] (Wiley, New York, 1984).
Blakley, R. L. Eukaryotic dihydrofolate reductase. Advan. Enzymol. Relat. Areas Mol. Biol. 70, 23–102 (1995).
Kumar, A. et al. High-throughput screening and sensitized bacteria identify an M. tuberculosis dihydrofolate reductase inhibitor with whole cell activity. Plos One. 7, e39961 (2012).
Robson, C. et al. Nonclassical 2,4-Diamino-5-aryl-6-ethylpyrimidine Antifolates: Activity as Inhibitors of Dihydrofolate Reductase from Pneumocystis carinii and Toxoplasma gondii and as Antitumor Agents. J. Med. Chem. 40, 3040–3048 (1997).
White, E. L., Ross, L. R., Cunningham, A. & Escuyer, V. Cloning, expression and characterization of Mycobacterium tuberculosis dihydrofolate reductase. FEMS Microbiol. Lett. 232, 101–105 (2004).
Brogden, R. N., Carmine, A. A., Heel, R. C., Speight, T. M. & Avery, G. S. Trimethoprim: a review of its antibacterial activity, pharmacokinetics and therapeutic use in urinary tract infections. Drugs. 23, 405–430 (1982).
Russell, P. B. & Hitchings, G. H. 2,4-Diaminopyrimidines as antimalarials. III. 5-Aryl derivatives. J. Am. Chem. Soc. 73, 3763–3770 (1951).
Sardarian, A. et al. Pyrimethamine analogs as strong inhibitors of double and quadruple mutants of dihydrofolate reductase in human malaria parasites. Org. Biomol. Chem. 1, 960–964 (2003).
Robson, C. et al. Nonclassical 2,4-diamino-5-aryl-6-ethylpyrimidine Antifolates: activity as inhibitors of dihydrofolate reductase from Pneumocystis carinii and Toxoplasma gondii and as antitumor agents. J. Med. Chem. 40, 3040–3048 (1997).
Fidock, D. A. & Wellems T. E. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA 94, 10931–10936 (1997).
Hekmat-Nejad, M. & Rathod, P. K. Plasmodium falciparum: kinetic interactions of WR99210 with pyrimethamine-sensitive and pyrimethamine-resistant dihydrofolate reductase. Expt. Parasitol. 87, 222–228 (1997).
Li, R. et al. Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs. J. Mol. Biol. 295, 307–323 (2000).
Cody, V. et al. Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes with reduced nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523. Biochemistry 36, 13897–13903 (1997).
El-Hamamsy, M. H. R. I., Smith, A. W., Thompson, A. S. & Threadgill, M. D. Structure-based design, synthesis and preliminary evaluation of selective inhibitors of dihydrofolate reductase from Mycobacterium tuberculosis. Bioorg. Med. Chem. 15, 4552–4576 (2007).
Case, D. A. et al. AMBER 12, University of California, San Francisco (2012).
Frisch, M. J. et al. Gaussian 09, Revision C.01, Gaussian, Inc., Wallingford CT (2009).
Bayly, C. I., Cieplak, P., Cornell, W.D. & Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 (1993).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Suling, W. J. et al. Susceptibilities of Mycobacterium tuberculosis and Mycobacterium avium complex to lipophilic deazapteridine derivatives, inhibitors of dihydrofolate reductase. J. Antimicrob. Chemother. 42, 811–815 (1998).
Li, R. et al. Three-dimensional structure of M-tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs. J. Mol. Biol. 295, 307–323 (2000).
Suling, W. J. & Maddry J. A. Antimycobacterial activity of 1-deaza-7,8-dihydropteridine derivatives against Mycobacterium tuberculosis and Mycobacterium avium complex in vitro. J. Antimicrob. Chemother. 47, 451–454 (2001).
Gerum, A. B. et al. Novel Saccharomyces cerevisiae screen identifies WR99210 analogues that inhibit Mycobacterium tuberculosis dihydrofolate reductase. Antimicrob. Agents Chemother. 46, 3362–3369 (2002).
Hawkins, P. C. D., Skillman, A. G. & Nicholls, A. Comparison of shape-matching and docking as virtual screening tools. J. Med. Chem. 50, 74–82 (2007).
McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 51, 578–596 (2011).
Hawkins, P. C. D., Skillman, A. G., Warren, G. L., Ellingson, B. A. & Stahl, M. T. Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and the cambridge structural database. J. Chem. Inf. Model. 50, 572–584 (2010).
Jones, G., Willett, P., Glen, R. C., Leach, A. R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
Nixon, M. R. et al. Folate pathway disruption leads to critical disruption of methionine derivatives in Mycobacterium tuberculosis. Chem. Biol. 21, 819–830 (2014).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem.Phys. 81, 3684–3690 (1984).
Humphrey, W., Dalke, A. & Schulten, K. VMD-Visual Molecular Dynamics. J. Mol. Graphics. 14, 33–38 (1996).
Kollman, P. A. et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 (2000).
Still, W. C., Tempczyk, A., Hawley, R. C. & Hendrickson, T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 112, 6127–6129 (1990).
Orozco, M. & Luque, F. J. Theoretical methods for the description of the solvent effect in biomolecular systems. Chem. Rev. 100, 4187–4226 (2000).
Wang, H. & Laughton, C. A. Molecular modelling methods for prediction of sequence-selectivity in DNA recognition. Methods. 42, 196–203 (2007).
Wang, H. & Laughton, C. A. Evaluation of molecular modelling methods to predict the sequence selectivity of DNA minor groove. Phys. Chem. Chem. Phys. 11, 10722–10728 (2009).
Acknowledgements
This work was supported by National Natural Science Foundation of China (81260481) given to H.W. and (81460538) given to W.H.; Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (2013-693) given to H.W. and University of Malaya-MOHE High Impact Research grant (UM.C/625/1/HIR/MOHE/DENT/22) given to I.C.P. The authors would like to thank Dr Charles Laughton at University of Nottingham, UK for offering the access to the computing facilities.
Author information
Authors and Affiliations
Contributions
W.H., Y.W. and Z.C. performed the virtual screening and molecular modeling work. Y.Y., J.P. and T.S. performed molecular work. S.K. and Y.F.N. performed antibacterial activity tests. H.J. and J.C.S. performed DHFR inhibition tests. W.L.W. and L.F.Y. performed human cells tests. W.H., I.C.P., Y.F.N., J.C.S. and H.W. developed the study strategy. I.C.P., Y.F.N., W.H. and H.W. contributed to the writing of the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hong, W., Wang, Y., Chang, Z. et al. The identification of novel Mycobacterium tuberculosis DHFR inhibitors and the investigation of their binding preferences by using molecular modelling. Sci Rep 5, 15328 (2015). https://doi.org/10.1038/srep15328
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15328
This article is cited by
-
TAGOPSIN: collating taxa-specific gene and protein functional and structural information
BMC Bioinformatics (2021)
-
A combined structure-based pharmacophore modeling and 3D-QSAR study on a series of N-heterocyclic scaffolds to screen novel antagonists as human DHFR inhibitors
Structural Chemistry (2021)
-
(±)-Japonones A and B, two pairs of new enantiomers with anti-KSHV activities from Hypericum japonicum
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.